Abstract
In recent years much attention has been given to the identification and characterisation of the key elements of the secretory machinery of Streptomyces lividans, a non-pathogenic filamentous Gram-positive soil bacterium, whose metabolism is relatively well characterised and capable of secreting large amounts of proteins when grown in laboratory conditions. The relevance of S. lividans from a commercial standpoint is due to its potential usefulness for the overproduction of secretory homologous and heterologous proteins of interest. Therefore, this review focuses on the knowledge already obtained on the S. lividans secretion pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomycetes are Gram-positive soil bacteria, whose complex morphological life cycle comprises the development of an aerial mycelium ending in the formation of spores, and during their different phases of growth endure morphological changes in combination with the production of secondary metabolites. They need to produce and secrete large quantities of proteins (Gilbert et al. 1995), such as the suitable and more prevalent hydrolytic enzymes, as well as enzyme inhibitors, in addition to antibiotics and signalling molecules, in order to adapt to their environment, largely formed by insoluble polymers, while undergoing differentiation (Chater 1998). It is precisely this extracellular production capacity that has aroused an interest in obtaining more in-depth information on how the streptomycetes secretion machinery functions, and whether this knowledge could be exploited to engineer Streptomyces strains as hosts for the synthesis of homologous or heterologous proteins of commercial interest.
Streptomyces lividans is a bacterium that can be cultivated and transformed in the laboratory with relative ease because of its relaxed restriction-modification capacity, since it can take up exogenous DNA without excessive degradation. Moreover, S. lividans has been found to be an ideal candidate as a bacterial host for extracellular protein overproduction, owing to the extensive homology of its genome to that of the widely studied and closely related bacterium, Streptomyces coelicolor, and its capacity to synthesise and secrete a significant number of extracellular proteins. Thus, S. lividans has frequently been used in experiments as a host for the secretory production of homologous and heterologous proteins (Gilbert et al. 1995; Anné and Mellaert 1993; Binnie et al. 1997; Lammertyn et al. 1997; Parro and Mellado 1994; Van Mellaert and Anné 1994; Isiegas et al.1999; Vrancken and Anné 2009).
The Sec pathway is the main route by which secretory proteins are released outside the cell across the cytoplasmic membrane (Driessen and Nouwen 2008). The second protein transport system found in streptomycetes is the Tat pathway that is able to transport already folded proteins across the cytoplasmic membrane (Berks et al. 2000). A third secretion system (Esx, or type VII) has been described in Gram-positive bacteria, where it seems to be required for the secretion of small extracellular proteins (Pallen et al. 2002). The function of this system in streptomycetes is currently unknown (Chater et al. 2010) and, therefore, cannot yet be considered as an essential system for protein secretion in streptomycetes.
Type I signal peptidases in S. lividans
Extracellular proteins are synthesised as pre-proteins carrying a signal peptide needed for proper targeting and translocation across the cellular membrane, and the signal peptide is removed by signal peptidases during the course of translocation or shortly after it (Van Welly et al. 2001). The majority of exported pre-proteins are processed by prokaryotic type I signal peptidases (SPases), also known as leader peptidases (Lep). Most organisms contain only one type I SPase, seemingly essential, as in E. coli (Dalbey and Wickner 1985; Tschantz et al. 1993) or yeast (Böhni et al. 1988); however, there are organisms containing two type I SPases such as the cyanobacterium Synechocystis PCC 6803 (Kaneko et al. 1996), the bacteria Bacillus amyloliquefaciens (Hoang and Hofemeister 1995; Meijer et al. 1995) and Staphylococcus aureus (Cregg et al. 1996). Most eukaryotic species also harbour two SPase isoforms (Dalbey et al. 1997). Three SPases have been found in the archaeon Archaeoglobus fulgidus (Klenk et al. 1997) and in Deinococcus radiodurans (White et al. 1999). Among the Gram-positive bacteria, seven SPases have been described for Bacillus subtilis, where the genes corresponding to five of these are widespread on the chromosome (van Dijl et al. 1995; Tjalsma et al. 1998), and two other genes have been found in plasmids (Meijer et al. 1995). Moreover, four adjacent genes (sipW, sipX, sipY and sipZ) encoding different type I signal peptidases have been identified in the S. lividans TK21 genome, where three of the sip genes (sipW, sipX and sipY) constitute an operon, and the fourth one (sipZ) is the first gene of another operon encompassing three additional unrelated genes (Parro et al. 1999). In accordance with the construction and in vivo phenotypical characterization of mutants in each of the four S. lividans sip genes, SipY appears to be the S. lividans SPase that plays a major role in pre-protein processing (Palacín et al. 2002).
Analysis of the S. lividans TK21 secretome by 2D-PAGE followed by mass-spectrometric analysis was of great assistance in identifying the S. lividans secretory proteins, the majority (84%) carrying typical export signals for the Sec route, and the rest being non-secretory proteins (Palacín et al. 2002). This proteomic analysis of the S. lividans secretome confirmed the differences in extracellular protein patterns between S. lividans TK21 and the different sip mutant strains, revealing a severe reduction in the accumulation of extracellular proteins in the sipY mutant compared to that of the remaining sip mutants, and additionally confirming the major role played by the SipY protein in secretion (Palacín et al. 2002). The SipY mutant also experiences intracellular retention of a model secretory protein (Palomino and Mellado 2005), consequently affecting the intracellular distribution of the secretion pathway components, blocking the translocon temporarily and triggering the accumulation of the model pre-protein at the membrane.
Upon complementation of the SipY mutant with each of the Sip proteins encoded in multicopy plasmids, the SipY mutant sporulation-delayed phenotype was repaired and the analysis of extracellular proteins indicated that the overall pattern of secretory proteins had recovered in all cases (Escutia et al. 2006). The conclusion reached is that individual mutations in the different S. lividans sip genes, except for sipY, do not seem to exert a severe effect on protein secretion, as observed previously (Palacín et al. 2002), due to the compensatory effects of the minor SPases. Thus, the existence of several type I SPases in bacteria reflects the need to ensure the secretion of the necessary extracellular proteins for their survival in their natural environment, affording the bacteria more fitness in changing environmental conditions, an objective that seems to be wisely achieved in the case of soil Gram-positive bacteria, by favouring compensatory effects over substrate specificity.
Transport to the membrane
The synthesis of secretory proteins takes place with a signal peptide at their amino end, which serves as a recognition signal for cytosolic chaperons, such as SecB in E. coli. The tight folding of the precursor protein is prevented by these chaperons, which help to direct the pre-secretory protein to the translocation sites at the membrane (Fekkes and Driessen 1999). This mechanism is called post-translational translocation and is mainly used for protein secretion in E. coli via the general secretion route (Sec pathway). The genome of the Gram-positive bacterium Bacillus subtilis does not have a gene encoding an equivalent SecB protein (Kunst et al. 1997), and the genome sequence of the S. coelicolor bacterium (Bentley et al. 2002), which is closely linked to S. lividans, as well as the available sequence of the S. lividans TK24 genome (http://www.ncbi.nlm.nih.gov) also revealed the absence of a secB gene, thereby rendering a particular appeal to the research of alternative means of transporting secretory proteins in these bacteria.
The co-translational protein targeting mechanism involving the signal recognition particle (SRP) seems to be conserved from bacteria to mammals (Keenan et al. 2001; Cao and Saier, 2003; Pohlschöder et al. 2004). In mammalian cells the translocation of proteins across the endoplasmic reticulum membrane occurs via the interaction of the SRP with the ribosome nascent chain complex (RNC), and the entire complex is subsequently targeted to the membrane by association with an SRP-receptor complex (Walter and Johnson 1994). Escherichia coli possess a similar SRP mechanism (Wolin 1994) comprising a 4.5 S RNA (scRNA), Ffh, a homologue of the mammalian SRP54 protein and FtsY, with a 300-residue long C-terminal domain resembling that of the mammalian SRP-receptor alpha-subunit, being all the bacterial SRP components essential for cell growth. Major interactions among SRP components are evolutionary conserved (Miller et al. 1994). The Ffh C-terminal M domain is involved in the interaction with the nascent polypeptide chain and the scRNA, representing the minimal functional SRP. The FtsY N-terminal A domain contains the membrane-targeting signal and varies among FtsY homologues, which is probably in line with the different modes for targeting FtsY to the membrane (Powers and Walter 1997; Millman and Andrews 1999; Herscovits et al. 2000). The SRP and the SecA/SecB-based secretion systems belong to two different targeting pathways in E. coli that function in a substrate-specific manner (Beck et al. 2000). Integral membrane proteins are targeted to the cytoplasmic membrane by SRP (de Gier et al. 1996; Ulbrandt et al. 1997). However, the postulated role of SRP in the export of E. coli SecB-independent secretory proteins has been ruled out (Beha et al. 2003).
A functional interaction has been detected between SecA and Ffh in B.subtilis (Bunai et al. 1999). This co-translational mechanism ought to be of greater importance in Gram-positive bacteria, where the SRP may interact with the signal sequence of the newly made protein and FtsY during targeting, as well as with the ribosome, and eventually with the translocon. The signal peptides of the B. subtilis secretory proteins seem to be more hydrophobic and longer than those of E. coli, and it has been claimed that these signal peptide characteristics may allow the B. subtilis SRP system to target both secretory and membrane proteins (Herscovits et al. 2000). Thus, the B. subtilis SRP system may be responsible for targeting both membrane and secretory proteins (Oguro et al. 1996; Bunai et al. 1996; Hirose et al. 2000).
The length and hydrophobicity of the signal peptides of S. lividans secretory proteins seem to be more similar to those of B. subtilis than to those of E. coli, therefore it seems reasonable to assume that the same intracellular assignments are conducted by the B. subtilis and S. lividans SRP systems. The S. lividans SRP system consists of a functional signal recognition particle, a ribonucleoprotein, composed of Ffh and an 82 nt long RNA (Palacín et al. 2003) and the small size of this RNA, as in E. coli, seems to leave no room for other proteins to adhere to the complex. The receptor protein, FtsY, also forms part of the S. lividans SRP system and its expression takes place throughout cellular growth, similarly to that of scRNA and Ffh. No mutants have been reported in any of the three corresponding genes, which may indicate the possible essential nature of the S. lividans SRP components. The ffh transcript seems to contain an internal downstream box (Wu and Janssen 1996) to ensure translation of the processed messenger, a feature also shared by the sip transcripts (Parro et al. 1999). Co-immunoprecipitation studies confirmed that the S.lividans SRP, despite being minimal in composition, is involved in targeting secretory proteins (Palacín et al. 2003).
A comparison of the hydrophobic profiles of FtsY proteins from E. coli, Mycobacterium leprae, M. tuberculosis and S. coelicolor (Bibi et al. 2001) suggested the presence of a hydrophobic N-terminal segment that could identify the three Actinobacteria FtsY homologs as integral membrane proteins. The use of an S. lividans strain lacking the gene coding for the major type I signal peptidase has permitted finding evidence of an in vivo interaction of the FtsY receptor with the SRP, as well as FtsY involvement in targeting secretory proteins to the membrane via the existence of a soluble FtsY, which acts as a functional cytoplasmic receptor for the SRP (Palomino and Mellado 2005). Inactivation of the eukaryotic translocon causes accumulation of membrane-bound SRP-RNC and its receptor, by diminishing the transfer of the signal peptide from the SRP54 to the vacant translocon complexes (Song et al. 2000). Similarly, the SipY mutation may temporarily block the translocon and trigger the accumulation of the SRP pre-protein at the membrane.
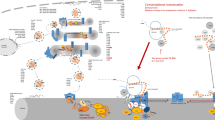
Thus, the S. lividans SRP also seems to evidence an ability to interact with the translocon by escorting the pre-protein to the translocation complex and possibly not being detached from the membrane until the cleavage of the signal peptide permits the complete release of the secretory protein. The whole process, which takes place efficiently in a sequential manner, is regularly coupled, and SRP as well as FtsY are released from the translocon almost simultaneously for their re-utilisation to target more pre-secretory proteins. An illustration of this model is depicted in Fig. 1. The SRP pathway is not affected by the overproduction of model secretory proteins, confirming that the respective level of SRP components is not a limiting factor for secretory protein overproduction in S. lividans (Palacín et al. 2002).
Secretion pathways in Streptomyces lividans. The left section of the figure schematises the major secretory route (Sec pathway), where the nascent polypeptide interacts with the minimal SRP to be targeted to the translocation complex at the membrane with the help of the FtsY receptor in a GTP-dependent process. Once the polypeptide is cleaved by the signal peptidase(s), the SRP components are released and the process is repeated. Improperly folded Sec-dependent mature proteins need the action of foldases or thiol-disulfide isomerases (Dsbs) to acquire their active conformation. Protein YidC has been included although its functionality has not been demonstrated in streptomycetes, as yet. The right section of the figure shows how the nascent polypeptide of a Tat-dependent secretory protein is targeted to the membrane. The mature protein is released fully folded to the cell wall after its leader peptide is cleaved by the signal peptidase(s). The figure is adapted from Fig. 4 (Palomino and Mellado 2005) inspired by Fig. 3 (Fekkes and Driessen 1999)
Translocation and membrane protein insertion
Once the secretory proteins have been targeted to the cytoplasmic membrane, translocation is believed to occur as in E. coli, where SecY, SecE and SecG form an heterotrimeric stable complex constituting the channel that conducts the protein to be translocated (Driessen and Nouwen 2008). From the genome sequence analysis it is known that streptomycetes seem to have only one gene for each of these Sec proteins (Bentley et al. 2002), as is also the case for SecA, somehow supporting the assumption of comparable functioning of their E. coli counterparts, as S. lividans SecA is required in vitro for the secretion of the Sec-dependent α-amylase (Blanco et al. 1998). Two secG homologous genes have been identified in S. coelicolor, S. lividans (SCO1944; Palomino and Mellado 2008) and B. subtilis (yvaL; Van Wely et al. 1999) chromosomes, and their encoded proteins are found to have a functional homology to that of E. coli. Depletion of the S. lividans secG does not seem to cause a cold-sensitive phenotype as it happens when the E. coli or B. subtilis are depleted of their corresponding secG genes, and, at standard growth temperature, SecG deficiency seems to have almost a negligible effect in these two bacteria, while this deficiency does confer a more noticeable effect in S. lividans where extracellular protein secretion is impaired and the overproduction of a model secretory enzyme is delayed (Palomino and Mellado 2008). In E. coli the SecYEG channel associates with the heterotrimeric membrane complex formed by SecD, SecF and YajC proteins, although YajC is not required for the complex function, and the complex itself does not seem to be needed for primary translocation (Driessen and Nouwen 2008). Genes equivalent to these have been found in the S. coelicolor genome, yet their functionality has not been determined experimentally. YidC has been identified as an essential membrane protein that assists in the insertion of some membrane proteins in E. coli (Driessen and Nouwen 2008), and two genes potentially encoding equivalent proteins have been found in the S. coelicolor genome (Bentley et al. 2002). Yet, again, the functionality of their encoded proteins still needs experimental assessment.
The twin-arginine translocation (Tat) pathway
The Tat pathway is an export pathway that has been described in several bacteria and in the thylakoid membranes of plant chloroplasts. Secretory proteins exported by the Tat route are transported across the cytoplasmic membrane in a folded state. This makes the Tat pathway particularly interesting as a potential secretory route to be exploited for the secretion of homologous or heterologous proteins in Streptomyces strains.
The Tat pathway consists of three proteins (TatA, TatB and TatC) in E. coli and in Streptomyces strains (Fig. 1), while B. subtilis and archaebacteria harbour Tat systems with only two Tat proteins (Dilks et al. 2005; Jongbloed et al. 2004; Pop et al. 2002). The N-terminal of the proteins targeted to the Tat route in eubacteria contains a conserved motif R-R-x-Φ-Φ, where x is usually a polar amino acid and Φ is a hydrophobic amino acid (Berks 1996; Stanley et al. 2000; Widdick et al. 2006). Different algorithms of a predictive nature have been designed to recognise Tat leader peptides, and the use of these algorithms has helped to predict the existence in S. coelicolor of a large number of Tat-dependent proteins (Bendtsen et al. 2005; Dilks et al. 2003). From the 145 to 189 Tat-dependent proteins predicted in silico in S. coelicolor, a total of 27 have been unequivocally established as Tat-dependent, when membrane proteins from TatC deficient strains were compared to those of the isogenic wild type strain (Widdick et al. 2006). Up to 127 proteins have been predicted to be Tat-dependent in the S. coelicolor closely-related strain, S. lividans (Schaerlaekens et al. 2004), however, experimentally, the final number has not yet been established. The annotated sequence of the S. coelicolor genome includes 819 secretory proteins, which clearly indicates that the 27 assigned Tat-dependent proteins constitute a reduced number when compared with that of the remaining of the secretory proteins, thereby confirming the subsidiary nature of the Tat route for secretion. Moreover, the experimentally identified 27 Tat-dependent proteins in S. coelicolor represent a significantly large number of proteins using the Tat pathway for secretion, when compared to the very few found in other bacteria. This substantiates the interest of this secretory route in streptomycetes for the secretion of proteins correctly folded at the bacterial supernatant, although the real usefulness of the Tat system for this purpose still needs to be experimentally assessed. In streptomycetes, the existence and mode of action of proteins with the capacity to intervene in the correct folding of extracellular proteins exported by the Sec route, such as foldases or thiol-disulfide isomerases, equivalent to those described in other eubacteria, still remains to be elucidated.
References
Anné J, Mellaert LV (1993) Streptomyces lividans as a host for heterologous protein production. FEMS Microbiol Lett 114:121–128
Beck K, Wu LF, Brunner J, Muller M (2000) Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J 19:134–143
Beha D, Deitermann S, Müller M, Koch H-G (2003) Export of beta-lactamase is independent of the signal recognition particle. J Biol Chem 278:22161–22167
Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S (2005) Prediction of twin-arginine signal peptides. BMC Bioinf 6:167–175
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Berks BC (1996) A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22:393–404
Berks BC, Sargent F, Palmer T (2000) The Tat protein export pathway. Mol Microbiol 35:260–274
Bibi E, Herskovits AA, Bochkareva E, Zelazny A (2001) Putative integral membrane SRP receptors. Trend Biochem Sci 26:15–16
Binnie C, Cossar JD, Stewart DI (1997) Heterologous biopharmaceutical protein expression in Streptomyces. Trend Biotechnol 15:315–320
Blanco J, Driessen AJ, Coque JJ, Martínn JF (1998) Biochemical characterization of the SecA protein of Streptomyces lividans interaction with nucleotides, binding to membrane vesicles and in vitro translocation of proAmy protein. Eur J Biochem 257:472–478
Böhni PC, Deshaies RJ, Schekman RW (1988) SEC11 is required for signal peptide processing and yeast cell growth. J Cell Biol 106:1035–1042
Bunai K, Takamatsu H, Horinaka T, Oguro A, Nakamura K, Yamane K (1996) Bacillus subtilis Ffh, a homologue of mammalian SRP54, can intrinsically bind to the precurssors of secretory proteins. Biochem Biophys Res Commun 227:762–767
Bunai K, Yamada K, Hayashi K, Nakamura K, Yamane K (1999) Enhancing effect of Bacillus subtilis Ffh, a homologue of the SRP54 subunit of the mammalian signal recognition particle, on the binding of SecA to precursors of secretory proteins in vitro. J Biochem 125:151–159
Cao T, Saier M (2003) The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim Biophys Acta 1609:115–125
Chater KF (1998) Taking a genetic scalpel to the Streptomyces colony. Microbiology 144:1465–1478
Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198
Cregg KM, Wilding I, Black MT (1996) Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococus aureus. J Bacteriol 178:5712–5718
Dalbey RE, Wickner W (1985) Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem 260:15925–15931
Dalbey RE, Lively MO, Bron S, van Dijl JM (1997) The chemistry and enzymology of the type I signal peptidases. Protein Sci 6:1129–1138
de Gier JW, Mansournia P, Valent QA, Phillips GJ, Luirink J, von Heijne G (1996) Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett 399:307–309
Dilks K, Rose RW, Hartmann E, Pohlschröder M (2003) Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol 185:478–1483
Dilks K, Giménez MI, Pohlschröder M (2005) Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol 187:8013–8104
Driessen AJ, Nouwen N (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667
Escutia MR, Val G, Palacín A, Geukens N, Anné J, Mellado RP (2006) Compensatory effect of the minor Streptomyces lividanstype I signal peptidases on the SipY major signal peptidase deficiency as determined by extracellular proteome analysis. Proteomics 6:4137–4146
Fekkes P, Driessen AJM (1999) Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Rev 63:161–173
Gilbert M, Morosoli R, Shareck F, Kluepfel D (1995) Production and secretion of proteins by streptomycetes. Crit Rev Biotechnol 15:13–19
Herscovits AA, Bochkareva E, Bibi E (2000) New prospects in studying the bacterial signal recognition particle pathway. Mol Microbiol 38:927–939
Hirose I, Sano K, Shioda I, Kumano M, Nakamura K, Yamane K (2000) Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65–75
Hoang V, Hofemeister J (1995) Bacillus amyloliquefaciens possesses a second type I signal peptidase with extensive sequence similarity to other Bacillus SPases. Biochim Biophys Acta 1269:64–68
Isiegas C, Parro V, Mellado RP (1999) Streptomyces lividansas a host to produce and secrete Escherichia coli TEM β-lactamase. Lett Appl Microbiol 28:321–326
Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM (2004) Two minimal Tat translocases in Bacillus. Mol Microbiol 545:1319–1325
Kaneko T, Sato S, Kotani H, Tanaka A et al (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement). DNA Res 3:185–209
Keenan RJ, Freymann DM, Stroud RM, Walter P (2001) The signal recognition particle. Annu Rev Biochem 70:755–775
Klenk HP, Clayton RA, Tomb JF, White O et al (1997) The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370
Kunst F, Ogasawara N, Moszer I, Albertini AM, et al. (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. 390:249–256
Lammertyn E, Mellaert LV, Schacht S, Dillen C, Sablon E, Broekhoven AV, Anné J (1997) Evaluation of a novel a novel subtilisin inhibitor gene and mutant derivative for the expression of mouse tumor necrosis factor alpha by Streptomyces lividans. Appl Environ Microbiol 63:1808–1813
Meijer WJ, de Jong A, Bea G, Wisman A, Tjalsma H, Venema G, Bron S, van Dijl JM (1995) The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol 17:621–631
Miller JD, Bernstein HD, Walter P (1994) Interaction of E.coli Ffh/4.5 Sribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature 366:351–354
Millman JS, Andrews DW (1999) A site-specific, membrane-dependent cleavage event defines the membrane binding domain of FtsY. J Biol Chem 274:33227–33234
Oguro A, Kakeshita H, Takamatsu H, Nakamura K, Yamane K (1996) The effect of Srb, a homologue of the mammalian SRP receptor alpha-subunit, on Bacillus subtilis growth and protein translocation. Gene 172:17–24
Palacín A, Parro V, Geukens N, Anné J, Mellado RP (2002) SipY is the Streptomyces lividanstype I signal peptidase exerting a major effect on protein secretion. J Bacteriol 184:4875–4880
Palacín A, de la Fuente R, Valle I, Rivas LA, Mellado RP (2003) Streptomyces lividans contains a minimal functional signal recognition particle that is involved in protein secretion. Microbiology 149:2435–2442
Pallen MJ (2002) The ESAT-6/WXG100 superfamily and a new gram-positive secretion system? Trends Microbiol 10:209–212
Palomino C, Mellado RP (2005) The Streptomyces lividans signal recognition particle receptor FtsY is involved in protein secretion. J Mol Microbiol Biotechnol 9:57–62
Palomino C, Mellado RP (2008) Influence of a Streptomyces lividans SecG functional analogue on protein secretion. Int Microbiol 11:25–31
Parro V, Mellado RP (1994) Effect of glucose on agarase overproduction by Streptomyces. Gene 145:49–55
Parro V, Schacht S, Anné J, Mellado RP (1999) Four genes encoding different type I signal peptidases are organized in a cluster in Streptomyces lividans TK21. Microbiology 145:2255–2263
Pohlschöder M, Dilks K, Hand N, Wesley Rose R (2004) Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol Rev 28:3–24
Pop O, Martin U, Abel C, Muller JP (2002) The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J Biol Chem 277:3268–3273
Powers T, Walter P (1997) Co-translational protein targeting catalised by the Escherichia coli signal recognition particle and its receptor. EMBO J 16:4880–4886
Schaerlaekens K, Van Mellaert L, Lammertyn E, Geukens N, Anné J (2004) The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome análisis. Microbiology 150:21–31
Song W, Raden D, Mandon E, Gilmore R (2000) Role of Sec61α in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell 100:333–343
Stanley NR, Palmer T, Berks BC (2000) The twin-arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem 275:11591–11596
Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhizen CP, Quax WJ, Venema G, Bron S, van Dijl JM (1998) Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes and Dev 12:2318–2331
Tschantz WR, Sung M, Delgado-Partin VM, Dalbey R (1993) A serine and a lysine residue implicated in the catalytic mechanims of the Escherichia coli leader peptidase. J Biol Chem 268:27349–27354
Ulbrandt ND, Newitt JA, Bernstein HD (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187–196
van Dijl JM, de Jong A, Venema G, Bron S (1995) Identification of the potential active site of the SPase SipS of Bacillus subtilis: structural and functional similarities with LexA-like proteases. J Biol Chem 270:3611–3618
Van Mellaert L, Anné J (1994) Protein secretion in gram-positive bacteria with high GC- content. Recent Res Dev Microbiol 3:324–340
Van Welly KHM, Swaving J, Freudl A, Driessen AJM (2001) Translocation of proteins across the cell envelope of Gram-positive bacteria. FEMS Microbiol Rev 25:437–454
Van Wely KHM, Swaving J, Broekhuizen CP, Rose M, Quax WJ, Driessen AJM (1999) Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol 181:1786–1792
Vrancken K, Anné J (2009) Secretory production of recombinant proteins by Streptomyces. Future Microbiol 4:181–188
Walter P, Johnson AE (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol 10:87–119
White O, Eisen JA, Heidelberg JF, Hickey EK et al (1999) Genome sequence of the radioresistant bacterium Deinococus radiodurans R1. Science 286:1571–1577
Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschröder M, Palmer T (2006) The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci USA 103:17927–17932
Wolin SL (1994) From the elephant to E. coli: SRP-dependent protein targeting. Cell 77:787–790
Wu CJ, Janssen GR (1996) Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of 5′ untranslated leader. Mol Microbiol 22:339–355
Acknowledgments
Grants BIO2003-03948 and BIO2006-12762 from the Spanish Ministry of Science and Innovation have supported this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mellado, R.P. Summing up particular features of protein secretion in Streptomyces lividans . World J Microbiol Biotechnol 27, 2231–2237 (2011). https://doi.org/10.1007/s11274-011-0709-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0709-3