Abstract
Superoxide dismutases (SODs; EC 1.15.1.1) are key enzymes in the cells protection against oxidant agents. Thus, SODs play a major role in the protection of aerobic organisms against oxygen-mediated damages. Three SOD isoforms were previously identified by zymogram staining from Allium sativum bulbs. The purified Cu, Zn-SOD2 shows an antagonist effect to an anticancer drug and alleviate cytotoxicity inside tumor cells lines B16F0 (mouse melanoma cells) and PAE (porcine aortic endothelial cells). To extend the characterization of Allium SODs and their corresponding genes, a proteomic approach was applied involving two-dimensional gel electrophoresis and LC–MS/MS analyses. From peptide sequence data obtained by mass spectrometry and sequences homologies, primers were defined and a cDNA fragment of 456 bp was amplified by RT-PCR. The cDNA nucleotide sequence analysis revealed an open reading frame coding for 152 residues. The deduced amino acid sequence showed high identity (82–87%) with sequences of Cu, Zn-SODs from other plant species. Molecular analysis was achieved by a protein 3D structural model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses represent a major cause of reactive oxygen species (ROS) overproduction by aerobic organisms, e.g., singlet oxygen, hydrogen peroxide, superoxide, and hydroxyl [1–3]. This phenomenon could lead to a disturbance in the normal redox state and cause a number of pathologic processes.
To repair damages initiated by ROS, cells have developed complex antioxidative systems [4–6]. Among antioxidants, superoxide dismutases (SODs) can convert superoxide anion into hydrogen peroxide, which is then degraded by catalases, giving water and molecular oxygen. SODs are considered as important enzymes to limit oxygen radical-mediated toxicity.
Four types of SODs have been characterized yet, based on the nature of the metal co-factor present at the catalytic site, i.e., copper/zinc (Cu, Zn-SOD), iron (FeSOD), manganese SODs (MnSOD), or nickel SOD. Cu, Zn-SOD is found in cytosol and plastids [7]. MnSOD is generally found in mitochondria, whereas FeSOD is present in prokaryotes [8] and within chloroplasts of some plants [9]. Nickel SOD was first purified from Streptomyces ssp. (Youn et al. 1996).
In the last few years, a wide range of clinical SOD applications has been reported. These include prevention of oncogenesis and tumor promotion [10, 11] and protection against reperfusion damages of ischemic tissue [12]. In addition, many interesting articles report on the effects of SOD in aging. Treatments using SODs seem to be promising alternatives for conventional therapies and some of them have been proposed in the treatment of oxidative stress-related diseases [13].
Recent studies dealing with SOD over expression in different systems obviously demonstrated some features of the recombinant SOD. Thus, the yeast become more resistant to environmental oxidative stress, e.g., heat shock or herbicide treatment, such as paraquat, when over expressing the human SOD1 [14]. The expression of the MnSOD in transgenic Alfalfa (Medicago sativa) reduced efficiently the damage caused by water deficit stress [15]. The over expression of the Cu, Zn-SOD also increased the resistance of Arabidopsis thaliana to salt stress by lignification of vascular structure [16] and its over expression in glioma human cells suppresses tumor growth [11].
Actually, it is admitted that SODs are indispensable for the control of the oxidative stress in plants [17]. All these data emphasize the high potential applications of SODs.
Molecular cloning of SODs has been reported in various prokaryotic and eukaryotic species and in particular in plants, e.g., Potentilla atrosanguinea [18], tobacco [19], tomato [20], maize [21], rice [7], and aspen [22], and fungi such as Beauveria bassiana [23]. However, no data was published on SOD sequences from Allium species and in particular Allium sativum, which played an important dietary and medicinal role throughout the history of mankind.
In a previous study, we demonstrated the great relevance of garlic SODs application. The native MnSOD enzyme has been shown to be able to activate the extra-regulated kinases (ERKs) in tumor cell lines and to influence the ability of cells to undergo mitosis, causing inhibition of tumor growth [24]. We showed also that the purified Cu, Zn-SOD2 from A. sativum was functional in an in vitro eukaryotic system and able to modulate the redox status of tumor cells [25].
The objective of this study is to investigate biochemical properties of Cu, Zn-SOD2 in cancer therapy. Indeed, since cytoplasmic Cu, Zn-SOD activity has a variable profile through cancer cells types [26] and the effect of over-expressed Cu, Zn-SOD has been examined in only a few cancers [10]. Some preliminary experiments were designed in this study, using pure enzyme to compare the effect of a common anticancer drug treatment, doxorubicin, on two tumor cell lines: B16F0 (mouse melanoma cells) and PAE (porcine aortic endothelial cells) in the presence or absence of pure Cu, Zn-SOD2.
In addition, a molecular characterization of the SOD from A. sativum has been performed by using a proteomic approach combining two-dimensional gel electrophoresis and LC–MS/MS. Biochemical data and bioinformatic study allowed to make out the coding region and therefore to determine the corresponding amino acid sequence of the Cu, Zn-SOD2 from A. sativum. Besides, we performed a model structure using the Swiss-model tool basing in the amino acid sequence homology with other characterized SOD structures.
Materials and Methods
MTT Reduction Test for Cell Toxicity
The cell lines B16F0 (mouse melanoma cells) and PAE (porcine aortic endothelial cells) were maintained in adequate media as described previously [25]. To assess cell survival after cytotoxic treatment, cells were seeded at 5 × 104/well (B16F0) or 6.25 × 104/well (PAE) in multi-well plate in the presence of 0.1 μg/ml. After 24 h, surviving cells were stained with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) as described by Mossman [27].
Preparation of Garlic Protein Extract
Bulbs of A. sativum L. were crushed in liquid nitrogen, stirred in a 50 mM Tris–HCl buffer pH 7.25, containing 0.1 mM ethylenediamine tetraacetic acid (EDTA), 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 0.5% (v/v) Triton X-100, then filtered and centrifuged. The supernatant was brought to 70% saturation with ammonium sulfate. The precipitate was resuspended in 50 mM, Tris–HCl buffer, pH 7.25, and desalted against the same buffer.
The crude protein extract (150 mg/ml) was loaded onto a column (100 × 2.2 cm) of Sephacryl S200-HR gel (Pharmacia, Uppsala, Sweden) for gel filtration chromatography. Fractions with SOD activity were pooled and used in further analyses.
All steps were carried out at 4°C, and SOD activity was measured at each step by using the epinephrine method [28]. For all experiments, protein amounts were measured using the Bio-Rad protein assay. SOD proteins (60 μg) were separated in 10% native PAGE using Laemmli’s Tris–glycine buffer system [29]. SOD activity on gels was observed by using the nitroblue tetrazolium (NBT) illumination method described by Beauchamp and Fridovich [30].
Two-Dimensional Gel Electrophoresis (2DE)
The active pool from gel filtration chromatography was precipitated by 100% trichloroacetic acid (TCA). The protein pellet (100 μg) was first dried then rehydrated in 400 μl of the first dimension buffer of the following composition: 7 M urea, 2 M thiourea, 65 mM 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 20 mM dithiothreitol (DTT), 2 mM tributylphosphine (TBP), 0.8% (w/v) carrier ampholytes (pH 3–11, Sigma®), and 0.01% (w/v) bromophenol blue. Immobiline DryStrips (18 cm, pH 3–10 nonlinear, from Amersham, Pharmacia Biotech) were rehydrated by protein samples. Isoelectric focusing (IEF) was performed by using the IEF cell apparatus (Bio-Rad) as follows: active rehydratation for 12 h at 50 V, 250 V for 15 min, gradient from 250 to 10,000 V for 3 h and final focusing for 12 h at 10,000 V. Strips were then stored at −20°C. Before the second dimension, strips were equilibrated for 10 min in a buffer containing 1% DTT. A second equilibration step was performed for 10 min in a buffer containing 4% iodoacetamide. The second dimension (SDS-PAGE) was carried out in Protean II Xi vertical system (Biorad), by using 12.5% polyacrylamide resolving gels (width 16 cm, length 20 cm, thickness 0.75 cm). The strips were embedded on the top of the gels and electrophoresis was carried out at 20 mA per gel.
After migration, proteins were visualized by silver nitrate staining (developing duration: 15 min) as described by Rabilloud et al. [31]. Spot quantification was achieved by computing scanning densitometry (ProXPRESS 2D, PerkinElmer Sciex). The digitized images were analyzed using the PerkinElmer ProFINDER 2D software version 2004.
Tryptic Digestion and Mass Spectrometry
Protein spots were excised from two-dimensional gels using an automatic spot cutter (ProXCISION, PerkinElmer) inside the chosen region (dashed square in Fig. 3). Excised plugs were washed and dried in a SpeedVac centrifuge for few minutes. Trypsin digestion (12.5 ng/ml trypsin dissolved in 0.1 M ammonium bicarbonate) was performed using an automatic digester (MultiPROBE II, PerkinElmer Sciex) as previously described [32]. After lyophilization, the peptide extracts were resuspended in 10 μl of 0.2% formic acid and 5% acetonitrile.
Peptides were enriched and separated using a lab-on-a-chip technology (Agilent, Massy, France) and fragmented using an on-line XCT mass spectrometer (Agilent). The fragmentation data were interpreted using the Data Analysis program (version 3.4, Bruker Daltonic, Billerica, MA, USA). For protein identification, MS/MS peak lists were extracted and compared with the protein database using the MASCOT Daemon (version 2.1.3; Matrix Science, London, UK) search engine. The searches were performed with no fixed modification and with variable modifications for oxidation of methionine, and with a maximum of one missed cleavage. MS/MS spectra were searched with a mass tolerance of 1.6 Da for precursor ions and 0.8 for MS/MS fragments.
If a protein was characterized by two peptides with a fragmentation profile score higher than 25 (default value of MASCOT), the protein was validated. When one of the criteria was not met, peptides were systematically checked and/or interpreted manually to confirm or cancel the MASCOT suggestion.
Molecular Study
Chemical Reagents and Media
Pfu DNA polymerase and primers were purchased from Bio basic Inc. IPTG (isopropyl β-D-1-thiogalactopyranoside), X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside), DTT, and ampicillin were from Sigma. Cloning procedure was carried out in Escherichia coli NEB5 (New England Bio Labs). T4 DNA ligase was obtained from Promega and the pUC19 plasmid from Invitrogen. E. coli cells were grown in LB supplemented with 1.5% agar for plate cultures.
Primers Conception
On the basis of the known N-terminal sequence [25], the internal peptide sequences obtained by LC–MS/MS analysis and the percentage of homology with plant SOD genes, a set of primers was chosen to amplify the garlic Cu, Zn-SOD2 gene. The primers composition and features were performed by using the Oligo-Analyser v3.1 software. Designing primers with degeneration was avoided. Thus, only nucleotides the most frequently found in conserved regions of plant Cu, Zn-SOD sequences was kept.
RNA Purification and RT-PCR Analysis
Fresh garlic bulb (1 g) was ground in liquid nitrogen to powder in a ceramic mortar. Total RNA was isolated using SV Total RNA Isolation System (Promega). An amount (5 μg) of total RNA was used for the first strand cDNA synthesis in a total reaction volume of 20 μl, in the presence of 200 U M-MLV reverse transcriptase (Invitrogen) and 0.5 μg of oligo dT primer.
The synthesized single strand cDNA served as a template for PCR amplification using primer sets: forward, (FN1) 5′-GTGAAGGCTGTTGCTGTT-3′ and reverse, (Y2) 5′-CCTTGGAGACCAATGATACC-3′. Thermal cycling reactions were carried out for 35 cycles (94°C for 1 min, hybridization at 52°C for 1 min, and 72°C for 1 min). The PCR was “hot-started” at 94°C for 10 min and completed by an elongation step of 10 min at 72°C. To confirm product synthesis and estimate DNA concentration by comparison with standard markers, 5 μl of each reaction was submitted to electrophoresis in a 1.2% agarose gel in Tris–acetate-EDTA buffer (TAE).
Identification of the SOD cDNA by Specific Nested PCR
The cDNA fragment obtained from the first PCR amplification was purified using Mini Elute gel extraction kit (Qiagen) and submitted to a PCR using the specific internal primers: forward, (X1) 5′-GCACCAGAGGATGAAAACCGC-3′ and reverse, (W2) 5′-CCTTGGAGGCCGATAATTCC-3′.
Cloning of the Amplified PCR Fragment
The PCR product was cloned in SmaI linearized pUC19-plasmid using T4 DNA ligase. Competent E. coli strain NEB5α was transformed with the ligation mix and plated onto selective medium supplemented with ampicillin (100 μg/ml), X-Gal (10 μg/ml), and IPTG (40 μM) for blue/white colonies selection. Plasmids were isolated from white colonies by alkaline lysis protocol according to Sambrook et al. [33], and then analyzed by PCR amplification. Recombinant vectors were sequenced from both ends, using the universal M13 forward and reverse primers (United States Biochemical, Cleveland, USA).
Sequence Analysis and Deduced Amino Acid Primary Structure
The cDNA was sequenced on automated DNA sequencer (Model 370A, Applied Bio systems). The corresponding amino acid sequence was deduced with DNAMAN program and then analyzed on the Expasy Swiss Prot Web Server (http://www.expasy.ch) [34]. Homology and identity analyses were carried out using BLAST through the NCBI server [35]. Nucleotide and amino acid sequences of SODs were derived from the EMBL/GenBank [36] and SwissProt databases, respectively. Multiple sequence alignment of the nucleotide sequences were generated with the DNAMAN program (v5.2.2; Lynnon Biosoft, Que., Canada) and the 450-bp regions were compared.
Structure Modeling
The 3D model structure of SOD was obtained using the online Swiss-Model program (http://swissmodel.expasy.org/) [37] based on homology modeling with Potentilla atrosanguina Cu, Zn SOD structure (Ref. PDB: 2Q2L). Visualization and analysis of the structure were performed with PyMol program (www.pymol.org) [38].
Results and Discussion
Cytoprotector Effect of Cu, Zn-SOD2
To understand whether the native Cu, Zn-SOD2 has an anti-proliferative effect on malignant cells as observed with MnSOD [24], experiments were carried on.
In a preliminary step, the results showed that pure enzyme addition does not affect tumoral cell lines viability. For this purpose, we determined the reaction of malignant cell lines treated by one of most used anticancer drugs: doxorubicin also named adriamycin (0.1 μg/ml). The results were compared to those obtained when adding 8 U of pure Cu, Zn-SOD2 (Fig. 1). In both doxorubicin-treated cell lines, we notated the decrease of cell survival of about 45 and 20% for B16F0 and PAE, respectively. These data correlate with the effective doxorubicin role since it is a widely used anthracycline class chemotherapeutic agent. This molecule is a quinone-containing antitumor antibiotic, which can be reduced to free radical semiquinone [39]. This semiquinone cannot induce DNA damage by itself but also redox cycles with O2 producing anion superoxide [40]. Superoxide radicals can react with hydrogen peroxide to form highly reactive hydroxyl radicals via the iron catalyzed Haber–Weiss reaction. The secondarily derived hydroxyl radicals can cause protein and DNA damage and initiate lipid peroxidation [41]. Here, we observed a resistance increase of tumor cells treated with 8 U of pure Cu, Zn-SOD2. Apparently, pure Cu, Zn-SOD2 alleviates anticancer drug cytotoxicity. The possible explanation is that the presence of Cu, Zn-SOD2 increases the dismutation rate of superoxide anion to hydrogen peroxide, which was scavenged by peroxide-removing enzymes (catalases and glutathione reductases). In particular, antioxidant’s defense system is usually changed during carcinogenesis or in tumors [42]. The difference in the activities of direct antioxidants enzymes (SOD and catalase) is generally lowered as compared to adjacent tumor free tissues [43]. As expected, SOD over-expressing cells cope with higher levels of oxygen radicals generated by redox-cycling agents, thereby preventing the initiation of apoptotic signaling pathways [44].
Increased resistance to doxorubicin of B16F0 and PAE cells in the presence of 8 U Cu, Zn-SOD2. Cells survival was evaluated by MTT reduction assay. B16F0 (5 × 104 cell/ml) and PAE (6.25 × 104 cell/ml) was treated with 0.1 μg/ml doxorubicin (24 h). All data were expressed by mean values ± SEM of duplicate samples. Statistical analysis was carried out using student’s t test. Statistical P value <0.05 was considered significant
From this finding, it can be concluded to the cytoprotector role of garlic Cu, Zn-SOD from doxorubicin. Many studies suggested that MnSOD could play a protective role against the cytotoxicity of a number of agents including UV radiation, anticancer drugs, and pro-oxidant chemicals in cell culture model [45, 46]. Thus, it has been reported that MnSOD over expression can improve the adriamycin-induced mitochondrial damage in the heart of transgenic mice [47]. Few data deals with copper–zinc SOD features.
Our current study shows that pure Cu, Zn-SOD2 maintains cellular integrity under adverse conditions, at the same manner that over expression of the Cu, Zn-SOD which, increased the resistance of Arabidopsis thaliana to salt stress by lignification of vascular structure [16].
Therefore, we started a protocol to clone and express these proteins to over produce enzymes and deepen our results.
Garlic Protein Extract
SOD activity was obtained from garlic protein extract (60 μg) obtained as shown in “Materials and Methods”. The NBT staining performed on native PAGE revealed the presence of three bands at pH (Fig. 2) [25]. They were designed as SOD1, 2, 3 according to their migration distance toward anode. For identification of various SOD isoforms, gel bands were treated with specific inhibitors as KCN (3 mM) or H2O2 (5 mM) in pre-equilibrating buffer before SOD activity staining. SOD1 was not sensitive to both reagents and was identified as manganese superoxide dismutase (MnSOD); SOD2 and SOD3 activities were both inhibited and consequently classified as two Cu, Zn-SODs (Cu, Zn-SOD2 and Cu, Zn-SOD3) [25].
Two-Dimensional Electrophoresis
Because of long, hard purification steps and low rate of protein extraction, 2DE analysis was successfully used to analyze the active SOD fraction directly after gel filtration chromatography. Electropherogram discriminated about 300 spots after silver staining (Fig. 3). For garlic SOD iso-enzymes identification, a region containing about 50 spots was selected on the basis of biochemical data deduced from Fig. 3. In addition, considered the subunit molecular weight known to be around 16 kDa for cytoplasmic SODs [48] and 25 kDa for manganese SOD [49] and second the isoelectric point known to range between 5 and 7 [25].
Identification of SODs
All chosen spots were subjected to in gel digestion and LC–MS/MS analysis. Trypsin produces a series of different molecular mass fragment of each particular protein. This analysis allowed to identify the spot 11 as a chloroplastic SOD (SOD3) with ion score equal to 75; the spot 20 as a MnSOD with ion score equal to 76 and the spot 34 corresponding to cytoplasmic Cu, Zn-SOD with ion score equal to 120 (Table 1). MnSODs are homotetrameric proteins. In our study, the molecular weight calculated for the subunit was evaluated to about 25 kDa, close to the mass average (25 kDa) of known MnSODs [49]. The isoelectric point of manganese isoforms is generally in the range of 7–8.5 [50, 51]. The molecular weight calculated for the chloroplastic SOD (SOD3) is about 22 kDa. Murai et al. [52] found that chloroplastic SOD of Solidago altissima shows as well as pI equal to 5.8.
The three internal peptides from Cu, Zn-SOD2 are shown in Table 2. The peptide sequence coverage observed is about 19%. Comparison of peptide sequences pointed out similarities with sequences of other plant Cu, Zn-SODs like those from Sandersonia aurantiaca, Nelumbo nucifera, and Populus tremuloides (Mascott data not shown).
Cu, Zn-SOD2 Gene Cloning and Sequencing
We attempted to identify the nucleotide sequence of the Cu, Zn-SOD2 gene. From peptide sequences obtained (see above) and homologies with known plant SODs nucleotide sequences, two primers FN1 and Y2 were generated and used for RT-PCR amplification. The amplified fragment displayed a length of 456 bp (Fig. 4) and matched with predicted fragment length. To confirm PCR specificity, two internal primers (X1, W2) were used for further PCR amplification. Result pointed out an amplicon of 250 bp as expected (Fig. 5).
Nested PCR amplification cDNA Cu, Zn-SOD2 analysis. Lane 1 negative control, lane 2 100 bp molecular weight marker (Promega). Lanes 3, 4, and 5 nested PCR occurred in different hybridization temperatures (54, 56, and 58°C). Lane 6 positive control (PCR with FN1/Y2 amplifying the cDNA total sequence)
Complete nucleotide and amino acid sequences of Cu, Zn-SOD2 are shown in Fig. 6. The open reading frame consisted of 456 bp (152 amino acids), and the N-terminal amino acid sequence deduced from the nucleotide sequence was consistent with the sequence determined from the purified protein except for the first methionyl residue, which was probably removed for maturation, as observed for other SOD proteins. The predicted molecular mass was 15.181 kDa, which was close to that estimated by SDS-PAGE for the Cu, Zn-SOD2 [25].
Nucleotide sequence of the garlic Cu, Zn-SOD2 cDNA and its deduced amino acid sequence. Numbers refer to nucleotide and its deduced amino acid residues. Asterisk indicates a stop codon. Bold letters indicate amino acids implicated in metal coordination. Underlined letters show the disulfur bridges. The dashed underlines show primers localization. The full lines with arrows indicate peptides obtained by LC–MS/MS
The predicted protein contains residues considered as essential for coordinating copper (His-45, -47, -62, and -119) and zinc (His-62, -70, -79, and Asp-82) atoms. Two cysteines (C-56 and C-145) that form a disulfide bond are also conserved. The sequence analysis shows that the functional residues conserved in all reported Cu, Zn-SOD sequences are present in SOD2 [53, 54].
The nucleotide sequence data of the Cu, Zn-SOD2 cDNA was deposited in the GenBank Database under accession number GU290312.2.
Investigation of protein sequence similarity revealed a high percentage of identity of the Allium Cu, Zn-SOD with those of other plant Cu, Zn-SODs. Highest identities were found with SODs from Sandersonia aurantiaca (gb/AAL85888.1) (87%), Oryza sativa [55], and Populus trichocarpa (gb/ABN58428.1) (85%). High identities were also found with those of Zea mays [56] and P. atrosanguina [18] of about 84 and 82%, respectively (Fig. 7).
Optimal alignment of Cu, Zn-SOD of different plant species. The alignment of amino acid sequences translated from cDNA sequences using the DNAMAN program. The plant names and the accession numbers in GenBank are: gar_SOD, A. sativum (gb/ADB28989), this study; Pot_SOD P. atrosanguina (gb/ACB38158); Sand_SOD, Sandersonia aurantiaca (gb/AAL85888.1); ory_SOD, Oryza sativa (gb/ABY52933.1); Zea_SOD, Zea mays (gb/AAB49913.1); PopT_SOD, Populus trichocarpa (gb/ABN58428). Asterisks refer to identities with garlic
Cu, Zn-SOD2 Structure Modeling
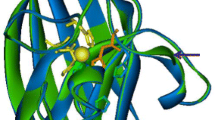
To identify the tri-dimensional structure of SOD2, we performed molecular modeling by Swiss-model. For that purpose, we used as template the X-ray crystallographic structure of P. atrosanguina Cu, Zn SOD (reference PDB: 2Q2L) [18].This model template presents the only available 3D structure having the closet identity with our SOD protein.
By using a modeling approach, a representation of the overall polypeptide chain fold was obtained for Cu, Zn-SOD2 and a high homology of structure with the protein from P. atrosanguina could be observed. The Cu, Zn-SOD2 displays a structure of eight-stranded antiparallel β-barrel with the well-studied Greek key fold connectivity (Fig. 8), which is observed in all (eukaryotic and prokaryotic) Cu, Zn-SOD structures [57]. As observed in other SODs, Cu, Zn-SOD2 contains an intra-subunit disulfide bond, and a zinc-binding site that contribute to its extreme thermo-chemical stability [58]. It should be noted that all residues interacting with the ligand are completely conserved between the model and the template. The RMSD between the interacting residues of model and template is lesser than two: 0.040. Given by the properties calculated in the ligand zZN was included in the final model of Potentilla SOD. Furthermore, the model structure of SOD2 was successfully performed with a dimer (data not shown) consistently with the dimer of two Potentilla SOD molecules in the crystallographic data. Besides, data revealed in native gel suggest that Cu, Zn-SOD2 oligomerises as an homodimer. These structural features may be correlated to the extreme thermo-chemical stability showed previously [25].The Cu, Zn-SOD2 enzyme activity was unaltered till 50°C and decrease with 20% at 70°C after 30 min incubation at pH 7.5.
Comparison of the model structure of Cu, Zn SOD2 (a) and the X-ray crystallography of P. atrosanguina SOD (b). The Ribbon model structure of the Cu, Zn-SOD2 of A. sativum was created based on the known structure of P. atrosanguina (PDB: 2Q2L). Structural views were performed via PyMol program. a Structure model of Cu, Zn-SOD2 from A. sativum. b Crystallographic structure of Potentiella atrosanguina SOD. The Zn metal ion is represented as a gray sphere (only one chain of the P. atrosanguina Cu, Zn-SOD is shown here). Histidine residues implicated into copper and zinc coordination are shown as sticks. The disulfide bonds are also shown with arrows
This was also observed in other SODs displaying such disulfide bonds, metal binding site and Greek key. The fully metallated and disulfide intact protein (Cu2, Zn2SODS-S) which is highly stable has a melting temperature (T m) of 95°C [58], while the metal-free and disulfide-reduced SOD protein (SODSH) is more unstable and exhibits a T m of 42°C [59].
In addition, the Greek key could also be a crucial element for the thermal stability as suggested from autoclavable P. atrosanguina Cu, Zn-SOD resistance [60].The conserved structure folding in SOD2 concomitant to its thermal stability support the hypothesis that Greek key together with disulfide bond and native metal binding participate to the thermal stability of SOD family.
Conclusions
In this study, we concluded first to the cytoprotector role of garlic copper, zinc SOD from cytotoxic stimuli. Then, we demonstrate that proteomic approach is a successful strategy to characterize internal protein sequences in the aim to clone respective genes. Such an approach combined with molecular and computational tools allowed to identify the nucleotide and amino acid sequences of SOD protein from A. sativum. The study of recombinant SOD action will be carried out and it is assumed that it should be the same as for native enzyme from garlic. This will allow us to more investigate in therapeutic application.
References
Hernández, J. A., Olmos, E., Corpas, F. J., Sevilla, F., & Del Rio, L. A. (1995). Salt induced oxidative stress in chloroplast of pea plants. Plant Science, 105, 151–167.
Foyer, C. H., & Noctor, G. (2000). Oxygen processing in photosynthesis: Regulation and signaling. New Phytologist, 14, 6359–6388.
Anderson, J. A. (2002). Catalase activity, hydrogen peroxide content and thermotolerance of pepper leaves. Scientia Horticulturae, 95, 277–284.
Foyer, C. H., Descourvières, P., & Kunert, K. J. (1994). Protein against oxygen radicals: An important defence mechanism studied in transgenic plants. Plant, Cell & Environment, 17, 507–523.
Smirnoff, N. (1998). Plant resistance to environmental stress. Current Opinion in Biotechnology, 9, 214–219.
Kuzniak, E., & Skodowska, M. (2004). The effect of Botrytis cinerea infection on the antioxidant profile of mitochondria from tomato leaves. Journal of Experimental Botany, 55, 605–612.
Sakamoto, A., Ohsuga, H., & Tanaka, K. (1992). Nucleotide sequences of two cDNA clones encoding different Cu/Zn superoxide dismutases expressed in developing rice seed. Plant Molecular Biology, 19, 323–327.
Pagani, S., Colnaghi, R., Palag, A., & Negri, A. (1995). Purification and characterization of an iron superoxide dismutase from the nitrogen-fixing Azotobacter vinelandii. FEBS Letters, 35, 779–782.
Bannister, J. V., Bannister, W. H., & Rotilio, G. (1987). Aspects of the structure, function and applications of superoxide dismutase. CRC Critical Reviews in Biochemistry, 22, 111–180.
Weydert, C. J., Waugh, T. A., Ritchie, J. M., Kanchan, S. I., Smith, J. L., & Li, L. (2006). Overexpression of manganese or copper–zinc superoxide dismutase inhibits breast cancer growth. Free Radical Biology and Medicine, 41, 226–237.
Zhang, Y., Zhao, W., Zhang, H. J., Domann, F. E., & Oberley, L. W. (2002). Overexpression of copper zinc superoxide dismutase suppresses human glioma cell growth. Cancer Research, 62, 1205–1212.
Kondo, T., Terajima, H., Todoroki, T., Hirano, T., Ito, Y., Usia, T., et al. (1999). Prevention of hepatic ischemia-reperfusion injury by SOD-DIVEMA conjugate. Journal of Surgical Research, 85, 26–36.
Muzykantov, V. R. (2001). Targeting of superoxide dismutase and catalase to vascular endothelium. Journal of Controlled Release, 71, 1–21.
Yoo, H. Y., Kim, S. S., & Rho, H. M. (1999). Overexpression and simple purification of human superoxide dismutase (SOD1) in yeast and its resistance to oxidative stress. Journal of Biotechnology, 68, 29–35.
McKersie, B. D., Bowley, S. R., Harjanto, E., & Leprince, O. (1996). Water-deficit tolerance and field performance of transgenic alfalfa overexpression superoxide dismutase. Plant Physiology, 111, 1177–1181.
Gill, T., Sreenivasulu, Y., Kumar, S., & Ahuja, P. S. (2010). Over-expression of superoxide dismutase exhibits lignifications of vascular structure in Arabidopsis thaliana. Journal of Plant Physiology, 167, 757–760.
Alsher, R. G., Erturk, N., & Heath, L. S. (2002). Role of superoxide dismutase (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany, 53, 1131–1141.
Yogavel, M., Gill, J., Mishra, P. C., & Sharma, A. (2007). SAD Phasing of a structure based on cocrystallized iodides using an in-house Cu Kalpha X-ray source: Effects of data redundancy and completeness on structure solution. Acta Crystallographica D, 63, 931–934.
Bowler, C., Aliotte, T., De Loose, M., Van Montagu, M., & Inze, D. (1989). The induction of manganese superoxide dismutase in response to stress in Nicotiana plumbaginifolia. EMBO Journal, 8, 31–38.
Perl-Treves, R., & Galun, E. (1991). The tomato Cu/Zn superoxide dismutase genes are developmentally regulated and respond to light and stress. Plant Molecular Biology, 17, 745–760.
Cannon, R. E., & Scandalios, J. G. (1989). Two cDNAs encode two nearly identical Cu/Zn superoxide dismutase proteins in maize. Molecular and General Genetics, 219, 1–8.
Akkapeddi, A. S., Shin, D. I., Stanek, M. T., Karnosky, D. F., & Podila, G. K. (1994). cDNA and derived amino acid sequence of the chloroplastic copper/zinc-superoxide dismutase from aspen (Poplus tremuloides). Plant Physiology, 106, 1231–1232.
Xie, X. Q., Ying, S. H., & Feng, M. G. (2010). Characterization of a new Cu/Zn-superoxide dismutase from Beauveria bassiana and two site-directed mutations crucial to its antioxidation activity without chaperon. Enzyme and Microbial Technology, 46, 217–222.
Hadji Sfaxi, I., Ferraro, D., Fasano, E., Pani, G., Limam, F., & Marzouki, M. N. (2009). Effects of a manganese superoxide dismutase purified from Allium sativum on tumoral cell culture. Biotechnology Progress, 25, 257–264.
Hadji, I., Marzouki, M. N., Ferraro, D., Fasano, E., Majdoub, H., Pani, G., et al. (2007). Purification and characterization of a Cu, Zn-SOD from garlic (Allium sativum L.). Antioxidant effect on tumoral cell lines. Applied Biochemistry and Biotechnology, 143, 129–141.
Oberley, L. W. (1982). Superoxide dismutase and cancer. In L. W. Oberley (Ed.), Superoxide dismutase, Vol. 2 (pp. 127–165). Boca Raton, FL: CRC Press.
Mossman, T. (1983). Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxic assays. Journal of Immunological Methods, 65, 55–63.
Misra, H. P., & Fridovich, I. (1972). The role of superoxide anion in autoxidation of epinephrine and a simple assay for SOD. Journal of Biological Chemistry, 247, 3170–3175.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Beauchamp, C., & Fridovich, I. (1971). Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry, 44, 276–287.
Rabillaud, T., Valette, C., & Lawrence, J. J. (1994). Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis, 15, 1552–1558.
Coquet, L., Vilain, S., Cosette, P., Jouenne, T., & Junter, G. A. (2006). A proteomic approach of biofilm cell physiology. In J. M. Guisan (Ed.), Methods in biotechnology: Immobilization of enzymes and cells, 3rd ed. (pp. 403–414). Totowa, NJ: Humana.
Sambrook, J., & Russel, D. W. (2001). Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Expasy Swiss Prot Web Server (http://www.expasy.ch).
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI BLAST: A new generation of protein database search programs. Nucleic Acid Research, 25, 3389–3402.
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., Rapp, B. A., & Wheeler, D. L. (2006). GeneBank. Nucleic Acid Research, 32, 23–26.
Swiss-Model program (http://swissmodel.expasy.org/).
PyMOL version 1.4.1 (www.Pymol.org/).
Gewirtz, D. A. (1999). A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochemical Pharmacology, 57, 727–741.
Kalyanaraman, B., Perez-Reyes, E., & Mason, R. P. (1980). Spin-trapping and direct electron spin resonance investigations of the redox metabolism of quinone anticancer drugs. Biochimica et Biophysica Acta, 630, 119–123.
Halliwell, B., & Gutteridge, J. M. C. (1989). Free radicals in biology and medicine, 2nd ed. (p. 543). New York: Oxford University Press.
Oberley, L. W., & Oberley, T. D. (1986). Free radicals, cancer, and aging. In J. E. Johnson Jr., R. Walford, D. Harmon, & J. Miquel (Eds.), Free radicals, aging, and degenerative diseases (pp. 325–381). New York: Alan R, Liss Inc.
Tsidale, M. J., & Mahmoud, M. B. (1983). Activities of free radical metabolizing enzymes in tumours. British Journal of Cancer, 47, 809–812.
Brockhaus, F., & Brune, B. (1999). Overexpression of CuZn superoxide dismutase protects RAW 264.7 macrophages against nitric oxide cytotoxicity. Biochemical Journal, 338, 295–303.
Hirose, K., Longo, D. L., Oppenhaim, J. J., & Matsushima, K. (1993). Over-expression of mitochondrial manganese superoxide dismutase promotes the survival of tumor cells exposed to interleukin 1, tumor necrosis factor, selected anticancer drugs and ionizing radiation. FASEB Journal, 7, 3361–3368.
Guo, H. L., Wolfe, D., Epperly, M. W., et al. (2003). Gene transfer of human manganese superoxide dismutase protects small intestinal villi from radiation injury. Journal of Gastrointestinal Surgery, 7, 229–235.
Yen, H. C., Oberley, T. D., Vichitbandha, S., Ho, Y. S., & St Clair, D. K. (1996). The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. Journal of Clinical Investigation, 98, 1253–1260.
Karpinska, B., Karlsson, M., Schinkel, H., Streller, S., Suss, K. H., Melzer, M., et al. (2001). A novel superoxide dismutase with a high isoelectric point in higher plants. Expression, regulation and protein localization. Plant Physiology, 126, 1668–1677.
Rodrıguez-Serrano, M., Romero-Puertas, M. C., Pastori, G. M., Corpas, F. J., Sandalio, L. M., del Rıo, L. A., et al. (2007). Peroxisomal membrane manganese superoxide dismutase: Characterization of the isozyme from watermelon (Citrullus lanatus Schrad.) cotyledons. Journal of Experimental Botany, 58, 2417–2427.
Kwon, S. I., & An, C. S. (1999). Isolation and characterization of mitochondrial manganese superoxide dismutase (MnSOD) from Capsicum annuum L. Molecules and Cells, 31, 625–630.
Vyas, D., & Kumar, S. (2005). Purification and partial characterization of a low temperature responsive Mn-SOD from tea (Camellia sinensis L.). Biochemical and Biophysical Research Communications, 15, 831–838.
Murai, R., & Murai, K. (1996). Different transcriptional regulation of cytosolic and plastidic Cu/Zn-superoxide dismutase genes in Solidago altissima (Asteraceae). Journal of Plant Sciences, 120, 71–79.
Liu, W., Zhu, R. H., Li, G. P., & Da-Cheng, W. (2002). cDNA cloning, high-level expression, purification and characterization of an avian Cu, Zn superoxide dismutase from Peking duck. Protein Expression and Purification, 25, 379–388.
Shin, S. Y., Lee, H. S., Kwon, S. Y., Kwon, S. T., & Kwak, S. S. (2005). Molecular characterization of a cDNA encoding copper/zinc superoxide dismutase from cultured cells of Manihot esculenta. Plant Physiology and Biochemistry, 43, 55–60.
Lu, L. M., Qin, M. L., Lan, H. H., Wang, P., Niu, X. Q., Wu, Z. J. & Xie, L. H. (2007) Construction and characterization of a yeast two-hybrid cDNA library from rice seedling leaves. Agriculture and Forestry University, Fuzhou, Fujian, China: Plant Protection, Plant Virology.
Gardiner, J., Schroeder, S., Polacco, M. L., Sanchez-Villeda, H., Fang, Z., Morgante, M., et al. (2004). Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiology, 134, 1317–1326.
Getzoff, E. D., Tainer, J. A., Stempien, M. M., Bell, G. I., & Hallewell, R. A. (1989). Evolution of Cu, Zn superoxide dismutase and the Greek key b-barrel structural motif. Proteins: Structure, Function, and Bioinformatics, 5, 322–336.
Hayward, L. J., Rodriguez, J. A., Kim, J. W., Tiwari, A., Goto, J. J., Cabelli, D. E., et al. (2002). Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. Journal of Biological Chemistry, 277, 15923–15931.
Rodriguez, J. A., Shaw, B. F., Durazo, A., Sohn, S. H., Doucette, P. A., Nersissian, A. M., et al. (2005). Destabilization of apoprotein is insufficient to explain Cu, Zn-superoxide dismutase-linked ALS pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 102, 10516–10521.
Kumar, S., Sahoo, R. & Ahuja, P. S. (2002). Isozyme of autoclavable superoxide dismutase (SOD), a process for the identification and extraction of the SOD in cosmetic, food and pharmaceutical compositions. United States Patent 6,485,950.
Acknowledgments
This study was sustained by a financial support of the Bioengineering Laboratory 99UR09-26 (INSAT) from the Tunisian Ministry of High Education Scientific Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hadji Sfaxi, I., Ezzine, A., Coquet, L. et al. Combined Proteomic and Molecular Approaches for Cloning and Characterization of Copper–Zinc Superoxide dismutase (Cu, Zn-SOD2) from Garlic (Allium sativum). Mol Biotechnol 52, 49–58 (2012). https://doi.org/10.1007/s12033-011-9473-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9473-8