Abstract
We present here the structural modeling and biochemical characterization of a recombinant superoxide dismutase (SOD) from Deschampsia antarctica E. Desv. [Poaceae] produced in Escherichia coli. The recombinant protein was purified by affinity chromatography nickel-nitrilotriacetic acid (Ni-NTA), and its identity was demonstrated by immunoblotting and inhibition by H2O2 and KCN. Inductively coupled plasma optical emission spectroscopy (ICP-OES) analysis confirmed the presence of Cu and Zn. Modeling of the D. antarctica Cu/Zn-SOD (DaSOD) amino acid sequence using the SWISS-MODEL and 2Q2L_B monomer of the psychrophilic Cu/Zu-SOD from Potentilla atrosanguinea (PaSOD) as template produced a structure similar to that of the typical eukaryotic Cu/Zn-SODs. Activity assays using the p-nitro blue tetrazolium chloride (NBT) solution method showed that the purified DaSOD had a specific activity of 5818 U/mg at 25 °C and pH 7.2 and that it was active in a pH interval of 5–8 and a temperature interval of 0–40 °C. Furthermore, DaSOD was still active at −20 °C as observed by a zymogram assay. We found 100 % activity when it was heated at 80 °C for 60 min, indicating a high thermostability. DaSOD properties suggest that this enzyme could be useful for preventing the oxidation of refrigerated or frozen foods, as well as in the preparation of cosmetic and pharmaceutical products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deschampsia antarctica E. Desv. [Poaceae] is one of the only two native vascular plants living in Antarctica [1]. Due to the conditions of its habitat, including frozen ground, ice/snow cover, deficient precipitation, incidence of low illumination during the winter, and high UV radiation during summer, high levels of reactive oxygen species (ROS) are produced; thus, as a response, high levels of peroxidase, glutathione reductase, and superoxide dismutase (SOD) activities are present in this plant [2].
Superoxide dismutase is the first line of defense against ROS; particularly, this converts the superoxide anion to molecular oxygen and hydrogen peroxide [3]. There are four types of SODs, and each possesses a distinct metal ion in its active site: Mn, Fe, Ni, or Cu/Zn [4, 5]. Eukaryotic Cu/Zn-SODs are highly conserved from primary to quaternary structure, and they are composed of two identical subunits. Each has a β-barrel of eight antiparallel β-chains forming a Greek key motif [6]. SODs are used in the preparation of a large variety of cosmetic and health-promoting supplements [7–9]; therefore, the search for SODs whose properties allow for better performance is of high biotechnological value.
A previous work has demonstrated the overexpression of superoxide dismutase reported in this work, in response to stress induced by cold and UV light in D. antarctica [10]. However, to our knowledge, this is the first report dealing with the structural modeling and biochemical characterization of the recombinant Cu/Zn-SOD from D. antarctica (DaSOD) expressed in Escherichia coli. Structural modeling was performed using the SWISS-MODEL and a psychrophilic Cu/Zu-SOD from Potentilla atrosanguinea (PaSOD) as template [11]. The recombinant protein was purified by affinity chromatography and its activity was assessed.
Materials and Methods
In Silico Modeling
The three-dimensional structure of the DaSOD was performed using its amino acid sequence (access number ACV65038.1) and the comparative protein modeling PROTEIN SWISS-MODEL server (http://swissmodel.expasy.org/), employing the 2Q2L_B monomer (access number ACB38158.1) belonging to the P. atrosanguinea Cu/Zn-SOD [11] as template.
Strain and Culture Media
E. coli BL21-SI (Gibco, Darmstadt, Germany) was transformed with the plasmid pDaSOD containing the DaSOD-His 6 gene under the control of the T7 promoter. A detailed description of the molecular vehicle used, the concentration of trace elements in culture medium, and procedures for strain preservation and inoculum preparation can be found elsewhere [12]. The production medium contained 5 g/L glucose, 3.5 g/L (NH4)2HPO4, 3.5 g/L KH2PO4, 1.0 g/L MgSO4, 40 μg/L thiamine, 35 mg/L kanamycin (Sigma-Aldrich, St. Louis, MO, USA), and trace elements. Before autoclaving, the pH of the medium was adjusted to 7.4 with 10 M NaOH. Preinocula were grown overnight in 100 mL of the production medium plus 5 g/L yeast extract (Difco Laboratories, Franklin Lakes, NJ, USA). Flasks were incubated in a shaker at 250 rpm and 37 °C.
Protein Expression
Batch cultures were performed in a 1.3 L bioreactor (Applikon) equipped with two six-blade Rushton turbines and stirred at 300 rpm. The cultures were started with 1 L of production medium at an initial optical density at 600 nm (OD600 nm) of 0.2. Batch cultures were performed at 37 °C until an OD600 nm of 0.6 was attained. Then, the expression was induced with 0.3 M NaCl, and the post-induction temperature was changed to 32.5 °C. The pH was maintained at 7.0 by automatic addition of a 2 N NaOH solution and dissolved oxygen at 20 % using an ADI-1030 Bio-controller (Applikon Biotechnology, Schiedam, Netherlands) and the BioXpert v1.3 software (Applikon).
DaSOD Purification and Quantification
Culture samples collected from the bioreactor were harvested at 16,000g for 2 min at 4 °C in a centrifuge model 5810R (Eppendorf, Hamburg, Germany), resuspended in 0.1 M phosphate buffer pH 7.8 (PBS), and sonicated in an Ultrasonic processor GE 505 (Sonics, Newtown, CT, USA) using ten pulses of 10 s at 25 % amplitude and 10 s resting between pulses. The soluble fraction was recovered by centrifugation at 8000g for 15 min at 4 °C. Protein purification was carried out using nickel-nitrilotriacetic acid (Ni-NTA) affinity columns with the ProBond Purification System (Invitrogen Corporation, Carlsbad, CA, USA) following manufacturer instructions. Protein was dialyzed in a 10 mM Tris-HCl buffer pH 7.5 using an Amicon cell (Millipore, Bedford, MA, USA) with ultrafiltration membranes Ultracel YM-10 (Millipore) for 3 h at 4 °C. The protein concentrations in the samples were quantified with Bradford protein assay using BSA as standard [13].
Analytical Procedures
Proteins were separated by 4–20 % gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a Mini-PROTEAN III System (Bio-Rad, Hercules, CA, USA). Proteins were visualized with Coomassie Blue R-250 (Bio-Rad) for the bioreactor production analysis or Silver Stain (Bio-Rad) after the purification process. For Western blot, proteins were transferred from the gel onto a nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ, USA) using a Semi-Dry Trans-Blot (Bio-Rad). The membrane was blocked with low-fat powder milk (3 %, w/v in PBS). The membrane was incubated with the mouse anti-His tag monoclonal antibody 0.2 μg/mL (AbD Serotec, Oxford, UK), followed by goat antimouse IgG antibody conjugated to alkaline phosphatase 1:3000 (Bio-Rad), and visualized with p-nitro blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP, Amersham Biosciences). Analysis of gels and nitrocellulose membranes was carried out using a photo-documenter Gel-Doc 2000 (Bio-Rad) and the Quantity OneTM v4.5 software (Bio-Rad).
Metal Quantification
The metal content in the purified DaSOD was analyzed in an inductively coupled plasma optical emission spectrometer (ICP-OES), Varian 730 (Palo Alto, CA, USA).
DaSOD Activity
DaSOD activity was visualized by the polyacrylamide method of Beauchamp and Fridovich [14] as follows: 3 μg of purified protein was electrophoresed by a 4–20 % SDS-PAGE. The electrophoresis was carried out at 4 °C and 100 V for 4 h. Then, the gels were washed twice at room temperature with 50 mL of 10 mM Tris-HCl buffer (pH 7.9) containing 25 % (v/v) of isopropanol for 25 min. The gels were incubated for 25 min in 50 mM phosphate buffer (pH 7.8) containing 2.5 mM NBT, washed briefly, and incubated for 15 min with 25 mL of 50 mM PBS (pH 7.8) with 1.1 mM riboflavin (Sigma, St. Louis, MO) and 1.4 mM TEMED (Sigma). Gels were washed three times with Milli-Q water and exposed to a 50-W white-light source (Phillips, IN) until bands were visualized. Bovine erythrocyte SOD (Sigma) was used as a control to show the activity. DaSOD activity was tested against 10 mM KCN and 5 mM H2O2 using zymograms. The specific DaSOD activity was measured by the free solution method of McCord and Fridovich [15]. In brief, 3 mL of reaction buffer contained 13 mM l-methionine, 0.1 mM EDTA, 75 mM NBT in 50 mM PBS (pH 7.8), and 3 μL purified enzyme or 3 μL water as the control. At time zero, 2 mM riboflavin was added and exposed to a 3800–4000-μCd LED white-light source (Steren, Mexico) located 3 cm above from a 3-mL cuvette. Reduction of NBT was monitored for 10 min at 560 nm using the Spectrophotometer Cary BIO-50 (Varian, Palo Alto, CA, USA) coupled with a cell-holder Peltier (Varian) with agitation and temperature control. Measurements were taken in triplicate and the average was used for results analysis. One unit of activity was defined as the amount of enzyme needed to attain half of the maximum inhibition of NBT reduction [15].
Effect of Temperature, pH, and Thermostability
To determine the effect of temperature on the activity of the DaSOD, 5-μL aliquots of the purified enzyme were subjected to temperatures from 0 to 70 °C with increments of 10 °C during the activity assay. The effect of pH was determined after incubating a 5-μL aliquot of the purified DaSOD in 145 μL of various buffers with a pH range from 2 to 10, for a period of 12 h. The buffers contained, respectively, 50 mM KCl/HCl (pH 2), 50 mM glycine/HCl (pH 3), 50 mM CH3COOH/CH3COONa (pH 4.5), 50 mM KH2PO4/NaOH (pH 6–8), and glycine/NaOH (pH 9–10). Thermostability was determined after incubating 5 μL of the enzyme at temperatures from 80 to 110 °C by intervals of 10 °C for 60 min and by increments of 10 min in a Peltier thermal plate (Bio-Rad). The relative activity of the enzyme subjected to the different treatments was determined by the free solution method described above.
Results and Discussion
In Silico Modeling
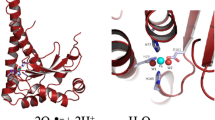
The DaSOD amino acid sequence was submitted to the SWISS-MODEL server for modeling using the automatic mode. The program constructed a tertiary structure using the 2Q2L_B biological unit as a template, which has a percentage identity of 82.237 %, with the DaSOD. This suggests that its tertiary and quaternary structures are similar to those of the template (Fig. 1). The 2Q2L_B unit belongs to one of the monomers of a Cu/Zn-SOD of the P. atrosanguinea plant induced by low temperatures [16]. This issue is interesting because P. atrosanguinea is a plant that grows in the western Himalayas, India, under extreme environmental conditions of high UV and low temperatures, similar to those supported by D. antarctica in the South Pole [17]. Additionally, we performed an Atomic Non-Local Environment Assessment (ANOLEA) diagram, which calculated the favorable energy of the predicted structure, to give reliability to the model [18]. As shown in Fig. 2, the key structural elements of the DaSOD activity were located in similar areas as PaSOD. The predicted DaSOD structure is similar to that of other Cu/Zn-SODs, which have a dimer structure comprising two identical subunits. Each monomer contains an eight-barrel chain with seven loops [19]. The structural differences between DaSOD and PaSOD are the disulfide loops and Greek key (Fig. 1).
Superimposition of DaSOD (green) and PaSOD (blue) obtained by SWISS-MODEL. The differences found in the Greek key and disulfide loops are shown with green and violet arrows, respectively. Histidine involved in the coordination of Zn+2 ion is shown in yellow, histidine bridge in orange, which coordinates both Zn+2 and Cu+2 ions. Green and blue histidines show those involved in the coordination of Cu+2 ion
Expression, Immunodetection, and Purification of the Recombinant DaSOD
DaSOD was produced in batch cultures, and the maximum protein production was reached at 5 h of culture after induction with 0.3 M NaCl (Fig. 3). Identification of recombinant DaSOD after purification is shown in Fig. 4. Recombinant DaSOD was purified through a column of Ni-NTA affinity under stringent conditions to prevent binding of contaminating proteins as indicated by the supplier. Western blot analysis was performed using a monoclonal antibody against the His6-tag. The assay detected a single band of 16 kDa corresponding to the fused DaSOD-His6 demonstrating the identity of the DaSOD (Fig. 4a, b). The purity of the DaSOD was determined by densitometry analysis of a SDS-PAGE with silver staining, showing that the protein had at least 97 % purity (Fig. 4c).
DaSOD-His6 identification after purification. a SDS-PAGE analysis of protein DaSOD purified by affinity chromatography on a column of Ni-NTA agarose. Lane 1 10 μL of sample eluted, lane 2 molecular weight marker. b Immunoblot of SDS-PAGE gel shown in a. c Analysis by SDS-PAGE of the purity of the DaSOD-His6. Ten microliters of various fractions eluted from the Ni-NTA column were electrophoresed, and the gel was stained with AgNO3. Lane 1 molecular weight marker, lanes 2–5 different fractions of the protein eluted with imidazole
Biochemical Characterization of the DaSOD
DaSOD assays in solution using 5 mM H2O2 or 10 mM KCN fully inhibited the enzyme activity, supporting the notion of DaSOD belonging to the Cu/Zn-SOD family. To corroborate the content of metal ions in the DaSOD, the metal content was determined by inductively coupled plasma optical emission spectroscopy (ICP-OES). The analysis showed that the DaSOD contains Cu and Zn in amounts of 0.1072 and 0.1791 μg-atom/mgprotein, respectively, confirming that it is a Cu- and Zn-dependent metalloprotein.
The enzymatic characterization of the DaSOD is summarized in Figs. 5, 6, 7, and 8. The enzyme activity was measured over a pH range of 2–10. As shown in Fig. 5, the enzyme was unaffected by pH in the range of 5–8, whereas a drop of the activity was observed above pH 8. The interval of pH insensibly depends on the protein origin. For instance, the black soybean Cu/Zn-SOD had maximum activity between 6 and 8 [20], whereas tomato Cu/Zn-SOD had an optimal pH of 7.8 [21]. As seen in Fig. 6, the optimum reaction temperature of DaSOD is between 10 and 30 °C with a maximum observed at 25 °C, which decreases as the temperature increases from 40 to 70 °C. It is noteworthy that the enzyme had only a 20 % reduction of the maximal activity when incubated at 0 °C and having detectable activity when assessed at −20 °C in SDS-PAGE as reported by Garcia-Echauri et al. [12]. These data clearly indicate that the DaSOD belongs to a very small group of SODs active at cold temperatures (0 °C) [22], and the second of its kind active at −20 °C [23]. The thermal stability of DaSOD was investigated by incubating at 80, 90, 100, and 110 °C, respectively. The results showed that DaSOD was not affected at 80 °C, and the half-life time was 35 min at 100 °C, whereas at 110 °C, the enzyme was completely inactivated in 20 min (Fig. 7). A hyper-thermostable SOD isolated from a polyextremophile higher plant, P. atrosanguinea, was engineered by mutation of a single amino acid that enhanced the thermostability of the enzyme twofold [11].
Under optimal conditions, the purified DaSOD had a specific activity of 5818 U/mg at 25 °C and pH 7.2. As seen in Fig. 8, the inhibition percentage of NBT photoreduction assay was not linear with the concentration of DaSOD, showing a typical Michaelis-Menten behavior. Under the conditions of our assay and using the equation of Asada et al. [24, 25], the K′ that is a function of the concentration of NBT and determines the affinities of DaSOD and NBT on the superoxide anion was 0.1719 μg/mL [24, 25].
Conclusions
We present the structural modeling and biochemical characterization of a recombinant superoxide dismutase (SOD) from D. antarctica E. Desv. produced in E. coli. DaSOD exhibits some properties similar to those of most plant Cu/Zn-SODs, such as molecular weight, thermal stability, and pH stability. However, DaSOD shows activity under freezing conditions and high thermoresistance. DaSOD properties suggest that this enzyme could be useful for preventing the oxidation of refrigerated or frozen foods, as well as in the preparation of cosmetic and pharmaceutical products. More studies on possible isoforms of this enzyme in response to variations in cold-stress in vivo plants are needed to understand the morpho-physiological response of this psychrophilic vascular plant.
References
Bravo, L. A., Ulloa, N., Zuniga, G. E., Casanova, A., Corcuera, L. J., & Alberdi, M. (2001). Cold resistance in Antarctic angiosperms. Physiologiae Plantarum, 111, 55–65.
Perez-Torres, E., Garcia, A., Dinamarca, J., Alberdi, M., Gutierrez, A., Gidekel, M., Ivanov, A. G., Huner, N. P. A., Corcuera, L. J., & Bravo, L. A. (2004). The role of photochemical quenching and antioxidants in photoprotection of Deschampsia antarctica. Functional Plant Biology, 31, 731–741.
Fridovich, I. (1997). Superoxide anion radical (O-2 radical anion), superoxide dismutases, and related matters. Journal of Biological Chemistry, 272, 18515–18517.
Alscher, R. G., Erturk, N., & Heath, L. S. (2002). Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany, 53, 1331–1341.
Youn, H. D., Kim, E. J., Roe, J. H., Hah, Y. C., & Kang, S. O. (1996). A novel nickel-containing superoxide dismutase from Streptomyces spp. Biochemical Journal, 318, 889–896.
Richardson, J. S. (1977). beta-Sheet topology and the relatedness of proteins. Nature, 268, 495–500.
Bafana, A., Dutt, S., Kumar, S., & Ahuja, P. S. (2011). Superoxide dismutase: an industrial perspective. Critical Reviews in Biotechnology, 31, 65–76.
Di Mambro, V. M., & Fonseca, M. J. (2007). Assessment of physical and antioxidant activity stability, in vitro release and in vivo efficacy of formulations added with superoxide dismutase alone or in association with alpha-tocopherol. European Journal of Pharmaceutics and Biopharmaceutics, 66, 451–459.
Mizushima, Y., Hoshi, K., Yanagawa, A., & Takano, K. (1991). Topical application of superoxide dismutase cream. Drugs Under Experimental and Clinical Research, 17, 127–131.
Sanchez-Venegas, J. R., Dinamarca, J., Moraga, A. G., & Gidekel, M. (2009). Molecular characterization of a cDNA encoding Cu/Zn superoxide dismutase from Deschampsia antarctica and its expression regulated by cold and UV stresses. BMC Research Notes, 2, 198.
Kumar, A., Dutt, S., Bagler, G., Ahuja, P. S., & Kumar, S. (2012). Engineering a thermo-stable superoxide dismutase functional at sub-zero to >50 degrees C, which also tolerates autoclaving. Scientific Reports, 2, 387.
Garcia-Echauri, S. A., Gidekel, M., Moraga, A. G., Ordonez, L. G., Contreras, J. A. R., Barba de la Rosa, A. P., & De Leon Rodriguez, A. (2009). Heterologous expression of a novel psychrophilic Cu/Zn superoxide dismutase from Deschampsia antarctica. Process Biochemistry, 44, 969–974.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Beauchamp, C., & Fridovich, I. (1971). Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Analytical Biochemistry, 44, 276–287.
McCord, J. M., & Fridovich, I. (1969). Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). Journal of Biological Chemistry, 244, 6049–6055.
Yogavel, M., Gill, J., Mishra, P. C., & Sharma, A. (2007). SAD phasing of a structure based on cocrystallized iodides using an in-house CuK alpha X-ray source: effects of data redundancy and completeness on structure solution. Acta Crystallography D, 63, 931–934.
Yogavel, M., Mishra, P. C., Gill, J., Bhardwaj, P. K., Dutt, S., Kumar, S., Ahuja, P. S., & Sharma, A. (2008). Structure of a superoxide dismutase and implications for copper-ion chelation. Acta crystallographica. Section D, Biological crystallography, D64, 892–901.
Chodanowski, P., Grosdidier, A., Feytmans, E., Michielin, O. (2008) Local alignment refinement using structural assessment. Plos One 3.
Perry, J. J. P., Shin, D. S., Getzoff, E. D., & Tainer, J. A. (2010). The structural biochemistry of the superoxide dismutases. Bba-Proteins Proteomics, 1804, 245–262.
Wang, S. Y., Shao, B., Liu, S. T., Ye, X. Y., & Rao, P. F. (2012). Purification and characterization of Cu, Zn-superoxide dismutase from black soybean. Food Research International, 47, 374–379.
Kumar, S., Dhillon, S., Singh, D., & Singh, R. (2004). Partial purification and characterization of superoxide dismutase from tomato (Lycopersicon esculentum) fruit. Journal of Food Sciences and Nutrition, 9, 283–288.
Zheng, Z., Jiang, Y. H., Miao, J. L., Wang, Q. F., Zhang, B. T., & Li, G. Y. (2006). Purification and characterization of a cold-active iron superoxide dismutase from a psychrophilic bacterium, Marinomonas sp NJ522. Biotechnology Letters, 28, 85–88.
Sahoo, R., Kumar, S., & Ahuja, P. S. (2001). Induction of a new isozyme of superoxide dismutase at low temperature in Potentilla astrisanguinea Lodd. variety argyrophylla (Wall. ex. Lehm) Griers. Journal of Plant Physiology, 158, 1093–1097.
Giannopolitis, C. N., & Ries, S. K. (1977). Superoxide dismutases. I: occurrence in higher plants. Plant Physiology, 59, 309–314.
Asada, K., Takahashi, M., & Nagate, M. (1974). Assay and inhibitors of spinach superoxide dismutase. Agricultural and Biological Chemistry, 38, 471–473.
Acknowledgments
This work was partially financed by CONACyT-Básicas Grant No. 178988. Juan Rojas thanks CONACyT for his scholarship No. 204213. The authors thank Leandro G. Ordoñez for technical support and Jennifer Ecklerly for English correction. We thank the Chilean Antarctic Institute (INACH) for the logistic support during the stay in the Scientific Base “Prof. Julio Escudero,” King George Island, Antarctic.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rojas-Contreras, J.A., de la Rosa, A.P.B. & De León-Rodríguez, A. Expression and Characterization of a Recombinant Psychrophilic Cu/Zn Superoxide Dismutase from Deschampsia antarctica E. Desv. [Poaceae]. Appl Biochem Biotechnol 175, 3287–3296 (2015). https://doi.org/10.1007/s12010-015-1496-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1496-3