Abstract
Somatic mutations in the PIK3CA gene are common in breast cancer and represent a clinically useful marker for prognosis and therapeutic target. Activating mutations in the PI3K p110 catalytic subunit (PIK3CA) have been identified in 18–40 % of breast carcinomas. In this study, we evaluated PIK3CA mutation in 185 Indian breast cancer patients by direct DNA sequencing. PIK3CA mutations were observed in 23.2 % (43/185) of breast tumor samples. PIK3CA mutations were more frequent exon 30 (76.8 %) than in exon 9 (23.2 %). Mutations were mostly clustered within two hotspot region between nucleotides 1624 and 1636 or between 3129 and 3140. Sequencing analysis revealed four different missense mutations at codon 542 and 545 (E542K, E545K, E545A and E545G) in the helical domain and two different amino acid substitutions at codon 1047 (H1047R and H1047L) in the kinase domain. None of the cases harbored concomitant mutations at multiple codons. PIK3CA mutations were more frequent in older patients, smaller size tumors, ductal carcinomas, grade II tumors, lymph node-positive tumors and non-DCIS tumors; however, none of the differences were significant. In addition, PIK3CA mutations were common in ER+, PR+ and HER2+ cases (30 %), and a comparatively low frequency were noted in triple-negative tumors (13.6 %). In conclusion, to our knowledge, this is the largest study to evaluate the PIK3CA mutation in Indian breast cancer patients. The frequency and distribution pattern of PIK3CA mutations is similar to global reports. Furthermore, identification of molecular markers has unique strengths and can provide insights into the pathogenic process of breast carcinomas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite several advancement in our current understandings of cancer in recent times, carcinoma of the breast still remains one of the leading malignancies of females worldwide. Although the highest incidence and mortalities occur in developed countries, as the western lifestyle is being adopted by increasing parts of the world, breast cancer cases continue to increase across the globe, including in India. In fact, it is the second most common cancer after cervical cancer in India with 115,251 new diagnoses, and almost 50 % of them succumb to the disease in 2008 [1]. Ironically, the average age of developing breast cancer in India has shifted over the last few decades and younger women are being affected. As we are entering into the era of personalized medicine, identification of prognostic marker and cancer therapeutics that specifically target a tumor’s genetic is the current unmet clinical need.
The phosphoinositide 3-kinase (PI3K) pathway plays an important and critical role in cell differentiation, cell proliferation, survival and migration [2–4]. PI3Ks have oncogenic potential and several components of these signaling pathways are altered in various human cancers [5]. Somatic mutations of PIK3CA gene represent the most common alterations currently known in breast cancer patients, occurring at a frequency of 20–45 % [6–8]. In vitro studies indicate that the expression of mutant PIK3CA that encodes the p110α catalytic subunit of the PI3K enzymes results in uncontrolled activation of the PI3K/AKT pathway and induces anchorage- and growth factor-independent proliferation, protection from apoptosis and drug resistance [9]. More than 80 % of these mutations occur at two “hotspots” in exon 9 and exon 20, which encode the helical (E542K and E545K) and kinase (H1047R) domains, respectively [10].
It is interesting to note that the available literature regarding the prognostic significance of PIK3CA mutations highlights conflicting results. Some study series reported negative clinical outcome in patients with PIK3CA mutation [11, 12], while other group of researchers demonstrated good clinical outcome in terms of significantly more relapse-free and metastasis-free survival in patients with PIK3CA mutant breast cancers [8, 13]. Notably, variable findings have also been reported in terms of association of PIK3CA with hormone and HER status in breast cancer. Several larger, population-based studies show a significant association between PIK3CA mutations and ER-/PR-positive, HER2-negative tumors [14, 15], while in contrast few reports suggest no significant correlation between PIK3CA mutations and hormonal status [16–18]. In addition, patients with PIK3CA mutation and HER2 amplified tumors are less responsive to combinations of HER2 inhibitors, adding to the prognostic and therapeutic significance of PIK3CA mutation testing [13, 19].
Most of the available reports on PIK3CA gene mutations in breast cancer come from western world [5, 7, 20] and Asian countries [21, 22], while there are only two small studies from India [6, 23]. However, no larger precise published data are available with respect to PIK3CA gene mutation in Indian breast cancer patients, despite the fact that carcinoma of breast is quite common disease in India. Notably, Indian patients appeared to develop breast cancer at much younger ages than those in the west, with the median age at onset ranging between 40 and 45 years only [24]. There is a need to investigate the presence of PIK3CA gene mutation in larger cohort, which will enhance our knowledge in further understanding the genetic heterogeneity of Indian breast cancer patients. Therefore, in the present study, we report mutational spectrum of PIK3CA mutations from Indian patients and set out to evaluate their frequencies, distribution pattern and association with the clinicopathological characteristics.
Materials and methods
The present study was conducted at the Research and Development Division of SRL Ltd., Mumbai, India. A total of 185 formalin-fixed paraffin-embedded (FFPE) breast carcinoma tissue samples were evaluated in the current study. Tumor histology, tumor grade and other histological findings were evaluated by two independent experienced histopathologists as per Scarff–Bloom–Richardson classification. Immunohistochemical analyses using antibodies against estrogen receptors (ER), progesterone receptors (PR) and human epidermal growth factor receptor 2 (HER2) were performed following optimized laboratory protocol. Briefly, slides with tumor section were stained with ER using the rabbit monoclonal antibody, SP1 (Thermo scientific) with 1:600 dilutions, PR using the rabbit monoclonal antibody, SP2 (Thermo scientific) with 1:600 dilutions and HER2 using the mouse monoclonal antibody, CB11 (Biogenix) with 1:600 dilutions. Scoring criteria for ER/PR and HER2 testing were as per CAP/ASCO guidelines [25, 26]. Appropriate positive and negative control samples were included in each run. Treatment and outcome was not evaluated. The study is in accordance with Declaration of Helsinki, with consent available from study participant. The current study being a retrospective study, it was presented to and was approved by Institutional Research Committee (IRC).
Genomic DNA extraction
Genomic DNA was extracted from FFPE tissue using Qiagen extraction kit as per manufacturer’s instruction with slight modification. Prior to DNA extraction, tumors were histologically evaluated on hematoxylin & eosin (H&E) sections for the presence of at least 40 % tumor in each case as suitable for DNA extraction. At least, five FFPE sections of 5 µm thickness were processed for genomic DNA extraction. The extracted DNA was checked for quality on 0.8 % agarose gel and quantitated using Qubit (Invitrogen).
PIK3CA mutation analysis for exon 9 and 20
Mutation analysis of PIK3CA exon 9 and 20 was performed using hemi-nested and nested PCR approach, respectively, using primers as per previous report with slight modifications [27–29]. The primer sequences used for PCR amplification were as follows: PIK3CA_Ex9_Outer_Forward 5′-TTGCTTTTCTGTAAATCATCTGTG-3′, PIK3CA_Ex9_Inner_Forward 5′-GGGAAAAATATGACAAAGAAAGC-3′ and Common_Ex9_Reverse 5′-GAATCTCCATTTTAGCACTTACCTGTGACT-3′, and for PIK3CA Exon 20, the primers were: Outer Forward 5′-TTTTCTCAATGATGCTTGGC-3′, Outer Reverse 5′-GGATTGTGCAATTCCTATGC-3′ and Inner Forward 5′-AATCTTTTGATGACATTGCATACATTCG-3′, Inner Reverse 5′-TCAGTTATCTTTTCAGTTCAATGCATG-3′. The total volume for both exon 9 and exon 20 nested PCR was 25 µl containing 50–100 ng of starting genomic DNA, 10 pmol of forward and reverse primers and 1X of HotStarTaq Master Mix (Qiagen). The PCR conditions consisted of an initial denaturation at 95 °C for 15 min followed by 25 cycles at 94 °C for 30 s, 60 °C for 30 s (exon 9, both rounds) and for exon 20, first round at 55 °C and second round at 64 °C for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 10 min. Nested PCR products were visualized on 2 % agarose gel.
Sequence analysis
The amplified products for PIK3CA exon 9 and 20 were purified using QIAquick PCR purification kit (Qiagen, Hilden, Germany) and directly sequenced in both the direction by Automated ABI 3500 dx Genetic Analyzer (Applied Biosystems Inc., Foster City, CA, USA) using ABI PRISM BigDye terminator kit. The abnormal results were reconfirmed by at least two repeated analysis right from PCR amplification. Furthermore, a wild-type sequencing control was run for comparison of abnormal results.
Statistical analysis
The data were analyzed by Chi-square or Fisher’s exact tests to study the correlation between the clinicopathological features and the occurrence of a particular mutation. All the p-values were two-tailed and the statistical significance was set at p < 0.05.
Result
Clinical characteristics of breast carcinoma cases
In the current study, we analyzed a total of 185 tumor tissues from breast carcinoma patients. The details of the clinical characteristics of all patients are depicted in Table 1. Almost equal numbers of breast cancer patients were in ≤50 and >50 years age groups. The median age of the cohort was 50 years ranging from 22 to 86 years. The size of the tumors in the majority of the cases were >2.5 cm (70.8 %), while some of them were <2.5 cm (29.2 %). Histopathological examination of the tumors revealed that that bulk of the cases were ductal in origin (n = 178, 96.1 %), followed by lobular (n = 5, 2.7 %) and other (n = 2, 1.2 %) category. Furthermore, the tumors were graded following the Scarff–Bloom–Richardson classification criteria, wherein ~50 % of the tumors were classified as grade III, 47 % as grade II and only 3.3 % as grade I. The presence of ductal carcinoma in situ, lymphovascular and perineural invasion was noted in 26.5, 36.8 and 10.8 % of the tumor samples, respectively. Most of the tumors were found to be associated with node positivity (~57 %) while in the remaining 43 % (80/185) cases, no node involvement was observed. Furthermore, tumor subtyping with immunohistochemical analysis for ER, PR and HER2 antibody revealed that 63.2, 49.2 and 28.2 % of the cases showed positive expression for the respective markers. Tumors were further classified into four different molecular types on the basis of IHC markers. Among these, HR+/HER2− group was the most predominant molecular type accounting for 48 % of the specimen, followed by HR−/HER2− (23.8 %), HR+/HER2+ (16.2 %) and HR−/HER2+ (12 %).
PIK3CA mutation type and its correlation with clinicopathological findings
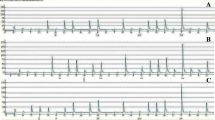
Of the 185 specimen analyzed by direct sequencing, 43 cases (23.2 %) harbored PIK3CA mutations while the remaining 142 cases were normal for both the exons. PIK3CA mutations were predominantly found in exon 20 (n = 33, 76.8 %) as compared to exon 9 (n = 10, 23.2 %) (Tables 2, 3). All the 43 mutations observed in both the exons were missense mutations as a result nucleotide substitution that predicted to change the amino acid at the respective codons (Fig. 1a–f). Most of the mutations detected in the current study occurred within two mutational hotspot regions: between nucleotide positions 1624 and 1636 (n = 10) or between 3129 and 3140 (n = 33) (Table 3).
We identified four different amino acid substitutions at codon 542 and 545 (E542K, E545K, E545A and E545G) in the helical domain of the PIK3CA gene as depicted in Tables 2 and 3(Fig. 1a–d). Missense mutation E542K and E545K were the most recurrent exon 9 mutations (4.4 %) followed by E545A and E545G (1 %) (Table 2). Similarly, two different amino acid substitutions were observed at codon 1047 (H1047R and H1047L) in the kinase domain (Fig. 1e–f). Of these, H1047R missense mutation was more prevalent (16.7 %) in exon 20 as comparison to H1047L (1.1 %) mutations (Table 2). All specimens with different mutations were heterozygous and retained a wild-type allele. None of the cases harbored concomitant mutations at multiple codons.
The correlation of clinicopathological data with PIK3CA mutation is summarized in Table 1. It can be noted that older patients tend to harbor marginally increased frequency of PIK3CA mutation in comparison with younger cases (24.2 vs. 23.3 %, p = 0.86); similarly, PIK3CA mutations were commonly found in smaller size tumors (≤2.5 cm) as compared to larger sized tumors (26 vs. 22 %, p = 0.7); however, these differences were not significant. On correlation with histopathological data, it was observed that PIK3CA mutation was predominantly found in ductal carcinomas while lobular carcinomas had lesser PIK3CA mutation (23.6 vs. 20 %, p = 0.7). Likewise, PIK3CA mutations were more prevalent in grade II tumors when compared to grade I and III tumors (25.3 vs. 22.8 %, p = 0.36). In addition, PIK3CA mutations were commonly observed in non-DCIS tumor as well as in tumors characterized by the presence of lymphovascular and perineural invasion. There were more cases with PIK3CA mutation in lymph node-positive tumors (26.6 %, 28/105), T4 tumors (44.4 %, 4/9), ER-positive tumors (25.6 %, 30/117), PR-positive tumors (28.6 %, 26/91) and HER2-positive tumors (28.8 %, 15/52). However, no statistical difference was observed between these parameters. Similarly, no association of PIK3CA mutations with molecular subtyping was observed, though a trend toward more frequent PIK3CA mutation in HR+/HER2+ (30 %, 9/30) subgroup was noted in comparison with other molecular subgroup (28 %, 34/121; p = 0.33).
Discussion
Carcinoma of breast is the second most common cancer after cervical cancer in India and is associated with high mortality rate. It is characterized by a heterogeneous group of tumors with great variability at the morphological and molecular levels. Activation of multiple signaling pathways specifically RAS–RAF–MAPK and PI3K–PTEN–AKT pathway plays an important role in regulating cell proliferation, angiogenesis, cell motility and apoptosis [30, 31].
In the current study, we evaluated the frequency and distribution patterns of PIK3CA somatic gene mutations in a cohort of 185 Indian breast carcinoma cases. Although PIK3CA gene mutations in breast cancer are extensively studied worldwide including two small studies in India (Table 4) [5–8, 13, 20–23, 32–43], to the best of our knowledge, this is one of the largest studies to evaluate the mutation status of PIK3CA gene in Indian patients. The frequency of PIK3CA mutations varies considerably across different parts of the globe, with reported incidence of about 14–34, 9–33, and 9–45 % in Asians, Americans and Europeans, respectively (Table 4). In this comprehensive study, we observed PIK3CA gene mutation in 23.2 % (43 of 185) of the breast carcinoma patients. Our frequency of 23.2 % is comparable to those reported from Germany, Japan and Greece (21–23 %) [22, 35, 39], higher than those from Netherlands, Korea and China (9–16 %) [21, 30, 43], while lower in comparison with Turkey, Australia and Denmark (31–45 %) [5, 8, 33]. In comparison with recent Indian studies, our frequency was pretty much similar to one study (20 %, 5/25) [6], though another study reported a higher mutation rate of 34 % (13/38) [23]. The differences in the frequencies across various studies can be attributed to sample size, sample selection, geographical distribution as well as use of sensitive techniques across some studies [32, 36].
The available literature suggests that over 90 % of the PIK3CA mutations are observed in exons 9 and 20, while only ~5–10 % of the mutations are found in other exons [32, 36]. Interestingly, we observed more mutations at exon 20 (helicase domain) than at exon 9 (kinase domain) which is consistent with most of the recent Asian and European studies [5, 33–36] but quite different from the result of an Australian study where in exon 9 was more frequently mutated [44]. In this study, in agreement with data from others [20, 37, 45], all PIK3CA mutations were of missense types with E542K/E545K and H1047R as the most recurrent mutation type. Nevertheless, irrespective of mutation type, our mutations were clustered within two well-known “hotspot” regions between nucleotides 1624 and 1636 (n = 10) or between 3129 and 3140 (n = 33) as it has been reported in other research studies [42, 45]. The exon 9 and 20 encodes the kinase and helicase domain of the PIK3CA gene, respectively, and provides auto-inhibition of the tyrosine kinases; however, mutations within this region initiate the process of constant auto-phosphorylation resulting in gain of function.
In the present series, the data show that somatic mutations in the PIK3CA gene were not significantly associated with several clinical and histopathological data which are consistent with previous findings [32, 35]. Concerning the age at diagnosis, a marginal higher PIK3CA mutation rate was noted in older patients which is in line with earlier reports [32, 46], but this trend was not observed in other series [16, 47]. The same is true for the histological type of breast cancer, when comparing ductal and lobular types. Some studies reported PIK3CA mutations to be more frequently observed in ductal than in lobular carcinomas [7, 13, 16] while others did not [15]. We found slightly more PIK3CA mutations in grade II tumors in comparison with grade III, suggesting that these mutations occur early in breast cancer development, which has also been shown in other studies [23, 36, 48]. In addition, the available literature suggests that the frequency of PIK3CA mutations in DCIS and non-DCIS is almost the same, and there is no significant difference among them [49]. The current study further corroborates the above findings wherein no statistical difference in the PIK3CA mutation rate was observed between DCIS and non-DCIS cases. Notably, a trend toward increased mutation frequency was also seen in node-positive cases, which is in contrast to a recent German study wherein more mutations were demonstrated in node-negative cases [36]. Furthermore, we found no association between mutations status and tumor size, tumor stage, lymphovascular invasion and perineural invasion.
Our data suggest that somatic mutations of PIK3CA gene are more common in ER+, PR+ and HER2+ cases, though no significant association was seen. Data from various research groups regarding the association of PIK3CA mutation with hormone receptor and HER2 status remain controversial till today. Some of them support positive association [7, 13], while in contrast some suggest no significant correlation between PIK3CA mutations and hormonal status [16–18] similarly to the results of the present study, or the association that is limited only to estrogen receptor status, but not to progesterone receptor status [15]. Similar observations have been demonstrated about the correlation between PIK3CA mutations and HER2 status [7, 13] but again are controversial in other series [15, 16] or observed when HER2 is determined using immunohistochemistry but not fluorescent in situ hybridization [17].
Molecular subtyping on the basis of hormone receptor and HER2 status such as basal-like, HER2+ and HR+ (luminal A and luminal B) are now being considered as an important aspect for effective patient management. We divided the cases into four different molecular types to understand the prevalence and association of PIK3CA mutation with these subtypes. PIK3CA mutations were more prevalent in HR+/HER2+ type (30 %) indicating that PIK3CA mutations might thus be characteristic of the luminal subtype which is in agreement previous finding [13]. Furthermore, a low frequency of PIK3CA mutations (13.6 %) in triple-negative tumors (HR−/HER2−), a subgroup reported to overlap with the basal-like subtype of breast cancer was noted which is in line with recent studies [13, 50]. As a matter of fact, another study from the USA reported a marked difference in PIK3CA mutation frequency across breast tumor subtypes, wherein PIK3CA mutations were more common in HR+ tumors (39 %) and ERBB2+ tumors (25 %) than in basal-like tumors (13 %) [50]. Nevertheless, despite these variations in frequencies, no significant association of PIK3CA mutation with molecular subtype was achieved (p = 0.34) which is consistent with earlier report [32].
As far as the prognostic significance of PIK3CA mutation is concerned, it is still debatable whether the presence of PIK3CA mutation is associated with good or poor clinical outcome. Some studies reported significantly longer metastasis-free survival and better clinical outcomes in patients with PIK3CA mutations [13, 51], while another series of studies reported lower survival and poor clinical outcomes [11, 12]. Interestingly, a recent systematic review of breast cancer clinical studies evaluation of 2587 breast cancer cases from 12 independent studies reported that patients with tumors harboring a PIK3CA mutation have a better clinical outcome than those with a wild-type PIK3CA gene [52]. Nevertheless, the impact of PIK3CA mutations on the clinical outcome of breast cancer seems to vary with the background of other genomic alterations such as HER2 status.
In summary, this is the first largest comprehensive study to report PIK3CA mutation in Indian breast cancer. The frequency and distribution pattern of PIK3CA mutations reported in the current study is similar to global reports. Similarities and dissimilarities of the present findings with those of other researchers may be attributed to the influence of different technologies, differences in ethnic origins as well as differential environmental exposure to unknown carcinogenic agents. Together, these data support the notion that the cancer-associated mutations in PIK3CA may significantly contribute to breast cancer pathogenesis and represent attractive targets for therapeutic inhibition.
References
Ferlay J, Shin HR, Bray F, et al. Cancer incidence and mortality worldwide: IARC cancer base no. 10. 2008. http://globocan.iarc.fr. Accessed 14 Feb 2016.
Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7.
Osaki M, Oshimura M, Ito H. PI3K–AKT pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–76.
Sui JQ, Xie KP, Zou W, et al. Emodin inhibits breast cancer cell proliferation through the ERα-MAPK/Akt-cyclin D1/Bcl-2 signaling pathway. Asian Pac J Cancer Prev. 2014;15:6247–51.
Li SY, Rong M, Grieu F, et al. PIK3CA mutations in breast cancer are associated with poor outcome. Breast Cancer Res Treat. 2006;96:91–5.
Sudhakar N, George Priya Doss C, Thirumal Kumar D, et al. Deciphering the impact of somatic mutations in exon 20 and exon 9 of PIK3CA gene in breast tumors among Indian women through molecular dynamics approach. J Biomol Struct Dyn. 2015;34:29–41.
Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049–59.
Dupont Jensen J, Laenkholm AV, Knoop A, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17:667–77.
Isakoff SJ, Engelman JA, Irie HY, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:992–1000.
Zhao L, Vogt PK. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–96.
Jensen JD, Knoop A, Laenkholm AV, et al. PIK3CA mutations, PTEN, and pHER2 expression and impact on outcome in HER2-positive early-stage breast cancer patients treated with adjuvant chemotherapy and trastuzumab. Ann Oncol. 2012;23:2034–42.
Baselga J, Cortés J, Im SA, et al. Biomarker analyses in CLEOPATRA: a phase III, placebo-controlled study of pertuzumab in human epidermal growth factor receptor 2-positive, first-line metastatic breast cancer. J Clin Oncol. 2014;32:3753–61.
Cizkova M, Susini A, Vacher S, et al. PIK3CA mutation impact on survival in breast cancer patients and in ER alpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28.
Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9.
Maruyama N, Miyoshi Y, Taguchi T, et al. Clinicopathological analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res. 2007;13:408–14.
Barbareschi M, Buttitta F, Felicioni L, et al. Different prog-nostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064–9.
Perez-Tenorio G, Alkhori L, Olsson B, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13:3577–84.
Ellis MJ, Lin L, Crowder R, et al. Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2010;119:379–90.
Baselga J, Cortés J, Im S-A, et al. Biomarker analyses in CLEOPATRA: A phase III, placebo-controlled study of pertuzumab in HER2-positive, first-line metastatic breast cancer (MBC). Thirty-fifth annual CTRC-AACR San Antonio breast cancer symposium, Dec 4–8, 2012, San Antonio, TX Cancer Res, 2012; 72 (24 Supplement): S5–S1.
Levine DA, Bogomolniy F, Yee CJ, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–8.
Lee JY, Park K, Lim SH, et al. Mutational profiling of brain metastasis from breast cancer: matched pair analysis of targeted sequencing between brain metastasis and primary breast cancer. Oncotarget. 2015;6:43731–42.
Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, et al. Prognostic role of PIK3CA mutations of cell-free DNA in early-stage triple negative breast cancer. Cancer Sci. 2015;106:1582–9.
Kandula M, Chennaboina KK, Ys AR, et al. Phosphatidylinositol 3-kinase (PI3KCA) oncogene mutation analysis and gene expression profiling in primary breast cancer patients. Asian Pac J Cancer Prev. 2013;14:5067–72.
Thakur NA, Humne AY, Godale LB. Delay in presentation to the hospital and factors affecting it in breast cancer patients attending tertiary care center in Central India. Indian J Cancer. 2015;52:102–5.
Hammond MEH, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohisto-chemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95.
Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013.
Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5.
Barbi S, Cataldo I, De Manzoni G, et al. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res. 2010;29:32.
Mao C, Zhou J, Yang Z, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS ONE. 2012;7:e36653.
Samuels Y, Diaz LA Jr, Schmidt-Kittler O, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–73.
Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62.
Harlé A, Lion M, Lozano N, et al. Analysis of PIK3CA exon 9 and 20 mutations in breast cancers using PCR-HRM and PCR-ARMS: correlation with clinicopathological criteria. Oncol Rep. 2013;29:1043–52.
Dirican E, Kaya Z, Gullu G, et al. Detection of PIK3CA gene mutations with HRM analysis and association with IGFBP-5 expression levels in breast cancer. Asian Pac J Cancer Prev. 2014;15:9327–33.
Castaneda CA, Lopez-Ilasaca M, Pinto JA, et al. PIK3CA mutations in Peruvian patients with HER2-amplified and triple negative non-metastaticbreast cancers. Hematol Oncol Stem Cell Ther. 2014;7:142–8.
Loibl S, von Minckwitz G, Schneeweiss A, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32:3212–20.
Arsenic R, Treue D, Lehmann A, et al. Comparison of targeted next-generation sequencing and Sanger sequencing for the detection of PIK3CA mutations in breast cancer. BMC Clin Pathol. 2015;15:20.
Ross JS, Ali SM, Wang K, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat. 2015;154:155–62.
Lips EH, Michaut M, Hoogstraat M, et al. Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res. 2015;17:134.
Papaxoinis G, Kotoula V, Alexopoulou Z, et al. Significance of PIK3CA mutations in patients with early breast cancer treated with adjuvant chemotherapy: a hellenic cooperative oncology group (HeCOG) study. PLoS ONE. 2015;10:e0140293.
Lee JW, Soung YH, Kim SY, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–80.
Karakas B, Colak D, Kaya N, et al. Prevalence of PIK3CA mutations and the SNP rs17849079 in Arab breast cancer patients’. Cancer Biol Ther. 2013;14:888–96.
Wang YL, Dai X, Li YD, et al. Study of PIK3CA, BRAF, and KRAS mutations in breast carcinomas among Chinese women in Qinghai. Genet Mol Res. 2015;14:14840–6.
Deng L, Chen J, Zhong XR, et al. Correlation between activation of PI3K/AKT/mTOR pathway and prognosis of breast cancer in Chinese women. PLoS ONE. 2015;10:e0120511.
Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81.
Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10(6):534–41.
Liedtke C, Cardone L, Tordai A, et al. PIK3CA activating mutations and chemotherapy sensitivity in stage II–III breast cancer. Breast Cancer Res. 2008;10:R27.
Lai YL, Mau BL, Cheng WH, et al. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064–9.
Dunlap J, Le C, Shukla A, et al. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res Treat. 2010;120:409–18.
Miron A, Varadi M, Carrasco D, et al. PIK3CA mutations in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–8.
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91.
Loi S, Haibe-Kains B, Majjaj S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc Natl Acad Sci. 2010;107:10208–13.
Dumont AG, Dumont SN, Trent JC. The favorable impact of PIK3CA mutations on survival: an analysis of 2587 patients with breast cancer. Chin J Cancer. 2012;31:327–34.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have any conflict of interest.
Rights and permissions
About this article
Cite this article
Ahmad, F., Badwe, A., Verma, G. et al. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol 33, 74 (2016). https://doi.org/10.1007/s12032-016-0788-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-016-0788-y