Abstract
Recent studies have reported that long noncoding RNAs (lncRNAs) play critical roles in carcinogenesis and progression. LncRNA-LET, a recently identified lncRNA, has been shown to be a tumor suppressor in hepatocellular carcinoma. However, the expression and functional of lncRNA-LET in other type of cancers remain largely unknown. In this study, we found that lncRNA-LET was significantly downregulated in nasopharyngeal carcinoma (NPC) tissues compared with corresponding normal tissues. Decreased LET expression is significantly correlated with advanced clinical stage, larger tumor size, increased lymph node tumor burden, and poor survival of NPC patients. Gain- and loss-of-function experiments demonstrated that enhanced LET expression inhibited NPC cells proliferation and induced cell apoptosis. By contrast, the knockdown of LET promoted NPC cells proliferation and inhibited cell apoptosis. Importantly, we found lncRNA-LET is transcriptional repressed by EZH2-mediated H3K27 histone methylation on the LET promoter. The expressions of EZH2 and lncRNA-LET are significantly inversely correlated in NPC tissues. Collectively, these findings indicate a pivotal role for lncRNA-LET in NPC cell proliferation and apoptosis, and reveal an epigenetic mechanism for lncRNA-LET dysregulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nasopharyngeal carcinoma (NPC), which arises from the nasopharyngeal epithelium, is one of the most common types of head and neck tumors [1–3]. It is vastly more prevalent in Southeast Asia, resulting in serious health problems in these regions [4]. Therefore, it is of great clinical value to further reveal the molecular mechanisms of NPC carcinogenesis, and develop effective therapeutic strategies [5–7].
Long noncoding RNA (lncRNA) is a novel type of RNA molecule, longer than 200 nucleotides in length, with no protein coding potential [8–10]. Recently, many studies have found lncRNAs are extensively transcribed from the genomic DNA [11–13]. They have been shown to be deregulated in many cancers and participate in carcinogenesis and cancer progression by modulating the expression of many oncogenes or tumor suppressor genes [14–16]. However, there are only several studies of lncRNAs in human NPC, and the expressions and functions of lncRNAs in NPC remain largely unknown [17–20]. LncRNA-LET, a recently identified lncRNA, has been reported to be downregulated and exhibit tumor-suppressive roles in hepatocellular carcinoma, cervical cancer, gastric cancer, and gallbladder cancer [21–24]. However, the expression, function, and the factors contributing to its abnormal expression remain unknown in human NPC.
In the present study, we investigated the expression levels of LET, its association with clinicopathological characteristics, and its biological functions in NPC cells. Our results show that LET is downregulated in NPC tissues, and its decreased expression is associated with advanced clinical stage, larger tumor size, increased lymph node tumor burden, and poor prognosis of NPC patients. Overexpression of LET inhibited NPC cells proliferation and induced cell apoptosis, while the knockdown of LET promoted NPC cells proliferation and inhibited cell apoptosis. Moreover, we also found that enhancer of zeste homolog 2 (EZH2) repressed LET expression via H3K27 histone methylation on the LET promoter.
Materials and methods
Tissue samples
A total of 68 NPC tissues and their pair-matched adjacent normal tissues were obtained with informed consent from patients who had been diagnosed at the Second Affiliated Hospital of Nanchang University, Nanchang, China. These patients were subjected to biopsy before standard chemoradiotherapy, and the survival of these patients was followed up with a median follow-up period of 56 months. The clinical stages of these patients were classified according to American Joint Committee on Cancer (AJCC). The clinicopathological information of these patients is described in Table S1. This study was performed with the approval of the Research Ethics Committee of the Second Affiliated Hospital of Nanchang University.
Cell cultures
The normal immortalized human nasopharyngeal epithelial cell line NP69, and the human NPC cell lines CNE2 and HNE2 were obtained from Sun Yat-sen University, Guangzhou, China. The cells were grown in RPMI 1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (Hyclone, Logan, UT, USA) and maintained at 37 °C in a humidified incubator containing 5 % CO2.
RNA extraction and qPCR analyses
Total RNAs were extracted using TRIzol (Life Technologies) according to the manufacturer’s protocol. First-strand cDNA was generated using a Reverse Transcription Kit (Takara, Dalian, China). Real-time PCR was performed on ABI 7500 real-time PCR system (Life Technologies). For each sample, gene expression was normalized to the level of GAPDH. The primer sequences used in this study were as follows: LET, 5′-GGAGTAAAGGGAAAGAGTTGC-3′ (forward) and 5′-GTGTCGTGGACTGGCAAAAT-3′ (reverse); EZH2, 5′-CAGCCTTGTGACAGTTCGT-3′ (forward) and 5′-AGATGGTGCCAGCAATAGA-3′ (reverse); GAPDH, 5′-GGAGCGAGATCCCTCCAAAAT-3′ (forward) and 5′-GGCTGTTGTCATACTTCTCATGG-3′ (reverse). The real-time PCRs were carried out in triplicate. The relative RNA expression was calculated using the comparative Ct method.
Vectors construction
The complementary DNA encoding LET or EZH2 was PCR-amplified by the prime STAR HS DNA polymerase (Takara) and subcloned into the BamH I and EcoR I, or Kpn I and BamH I sites of pcDNA3.1 vector (Life Technologies), respectively. The primers used were as follows: 5′-CGGGATCCGAAAGAATGTGGCCCCAGAGG-3′ (forward) and 5′-GGAATTCAGGAAGCAGCTAAACCTCAATCCTA-3′ (reverse) for LET; 5′-GGGGTACCGAAGAATAATCATGGGCCAGACTGG-3′ (forward) and 5′-CGGGATCCAGATGTCAAGGGATTTCCATTTCTC-3′ (reverse) for EZH2.
Small interfering RNA synthesis and transfection
Small interfering RNAs (siRNAs) specifically targeting LET and EZH2 were purchased from GenePharma company (Suzhou, China). The siRNA sequences were 5′-GTGCATGTGGTAGGTTAGATT-3′ for LET; 5′-AAGACTCTGAATGCAGTTGCT-3′ for EZH2. Transfections were performed using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s instructions.
Cell proliferation assay
A total of approximately 1000 transfected cells were plated in 96-well plates. Cell proliferation was assessed every 24 h using Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. The absorbencies at each time point were used to plot cell proliferation curves. Ethynyl deoxyuridine (EdU) immunofluorescence staining was carried out using an EdU Kit (Roche, Mannheim, Germany). All experiments were performed in triplicate.
Western blotting
Protein extracts were prepared in a 1× sodium dodecyl sulfate buffer. Identical quantities of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. After incubation with antibodies specific for cleaved caspase-3 (Cell Signaling Technology, Boston, USA) or β-actin (Sigma-Aldrich, Saint Louis, MO, USA), the blots were incubated with goat anti-rabbit or anti-mouse secondary antibody (Cell Signaling Technology) and visualized with enhanced chemiluminescence.
TdT-mediated dUTP nick end labeling assay
TdT-mediated dUTP nick end labeling (TUNEL) assays were performed using the Dead End™ Fluorometric TUNEL System (Promega, Madison, WI, USA) to measure the fragmented DNA of apoptotic cells according to the manufacturer’s instructions. TUNEL-positive cells were defined as cells with nuclear green fluorescence labeling.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were carried out using the EZ ChIP™ Chromatin Immunoprecipitation Kit (Millipore, Bedford, MA, USA) according to the manufacturer’s instructions. An anti-EZH2 antibody (Millipore) or anti-H3K27me3 antibody (Millipore) was used to precipitate DNA–protein complexes. ChIP-derived DNA was quantified using qPCR. The primer sequences specific for the LET promoter were as follows: 5′-CCTTGGTTGGTCCCTTTGA-3′ (forward) and 5′-TTTCTGTACGCCGGTCATG-3′ (reverse).
Generation of cells stably overexpressing LET
To generate the cell line stably overexpressing LET, CNE2 cells were transfected with the plasmid pcDNA3.1-LET and selected with neomycin (800 µg/ml) for 4 weeks.
In vivo tumorigenesis assay
LET stably overexpressed CNE2 cells (1.0 × 106) were implanted subcutaneously into the flanks of nude mice. Primary tumor growth was detected by measuring the tumor length (L) and width (W), and tumor volume was calculated according to the equation V = 0.5 × LW 2. The animal studies were approved by the Institutional Animal Care and Use Committee of the Second Affiliated Hospital of Nanchang University.
Statistical analysis
For comparisons, Wilcoxon signed-rank test, log-rank test, Pearson Chi-square test, Student’s t test, and Pearson correlation analysis were carried out using the SPSS 18.0 software package (SPSS, Chicago, IL, USA). Differences were defined as statistically significant for P values <0.05.
Results
LET expression in NPC tissues and its association with clinicopathological characteristics
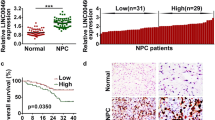
The expression level of LET was examined using qRT-PCR in 68 pairs of NPC tissues and pair-matched adjacent normal tissues. LET transcript expression was significantly downregulated in NPC tissues compared with corresponding normal tissues (P < 0.001, Fig. 1a).
LET expression is downregulated in NPC tissues and cell lines. a The expression level of LET was analyzed by qRT-PCR in 68 pairs of NPC tissues and pair-matched adjacent normal tissues. The horizontal lines in the box plots represent the medians, the boxes represent the interquartile range, and the whiskers represent the minimum and maximum values. The significant differences between samples were analyzed using the Wilcoxon signed-rank test. ***P < 0.001. b LET expression level in immortalized normal nasopharyngeal epithelial cell line NP69 and NPC cell lines CNE2 and HNE2 was analyzed by qRT-PCR. Data are presented as the mean ± SE based on at least three independent experiments. *P < 0.05; **P < 0.01
We next examined the correlation between the expression of LET and clinicopathological characteristics of the 68 NPC samples. The 68 NPC samples were classified into two groups according to the median expression level of LET. Correlation regression analysis showed that decreased LET expression is significantly correlated with advanced clinical stage (P = 0.026), larger tumor size (P = 0.008), and increased lymph node tumor burden (P = 0.049; Table 1).
Next, we detected the expression of LET in immortalized normal nasopharyngeal epithelial cell line NP69 and NPC cell lines CNE2 and HNE2. LET is significantly downregulated in NPC cell lines, compared with normal nasopharyngeal epithelial cell line (Fig. 1b).
Low LET expression predicts poor prognosis of NPC patients
We further examined whether the LET expression level was correlated with the outcome of NPC patients. Kaplan–Meier survival analysis and log-rank tests showed that low LET expression in NPC tissues is significantly associated with poor recurrence-free survival (P = 0.0151) and overall survival (P = 0.0214; Fig. 2a, b). These results indicate that decreased expression of LET may play a pivotal role in the progression of NPC.
Low LET expression predicts poor prognosis of NPC patients. a Kaplan–Meier analyses of correlations between the LET expression level and recurrence-free survival of 68 NPC patients. b Kaplan–Meier analyses of correlations between the LET expression level and overall survival of 68 NPC patients. LET expression was detected by qRT-PCR. The median expression level was used as the cutoff, and each group has 34 cases
Overexpression of LET inhibits cell proliferation and induces apoptosis of NPC cell
Based on the results that LET is downregulated in NPC and associated with the progression of NPC, we next examined the biological function of LET in NPC. As shown in Fig. 3a, we enhanced LET expression in CNE2 cells by transfecting a LET expression vector (pcDNA3.1-LET). CCK-8 assays indicated that enhanced LET expression significantly inhibited cell proliferation of CNE2 cells compared with the control cells (Fig. 3b). EdU incorporation assays showed that overexpression of LET in CNE2 cells significantly decreased the ratio of EdU-positive cells (Fig. 3c). These results confirm that LET could inhibit cell proliferation. Next, we investigated whether the proliferation inhibition effect of LET is through apoptosis pathway. As shown in Fig. 3d, enhanced LET expression in CNE2 cells upregulated the level of cleaved caspase-3. TUNEL assays showed that overexpression of LET in CNE2 cells significantly increased the ratio of TUNEL-stained apoptotic cells (Fig. 3e). These results demonstrated that LET induces NPC cells apoptosis. To examine the effects of LET in NPC cell proliferation in vivo, we stably overexpressed LET in CNE2 cells, and subcutaneously injected the LET-overexpressed CNE2 and control CNE2 cells into nude mouse. CNE2 cells overexpressing LET showed significantly reduced tumor growth (Fig. 3f).
LET inhibits NPC cell proliferation and induces apoptosis. a LET expression levels after transfection of the LET expression vector (pcDNA3.1- LET) or control vector into CNE2 cells. b The effects of LET on CNE2 cells proliferation were determined by the CCK-8 assay, and the relative number of cells to 0 h is presented. c The effects of LET on CNE2 cell proliferation were detected using EdU immunofluorescence staining. The blue color indicates the nuclei, and the red color represents EdU-positive nuclei. Scale bars 100 µm. Right percentages of EdU-positive cells. d The effects of LET on CNE2 cells apoptosis were determined by detecting cleaved caspase-3. e The effects of LET on CNE2 cells apoptosis were detected by TUNEL assay. Scale bars 100 µm. Right percentages of TUNEL-positive cells. For a–e, data are presented as the mean ± SE based on at least three independent experiments. **P < 0.01. f LET-overexpressed and control CNE2 cells were injected into nude mice subcutaneously, and tumor volume was determined. n = 5, ***P < 0.001
Repression of LET promotes cell proliferation and inhibits apoptosis of NPC cell
To further confirm the biological function of LET in NPC, we inhibited LET expression in HNE2 cells by transfecting LET-specific siRNAs (Fig. 4a). CCK-8 assays indicated that knockdown of LET significantly promoted cell proliferation of HNE2 cells compared with the control cells (Fig. 4b). EdU incorporation assays showed that knockdown of LET in HNE2 cells significantly increased the ratio of EdU-positive cells (Fig. 4c). These results suggest that knockdown of LET promotes cell proliferation. Knockdown of LET in HNE2 cells downregulated the level of cleaved caspase-3 (Fig. 4d). TUNEL assays showed that knockdown of LET in HNE2 cells significantly decreased the ratio of TUNEL-stained apoptotic cells (Fig. 4e). These data demonstrated that LET is involved in the progression of NPC through its effect on NPC cells proliferation and apoptosis.
Knockdown of LET promotes NPC cell proliferation and inhibits apoptosis. a LET expression levels after transfection of LET siRNA or control siRNA into HNE2 cells. b The effects of LET inhibition on HNE2 cells proliferation were detected by the CCK-8 assay, and the relative number of cells to 0 h is presented. c The effects of LET inhibition on HNE2 cell proliferation were detected using EdU immunofluorescence staining. The blue color indicates the nuclei, and the red color represents EdU-positive nuclei. Scale bars 100 µm. Right percentages of EdU-positive cells. d The effects of LET inhibition on HNE2 cells apoptosis were determined by detecting cleaved caspase-3. e The effects of LET inhibition on HNE2 cells apoptosis were detected by TUNEL assay. Scale bars 100 µm. Right percentages of TUNEL-positive cells. For all panels, data are presented as the mean ± SE based on at least three independent experiments. **P < 0.01
LET is repressed by EZH2 and is inversely correlated with EZH2 levels in NPC tissues
We next sought to investigate the mechanism responsible for the downregulation of LET in NPC. We examined the promoter region of LET and identified consensus polycomb response elements (PRE) (Fig. 5a). Enhancer of zeste homolog 2 (EZH2), which induces histone H3K27me3 modification, is the catalytically active component of the polycomb response complex [25, 26]. In previous studies, EZH2 is shown to be upregulated in NPC tissues and promote NPC cells proliferation [27]. So we investigate whether EZH2 is responsible for the downregulation of LET in NPC. We performed ChIP assays to examine whether EZH2 could bind the promoter region of LET. As shown in Fig. 5b, EZH2 immunoprecipitates were highly enriched in the promoter region of LET compared with control IgG immunoprecipitates. Next, we inhibited EZH2 expression in CNE2 cells by transfecting EZH2-specific siRNAs (Fig. 5c). ChIP assay results showed that knockdown of EZH2 reduced H3K27me3 level at the promoter region of LET (Fig. 5d). We further test whether EZH2 regulate the expression of LET. Our results showed that knockdown of EZH2 significantly increased LET expression in CNE2 cells (Fig. 5e). We also overexpressed EZH2 in CNE2 cells by transfecting pcDNA3.1-EZH2 (Fig. 5f), and the results showed that enhanced expression of EZH2 decreased LET expression (Fig. 5g). These data demonstrated that LET is repressed by EZH2-mediated histone H3 methylation. Next, we examined whether LET is coexpressed with EZH2 in human NPC tissues. We measured the expression levels of EZH2 in the same set of 68 NPC tissues shown in Fig. 1 and then performed a correlation analysis of the EZH2 mRNA and LET expression levels. A statistically significant inverse correlation was found between EZH2 mRNA and LET expression levels (r = −0.481, P < 0.001, Pearson’s correlation, Fig. 5h), supporting the role of EZH2 in repressing LET.
LET is repressed by EZH2-mediated histone H3 methylation. a Illustration of polycomb response element (PRE) within the LET promoter. b Products of ChIP using an EZH2 antibody were amplified by qPCR in CNE2 cells. Data in the right panel are expressed as a percent of input DNA. c EZH2 mRNA levels after the transfection of control siRNA or EZH2 siRNA into CNE2 cells. d Products of ChIP using an H3K27me3 antibody were amplified by qPCR in CNE2 cells after transfection with EZH2 siRNA for 48 h. Data in the right panel are expressed as a percent of input DNA. e LET transcript levels after the transfection of control siRNA or EZH2 siRNA into CNE2 cells. f EZH2 mRNA levels after the transfection of pcDNA3.1-EZH2 or pcDNA3.1 into CNE2 cells. g LET transcript levels after the transfection of pcDNA3.1-EZH2 or pcDNA3.1 into CNE2 cells. For b–g, data are presented as the mean ± SE based on at least three independent experiments. **P < 0.01; ***P < 0.001. h The correlation between EZH2 mRNA level and LET transcript level was measured in 68 NPC tissues. The respective expression values were subjected to Pearson correlation analysis (r = −0.481, P < 0.001, Pearson’s correlation)
Discussion
In this study, we examined the expression of long noncoding RNA-LET in NPC tissues and their adjacent normal tissues. We also detected the function of LET in NPC cells by applying gain-of-function and loss-of-function approaches. Our results demonstrate that LET is downregulated in NPC tissues in comparison with adjacent normal tissues, and that decreased LET expression is positively correlated with advanced clinical stage, larger tumor size, and increased lymph node tumor burden. Furthermore, we also found that low LET expression in NPC tissues indicates poor recurrence-free survival and overall survival of NPC patients. Enhanced LET expression inhibits cell proliferation and induces apoptosis of NPC cells, while the inhibition of LET promotes cell proliferation and inhibits apoptosis of NPC cells. Therefore, our results demonstrated that LET functions as a tumor suppressor in NPC.
The molecular mechanisms for the dysregulation of lncRNAs in cancer are not yet completely understood [9]. They can be regulated by cytokines and transcription factors, such as lncRNA-ATB activated by transforming growth factor beta, and lncRNA-CCAT1 and H19 induced by c-Myc [28–31]. Similar to mRNAs, lncRNAs could also be controlled by epigenetic regulation, including DNA methylation, histone acetylation, and histone methylation [32]. LncRNA-LET has been reported to be repressed by histone deacetylase 3-mediated histone deacetylation in hepatocellular carcinoma [21]. In this study, we further demonstrated that LET could also be regulated by histone methylation in NPC. Suppression of EZH2 induced a significant increase in LET expression, accompanied by a reduction of H3K27 methylation level at the promoter of LET. The inverse correlation between EZH2 mRNA and LET expression levels in NPC tissues supports EZH2’s regulation of LET expression.
LncRNAs may exert their biological functions via regulating expression of cancer-associated genes in transcriptional level or post-transcriptional level [33–36]. However, LET has been reported to bind and downregulate nuclear factor 90 protein abundance in hepatocellular carcinoma [21]. The molecular mechanisms responsible for LET’s function in NPC require further investigation.
In conclusion, our studies demonstrate that LET, which is repressed by EZH2-mediated histone H3K27 methylation, could regulate NPC cell proliferation and apoptosis, and is a prognosis factor in patients with NPC. Our findings indicate that LET is an important NPC biomarker for determining prognosis and a potential NPC therapy target.
References
Lin DC, Meng X, Hazawa M, Nagata Y, Varela AM, Xu L, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46(8):866–71.
Burgos JS. Involvement of the Epstein-Barr virus in the nasopharyngeal carcinoma pathogenesis. Med Oncol. 2005;22(2):113–21.
Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5(5):423–8.
Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365(9476):2041–54.
Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei RR, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13(6):633–41.
Acikalin MF, Etiz D, Gurbuz MK, Ozudogru E, Canaz F, Colak E. Prognostic significance of galectin-3 and cyclin D1 expression in undifferentiated nasopharyngeal carcinoma. Med Oncol. 2012;29(2):742–9.
Xiong S, Wang Q, Zheng L, Gao F, Li J. Identification of candidate molecular markers of nasopharyngeal carcinoma by tissue microarray and in situ hybridization. Med Oncol. 2011;28(Suppl 1):S341–8.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9.
Muers M. RNA: genome-wide views of long non-coding RNAs. Nat Rev Genet. 2011;12(11):742.
Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–37.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Krzyzanowski PM, Muro EM, Andrade-Navarro MA. Computational approaches to discovering noncoding RNA. Wiley Interdiscip Rev RNA. 2012;3(4):567–79.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26(3):344–57.
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31(7):31.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):253.
Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X, et al. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44(5):545–54.
Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104(4):458–64.
Gao W, Chan JY, Wong TS. Differential expression of long noncoding RNA in primary and recurrent nasopharyngeal carcinoma. Biomed Res Int. 2014;2014:404567.
Yang QQ, Deng YF. Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology. 2015;. doi:10.1111/his.12616.
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49(6):1083–96.
Jiang S, Wang HL, Yang J. Low expression of long non-coding RNA LET inhibits carcinogenesis of cervical cancer. Int J Clin Exp Pathol. 2015;8(1):806–11.
Zhou B, Jing XY, Wu JQ, Xi HF, Lu GJ. Down-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol. 2014;7(12):8893–8.
Ma MZ, Kong X, Weng MZ, Zhang MD, Qin YY, Gong W, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog. 2014;. doi:10.1002/mc.22215.
Heyn H, Esteller M. EZH2: an epigenetic gatekeeper promoting lymphomagenesis. Cancer Cell. 2013;23(5):563–5.
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298.
Yan M, Zhang Y, He B, Xiang J, Wang ZF, Zheng FM, et al. IKKalpha restoration via EZH2 suppression induces nasopharyngeal carcinoma differentiation. Nat Commun. 2014;5:3661.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–81.
Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31(5):914.
He X, Tan X, Wang X, Jin H, Liu L, Ma L, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35(12):12181–8.
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, et al. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139(3):437–45.
Liu J, Zhu X, Xu X, Dai D. DNA promoter and histone H3 methylation downregulate NGX6 in gastric cancer cells. Med Oncol. 2014;31(1):817.
Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34(1):18.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–81.
Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58.
Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;. doi:10.1007/s13277-015-3465-4.
Acknowledgments
This work was funded by supporting program of Jiangxi Provincial Science and Technology Department (No. 2010BSB00205).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Qiuzhen Sun and Hongbing Liu contributed equally to this work and should be regarded as joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Q., Liu, H., Li, L. et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol 32, 226 (2015). https://doi.org/10.1007/s12032-015-0673-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-015-0673-0