Abstract
Long noncoding RNA (lncRNA) CRNDE has been broadly implicated in many malignancies. The aim of this study was to explore the function and potential mechanisms of CRNDE in nasopharyngeal carcinoma (NPC). Here, we discovered that CRNDE level was increased in NPC tissues and cell lines. Additionally, elevated CRNDE positively correlated with large tumor size, advanced TNM stage, distant metastasis, EBV infection and worse prognosis. Furthermore, depletion of CRNDE significantly impaired the capacity of proliferation, migration and invasion in NPC cells. Mechanically, CRNDE could serve as a molecular sponge of miR-3163 to regulate the expression of TWIST1. Importantly, the inhibitory effects of CRNDE knockdown on cell proliferation and metastasis were blocked by silence of miR-3163 or restoration of TWIST1 expression. Overall, our data highlighted that CRNDE could promote NPC progression via altering miR-3163/TWIST1 axis, suggesting CRNDE as a potential prognostic biomarker and therapeutic target for NPC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common carcinomas in Southern China [1, 2]. With the advancement of technologies for the treatment and intervention, the survival rate of patients with NPC has been considerable improved. However, approximately 30–40% cases eventually develop distant metastasis, which becomes a major barrier to successful treatment [3, 4]. In this regard, there is an urgent need to study the mechanism involved in NPC tumorigenesis and metastasis.
Long noncoding RNAs (lncRNAs) are a class of non-coding RNAs comprising longer than 200 nt [5]. Recently, compelling evidence suggested that lncRNAs have major roles in various biological processes, such as cell growth, proliferation, invasion, autophagy and drug resistance [6, 7]. They can work as oncogenes or tumor suppressors to regulate gene expression by competitively sponging microRNAs [8]. Hence, understanding the expression and biological function of individual lncRNA might provide diagnostic markers and therapeutic targets for cancer treatment.
Colorectal neoplasia differentially expressed (CRNDE) is a lncRNA that was originally discovered in colorectal cancer [9]. Recently, CRNDE showed a potential prognostic value in a number of cancer types, such as ovarian cancer, clear cell renal cell carcinoma, acute myeloid leukemia, cervical cancer and glioma [10,11,12,13,14]. CRNDE has also been identified as an oncogene in many cancers to facilitate tumor progression by interacting with various miRNAs, such as miR-203, miR-338-3p and miR-181a-5p [15,16,17]. However, the role of CRNDE in NPC pathogenesis remains to be elucidated.
Twist basic helix-loop-helix transcription factor 1 (TWIST1) is highly expressed in various cancers and associated with poor prognosis, including NPC [18]. Previous studies indicated that upregulated TWIST1 could promote NPC cell proliferation, migration and invasion [19, 20]. In NPC, recent studies also showed an involvement of TWIST1 in taxol resistance and radioresistance [21, 22]. However, a more detailed role of TWIST1 promoting NPC metastasis is still unclear.
In this study, we first determined CRNDE expression in NPC tissues and cell lines. Then, the associations between CRNDE expression and clinical characteristics as well as the overall survival were analyzed. Furthermore, the effects of CRNDE on NPC cell proliferation, migration and invasion were assessed by loss-of-function experiments in vivo and in vitro.
Materials and Methods
Clinical Specimens and Cell Culture
Seventy-three paired of NPC tissues and adjacent normal nasopharynx tissues were collected from patients during operations at Shanghai Ninth People’s Hospital. No patient received any pre-operative chemotherapy and radiotherapy. Collections and use of human tissues were approved by the Ethics Committee of Shanghai Ninth People’s Hospital and written informed consents were obtained. Five NPC cell lines (SUNE-1, 5-8F, CNE2, C666-1 and HONE-1) were purchased from ATCC and cultured in RPMI-1640 (Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Invitrogen). NP69 cell was purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai) and used as the control cell line. Cell lines were cultured in a humidified chamber at 37° C and 5% CO2.
Oligoribonucleotides, Plasmids and Transient Transfection
The miR-3163 mimics or inhibitor, small interfering RNA (siRNA) targeting CRNDE (si-CRNDE), short hairpin RNA (shRNA) targeting CRNDE (sh-CRNDE) and negative controls were synthesized by GenePharma (Shanghai, China) according to the sequences shown in Supplementary Table 1. The full-length TWIST1 cDNA sequence was PCR amplified and then subcloned into the vector pcDNA3.1 (Invitrogen). The transfections were conducted using lipofectamine 2000 (Invitrogen).
Cell Viability and Colony Formation Assay
Cell viability was measured using CCK-8 assay. Briefly, 3 × 103 cells/well were seeded onto 96-well plates and incubated for various times. Then, 10 µl of CCK-8 reagent was introduced into each well, and incubation was continued for 1 h. A microplate reader was used to measure the absorbance values at 450 nm. For colony formation assay, 500 cells/well were seeded onto 6-well plates. After two weeks later, the colonies were fixed using methanol and dyed with 0.1% crystal violet (Invitrogen). The total number of visible colonies were counted.
Transwell Migration and Invasion Assay
Cell migration and invasion ability was assessed using transwell assays. A total of 1 × 105 cells were placed onto the upper chambers of each insert pre-coated with the Matrigel (Corning, without Matrigel for migration) and the lower chambers contained medium with 10% fetal bovine serum. After incubation of 24 h, cells were fixed and stained with Crystal Violet Staining Solution (Beyotime) and then counted under a light microscope.
Total RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted by TRIzol reagent (Invitrogen) and then reversely transcribed to cDNA using PrimeScript RT reagent Kit (Takara, Tokyo, Japan). qRT-PCR was performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on an ABI StepOnePlus Real-Time PCR System (Applied Biosystems, CA, USA). The standard control for mRNA/lncRNA and miRNA was β-actin or U6 and calculated using the 2−ΔΔCt method. The primers used in this study were as follows:
CRNDE, 5’-TGAAGGAAGGAAGTGGTGCA-3’ (forward) and 5’-TCCAGTGGCATCCTACAAGA-3’ (reverse);
miR-3163, 5’-GGAATGTTCCTTCTTTGC-3’ (forward) and 5’-GAACATGTCTGCGTATCTC-3’ (reverse);
TWIST1, 5’-GCCAGGTACATCGACTTCCTCT-3’ (forward) and 5’-TCCATCCTCCAGACCGAGAAGG-3’ (reverse).
Western Blot Analysis
Total protein from cells were extracted by RIPA buffer (Beyotime) and quantified by BCA Protein Assay Kit (Beyotime). Equal amount of protein (30 μg) was resolved by 10% SDS-PAGE gels and then transferred to PVDF membranes (Millipore). After, the membranes were blocked with 5% nonfat milk and then incubated with primary antibodies against TWIST1 (1:1000; #69,366, Cell Signaling Technology) and β-actin (1:2000; ab8227, Abcam) overnight at 4 °C. Following incubation with secondary antibodies (1:5000; ab205718, Abcam), the blots were visualized using enhanced chemiluminescence (Millipore).
Luciferase Reporter Assay
The wild-type (wt) and mutant (mut) sequences of CRNDE or TWIST1 containing putative binding site of miR-3163 were synthesized (GenePharma) and subcloned into the pMIR-REPORT luciferase reporter vector (Ambion, Austin, TX, USA). Then, wt or mut reporter vectors with miR-3163 mimics was cotransfected into NPC cells. After 48 h transfection, luciferase activity was measured using a Dual Luciferase Assay kit (Promega).
Tumor Xenograft in Nude Mice
The SUNE-1 cells stably transfected with sh-CRNDE or shRNA control (sh-NC) were selected using puromycin (1 mg/mL) and validated by qRT-PCR. For the tumor growth model, 1 × 106 SUNE-1 cells stably expressing sh-CRNDE or sh-NC were injected into each flank of the 4–6-week-old BALB/c nude mice (n = 6 each group). Tumor volumes were monitored (0.5 × length × width2) every week. All mice were sacrificed for measurement of tumor weights 4 weeks after establishment of the model. All animal protocols were performed with the approval of the Ethics Committee of Shanghai Ninth People’s Hospital.
Immunohistochemical (IHC) Staining
IHC staining was performed to detect Ki-67 expression of tumor tissues from the mouse model experiment. Briefly, tissues were fixed in 10% formaldehyde in PBS, fixed, embedded in paraffin and cut into 4 μm-thick sections. After de-waxing and hydration, antigen retrieval by citrate buffer and blocking with 1% bovine serum albumin, the sections was incubated with primary antibody against Ki-67 (1:50; ab15580, Abcam) overnight at 4 °C and then treated with secondary antibody (1:1000; ab6721, Abcam) at room temperature for 30 min. Finally, the sections were stained with diaminobenzidine, counterstaining with 10% Mayer’s hematoxylin, dehydration, and mounting. Light microscopy was used to calculate positive cells in 5 random fields.
Statistical Analysis
Statistical calculations were performed with the GraphPad Prism 6 (GraphPad Software) using Student’s t test or one-way ANOVA. The data from independently repeated at least three times are expressed as the mean ± SD. The overall survival rate was evaluated using Kaplan–Meier method and log rank test according to the median expression level of genes. The criterion of statistical significance was P-value < 0.05.
Results
CRNDE Expression is Upregulated and Associated with Poor Overall Survival in NPC
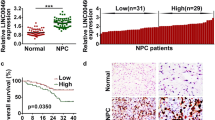
The expression of CRNDE were higher in NPC cell lines (SUNE-1, 5-8F, CNE2, C666-1 and HONE-1) compared to NP69 cells (Fig. 1A). Furthermore, CRNDE in NPC tissues was increased 2.2-fold compared with adjacent normal tissues (Fig. 1B). Moreover, patients with high CRNDE expression showed a poor prognosis (Fig. 1C; P = 0.0007). To evaluate the correlations between CRNDE expression and clinicopathological factors, the 73 NPC patients were divided into high and low expression of CRNDE based on the median value of CRNDE. As shown in Table 1, CRNDE expression was dramatically associated with large tumor size (P = 0.013), advanced TNM stage (P = 0.006), distant metastasis (P = 0.003) and EBV DNA (P = 0.003). These results indicate that CRNDE might serve as a prognostic biomarker for NPC patients.
CRNDE expression is upregulated and associated with poor overall survival in NPC. (A) Relative CRNDE expression in NPC cells and control cell NP69 was analyzed by qRT-PCR. (B) Relative CRNDE expression in 73 NPC tissues and 73 adjacent normal tissues analyzed by qRT-PCR. (C) The Kaplan–Meier overall survival curve of 73 NPC patients with high CRNDE level (n = 37) and low CRNDE level (n = 36) according to the median expression level of CRNDE. *P < 0.05; **P < 0.01
Knockdown of CRNDE Suppresses Proliferation and Metastasis of NPC Cells
Then, loss-of-function assays were performed in SUNE-1 and HONE-1 cells by transfecting si-CRNDE or negative control, and the efficiency of CRNDE downregulation was first validated (Fig. 2A). CRNDE knockdown obviously inhibited the growth of NPC cells (Fig. 2B, C). Similarly, the capacity to form colonies in the CRNDE-knockdown cells was also restrained (Fig. 2D, E). The effects of CRNDE on tumor growth was further assessed in vivo. The tumor volume and weight in sh-CRNDE group were decreased than that of control group (Fig. 2F–H). IHC staining analysis revealed that xenograft tumors derived from sh-CRNDE group had lower expression of Ki-67 than the sh-NC group (Fig. 2I, J). Transwell assays showed that the number of migratory cells in the CRNDE siRNA group was significantly reduced compared with that in the non-specific siRNA group (Fig. 2K, L). In consistent with migration, CRNDE knockdown suppressed the cell invasion ability in both SUNE-1 and HONE-1 cells (Fig. 2M, N).
Knockdown of CRNDE suppresses proliferation and metastasis of NPC cells. A Relative CRNDE expression was detected using qRT-PCR. B, C CRNDE knockdown decreased cell growth of SUNE-1 (B) and HONE-1 (C) using CCK-8 assays. D, E CRNDE knockdown attenuated the capacity of colony formation in NPC cells. F The representative tumor images of nude mice treated with sh-NC and sh-CRNDE (n = 6 each group). G Tumor growth curve. H Tumor weight. I, J IHC staining analysis for Ki-67 in xenograft tumors. K–N Representative migratory (K–L) and invasive (M–N) SUNE-1 and HONE-1 cells after CRNDE knockdown. *P < 0.05; **P < 0.01
CRNDE Binds to miR-3163 and Suppresses Its Expression
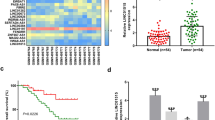
By using online tool starBase, we found that CRNDE contains putative binding sites for miR-3163 (Fig. 3A). miR-3163 overexpression caused a significant decrease in luciferase activity in the wild-type CRNDE, but not in the mutant form (Fig. 3B, C). Meanwhile, CRNDE knockdown markedly increased miR-3163 expression (Fig. 3D). Surprisingly, miR-3163 mimics did not affect CRNDE level (Fig. 3E). Compared with NP69 cells, NPC cell lines displayed lower expression of miR-3163 (Fig. 3F). As expected, miR-3163 expression was also remarkably decreased (Fig. 3G) and exhibited a negative correlation with CRNDE expression in NPC tissues (Fig. 3H). These results indicate that CRNDE could directly sponge miR-3163 and decrease its expression.
CRNDE binds to miR-3163 and suppresses its expression. A Putative biding sites of miR-3163 on CRNDE predicted using starBase. B, C Luciferase reporter assays were performed to confirm miR-3163 binding to the CRNDE in SUNE-1 (B) and HONE-1 (C) cells. D qRT-PCR analysis showed that CRNDE knockdown reduced miR-3163 expression in SUNE-1 and HONE-1 cells. E qRT-PCR analysis showed that miR-3163 overexpression did not alter CRNDE expression in SUNE-1 and HONE-1 cells. F Relative miR-3163 expression in NPC cells and control cell NP69 was analyzed by qRT-PCR. G Relative miR-3163 expression in 73 NPC tissues and 73 adjacent normal tissues analyzed by qRT-PCR. H miR-3163 was negatively correlated with CRNDE expression in NPC tissues. *P < 0.05; **P < 0.01
CRNDE Regulates NPC Cell Proliferation and Metastasis Through miR-3163
To explore whether CRNDE exerted its oncogenic role through sponging miR-3163, miR-3163 inhibitor and si-CRNDE were cotransfected into SUNE-1 and HONE-1 cells. We observed that downregulation of miR-3163 reversed the suppression of CRNDE knockdown on the cell proliferation (Fig. 4A, B). Colony formation assay demonstrated that miR-3163 inhibitor transfection counteracted CRNDE knockdown-induced decrease of colony numbers (Fig. 4C, D). Transwell assays revealed that miR-3163 inhibitor transfection also abolished the suppressive effects of CRNDE knockdown-induced on the migrative (Fig. 4E, F) and invasive (Fig. 4G, H) abilities of NPC cells. The data imply that miR-3163 is a direct target of CRNDE.
CRNDE regulates NPC cell proliferation and metastasis through miR-3163. A, B CCK-8 assays revealed that miR-3163 downregulation reversed the influence of CRNDE knockdown on cell growth in SUNE-1 (A) and HONE-1 (B) cells. C, D Colony formation assays were performed to evaluate the effects of miR-3163 downregulation on the capacity of form colonies-induced by CRNDE knockdown in NPC cells. E–H Transwell assays results demonstrated that the suppressive migration (E–F) and invasion (G–H) abilities-induced by CRNDE knockdown were weakened after miR-3163 downregulation in NPC cells. *P < 0.05; **P < 0.01
CRNDE Facilitates TWIST1 Expression Through Serving as a ceRNA for miR-3163
TWIST1 is a classical oncogene involved in NPC pathogenesis [19, 20]. To verify whether miR-3163 could target TWIST1, bioinformatics analysis was first performed using starBase. TWIST1 3’UTR harbored two potential binding sites of miR-3163 (Fig. 5A). Compared with TWIST1-mut group, luciferase activity was dramatically decreased in cells cotransfected with miR-3163 and TWIST1-wt (Fig. 5B, C). miR-3163 inhibitor transfection could rescue the TWIST1 mRNA and protein level decrease induced by CRNDE knockdown (Fig. 5D, E). Moreover, TWIST1 was significantly upregulated in NPC tissues and positively correlated with CRNDE expression as well as poor overall survival (Fig. 5F-H). The above results suggest that CRNDE regulates TWIST1 expression by sponging miR-3163.
CRNDE facilitates TWIST1 expression through serving as a ceRNA for miR-3163. A Putative biding sites of miR-3163 on TWIST1 3’UTR predicted using starBase. B, C Luciferase reporter assays were performed after SUNE-1 (B) and HONE-1 (C) cells cotransfected with wt- or mut-TWIST1 3’UTR luciferase reporter vectors miR-3163 mimics or miRNA negative control. D, E miR-3163 inhibitor increased TWIST1 mRNA (D) and protein (E) level in SUNE-1 and HONE-1 cells. F Relative TWIST1 mRNA expression in 73 NPC tissues and 73 adjacent normal tissues analyzed by qRT-PCR. G Correlation analysis between CRNDE and TWIST1 in NPC tissues. H The Kaplan–Meier overall survival curve of 73 NPC patients with high TWIST1 level (n = 37) and low TWIST1 level (n = 36) according to the median expression level of CRNDE. **P < 0.01
TWIST1 is Responsible for CRNDE-Mediated Cell Proliferation, Migration and Invasion
To further confirm whether CRNDE caused NPC malignant phenotypes via TWIST1-dependent manner, pcDNA-TWIST1 and si-CRNDE were cotransfected into SUNE-1 and HONE-1 cells. First, the overexpression of TWIST1 was validated by Western blot (Fig. 6A). Intriguingly, CCK-8 and colony formation assays results showed that the inhibitory effects of si-CRNDE on SUNE-1 and HONE-1 cell growth (Fig. 6B, C) and colony formation capacity (Fig. 6D, E) were partly restored by TWIST1 overexpression. Similar results were also obtained for cell migration (Fig. 6F, G) and invasion (Fig. 6H, I) using Transwell assays. These results indicate that CRNDE promotes NPC proliferation and metastasis via a TWIST1-dependent way.
TWIST1 is responsible for CRNDE-mediated cell proliferation, migration and invasion. A Western blot was performed to validate the overexpression of TWIST1 in SUNE-1 and HONE-1 cells after transfection with pcDNA-TWIST1 or pcDNA-NC. B, C TWIST1 overexpression counteracted the suppression of cell growth-induced by CRNDE knockdown in SUNE-1 (B) and HONE-1 (C) cells by CCK-8 assay. D, E Colony formation assays indicated that TWIST1 overexpression reversed the inhibitory capacity of form colonies-induced by CRNDE knockdown in NPC cells. F-I Effects of CRNDE knockdown on the migrative (F–G) and invasive (H–I) abilities were diminished by TWIST1 overexpression in NPC cells using Transwell assays. *P < 0.05; **P < 0.01
Discussion
A large body of studies have reported that many dysregulated lncRNAs participated in NPC pathogenesis. For example, lncRNA MACC1-AS1 was highly expressed in NPC and promoted cell stemness through miR-145/Smad2 negative feedback loop [23]. Yang et al. reported that lncRNA DLX6-AS1 increased proliferation, migration and invasion of NPC cells via miR-199a-5p/HIF-1α axis [24]. Although lncRNA CRNDE has been broadly implicated in carcinogenesis, yet its functional roles in NPC is unclear [6,7,8,9,10]. Here, we identified that CRNDE was upregulated in NPC tissues and cell lines. Additionally, highly expressed CRNDE was associated with tumor progression and worse prognosis of patients with NPC, implying that CRNDE expression may be a useful prognostic biomarker for NPC patients.
Utilizing in vitro and in vivo assays, we investigated the biological functions of CRNDE in NPC tumorigenesis. Loss-of-function assays of CRNDE by its specific siRNA and shRNA showed that CRNDE knockdown suppressed cell growth, migration and invasion, which was consistent with the clinicopathological observations of CRNDE upregulation correlating NPC metastasis. These results indicate that CRNDE might be a potential therapeutic target for NPC treatment, especially for patients with metastasis.
Previous studies have suggested that CRNDE could exerts its biological function by interacting with many proteins, such as Bcl-3, PUMA, EZH2 and hnRNPUL2 [25,26,27,28]. More importantly, CRNDE was also reported to post-transcriptionally regulate gene expression through sponging microRNAs [11,12,13]. Our bioinformatics analysis and luciferase reporter assay demonstrated that CRNDE could directly bind miR-3163 and decrease its expression in NPC cells. Besides, miR-3163 downregulation reversed the inhibitory effects of CRNDE knockdown on malignant phenotypes. These findings suggest that miR-3163 is a direct target of CRNDE in NPC.
In this study, we observed that miR-3163 was obviously downregulated and functioned as a tumor suppressor in NPC progression, which was similar to Shi and Ren’s reports of miR-3163 inhibiting cell proliferation and migration in hepatocellular carcinoma and colorectal cancer [29, 30]. Studies also showed a regulatory role of miR-3163 in the chemoresistance. For instance, overexpression of miR-3163 could restrain multidrug resistance of retinoblastoma cancer stem cells by reducing ABCG2 [31]. In hepatocellular carcinoma, miR-3163 has been shown to sensitize cells to sorafenib via targeting ADAM-17 and inhibiting the activation of the Notch signaling pathway [32]. Thus, the present findings extend the current knowledge of miR-3163 function.
Given the critical roles of TWIST1 in tumor growth and metastasis, we investigated its biological activities in CRNDE-mediated NPC progression. Consistently, our results showed that TWIST1 overexpression counteracted CRNDE knockdown-induced suppression of malignant phenotypes. Our data also revealed that CRNDE upregulated TWIST1 expression through interacting with miR-3163. Besides, TWIST1 positively correlated with CRNDE and highly expressed TWIST1 was strikingly associated with unfavorable survival in NPC. Thus, CRNDE exerts its oncogenic function in a TWIST1-dependent way.
Collectively, our study established an oncogenic role for CRNDE in NPC. We also demonstrated the involvement of CRNDE as a miRNA sponge of miR-3163 to upregulate TWIST1. Overall, our data suggest that CRNDE may represent a useful prognostic biomarker and potential therapeutic target for NPC treatment.
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-A Cancer Journal for Clinicians, 68, 394–424.
Chua, M. L. K., Wee, J. T. S., Hui, E. P., & Chan, A. T. C. (2016). Nasopharyngeal carcinoma. Lancet, 387, 1012–1024.
Zhang, L., Huang, Y., Hong, S., Yang, Y., Yu, G., Jia, J., et al. (2016). Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: A multicentre, randomised, open-label, phase 3 trial. Lancet, 388, 1883–1892.
Lai, S. Z., Li, W. F., Chen, L., Luo, W., Chen, Y. Y., Liu, L. Z., et al. (2011). How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? International Journal of Radiation Oncology, Biology, Physics, 80, 661–668.
Mercer, T. R., Dinger, M. E., & Mattick, J. S. (2009). Long non-coding RNAs: Insights into functions. Nature Reviews Genetics, 10, 155–159.
Cheng, G., Yu, Y., Wang, L., & Pan, Q. (2020). Overexpression of LINC00160 predicts poor outcome and promotes progression of clear cell renal cell carcinoma. Aging (Albany NY), 12, 7448–7464.
Shi, R., Wu, P., Liu, M., Chen, B., & Cong, L. (2020). Knockdown of lncRNA PCAT6 enhances radiosensitivity in triple-negative breast cancer cells by regulating miR-185-5p/TPD52 Axis. OncoTargets and Therapy, 13, 3025–3037.
Ming, X. L., Feng, Y. L., He, D. D., Luo, C. L., Rong, J. L., Zhang, W. W., et al. (2019). Role of BCYRN1 in hepatocellular carcinoma pathogenesis by lncRNA-miRNA-mRNA network analysis and its diagnostic and prognostic value. Epigenomics, 11, 1209–1231.
Graham, L. D., Pedersen, S. K., Brown, G. S., Ho, T., Kassir, Z., Moynihan, A. T., et al. (2011). Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes & Cancer, 2, 829–840.
Szafron, L. M., Balcerak, A., Grzybowska, E. A., Pienkowska-Grela, B., Podgorska, A., Zub, R., et al. (2015). The putative oncogene, CRNDE, is a negative prognostic factor in ovarian cancer patients. Oncotarget, 6, 43897–43910.
Ding, C., Han, F., Xiang, H., Xia, X., Wang, Y., Dou, M., et al. (2018). LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. Journal of Cellular Biochemistry, 119, 10406–10414.
Wang, Y., Zhou, Q., & Ma, J. J. (2018). High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. European Review for Medical and Pharmacological Sciences, 22, 763–770.
Yang, H. Y., Huang, C. P., Cao, M. M., Wang, Y. F., & Liu, Y. (2018). Long non-coding RNA CRNDE may be associated with poor prognosis by promoting proliferation and inhibiting apoptosis of cervical cancer cells through targeting PI3K/AKT. Neoplasma, 65, 872–880.
Jing, S. Y., Lu, Y. Y., Yang, J. K., Deng, W. Y., Zhou, Q., & Jiao, B. H. (2016). Expression of long non-coding RNA CRNDE in glioma and its correlation with tumor progression and patient survival. European Review for Medical and Pharmacological Sciences, 20, 3992–3996.
Ji, D., Jiang, C., Zhang, L., Liang, N., Jiang, T., Yang, B., et al. (2019). LncRNA CRNDE promotes hepatocellular carcinoma cell proliferation, invasion, and migration through regulating miR-203/ BCAT1 axis. Journal of Cellular Physiology, 234, 6548–6560.
Jing, H., Xia, H., Qian, M., & Lv, X. (2019). Long noncoding RNA CRNDE promotes non-small cell lung cancer progression via sponging microRNA-338-3p. Biomedicine & Pharmacotherapy, 110, 825–833.
Han, P., Li, J. W., Zhang, B. M., Lv, J. C., Li, Y. M., Gu, X. Y., et al. (2017). The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Molecular Cancer, 16, 9.
Song, L. B., Liao, W. T., Mai, H. Q., Zhang, H. Z., Zhang, L., Li, M. Z., et al. (2006). The clinical significance of twist expression in nasopharyngeal carcinoma. Cancer Letters, 242, 258–265.
Zhou, J., Zhang, J., Xu, M., Ke, Z., Zhang, W., & Mai, J. (2019). High SRC-1 and Twist1 expression predicts poor prognosis and promotes migration and invasion by inducing epithelial-mesenchymal transition in human nasopharyngeal carcinoma. PLoS ONE, 14, e0215299.
Li, L., & Zhang, F. (2020). Novel long noncoding RNA LINC01385 promotes nasopharyngeal carcinoma proliferation via the miR-140-3p/Twist1 signaling pathway. Cell Cycle, 19, 1352–1362.
Wang, X., Ling, M. T., Guan, X. Y., Tsao, S. W., Cheung, H. W., Lee, D. T., et al. (2004). Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene, 23, 474–482.
Zhang, L., Su, B., Sun, W., Li, W., Luo, M., Liu, D., et al. (2016). Twist1 promotes radioresistance in nasopharyngeal carcinoma. Oncotarget, 7, 81332–81340.
Chen, S., Luo, X., Wu, W., Li, Y., Yu, H., Wang, Y., et al. (2020). The long non-coding RNA MACC1-AS1 promotes nasopharyngeal carcinoma cell stemness via suppressing miR-145-mediated inhibition on SMAD2/MACC1-AS1 axis. Biomedicine & Pharmacotherapy, 125, 109986.
Yang, B., Jia, L., Ren, H., Jin, C., Ren, Q., Zhang, H., et al. (2020). LncRNA DLX6-AS1 increases the expression of HIF-1α and promotes the malignant phenotypes of nasopharyngeal carcinoma cells via targeting MiR-199a-5p. Molecular Genetics & Genomic Medicine, 8, e1017.
Li, K., Cui, M., Zhang, K., Wang, G., & Zhai, S. (2020). LncRNA CRNDE affects the proliferation and apoptosis of vascular smooth muscle cells in abdominal aortic aneurysms by regulating the expression of Smad3 by Bcl-3. Cell Cycle, 19, 1036–1047.
Zhang, J. J., & Fan, L. P. (2019). Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomedicine & Pharmacotherapy, 117, 108726.
Ding, J., Li, J., Wang, H., Tian, Y., Xie, M., He, X., et al. (2017). Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death & Disease, 8, e2997.
Jiang, H., Wang, Y., Ai, M., Wang, H., Duan, Z., Wang, H., et al. (2017). Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death & Disease, 8, e2862.
Shi, C., Yang, Q., Pan, S., Lin, X., Xu, G., Luo, Y., et al. (2020). LncRNA OIP5-AS1 promotes cell proliferation and migration and induces angiogenesis via regulating miR-3163/VEGFA in hepatocellular carcinoma. Cancer Biology & Therapy, 21, 604–614.
Ren, H., Li, Z., Tang, Z., Li, J., & Lang, X. (2020). Long noncoding MAGI2-AS3 promotes colorectal cancer progression through regulating miR-3163/TMEM106B axis. Journal of Cellular Physiology, 235, 4824–4833.
Jia, M., Wei, Z., Liu, P., & Zhao, X. (2016). Silencing of ABCG2 by MicroRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. Journal of Korean Medical Science, 31, 836–842.
Yang, B., Wang, C., Xie, H., Wang, Y., Huang, J., Rong, Y., et al. (2019). MicroRNA-3163 targets ADAM-17 and enhances the sensitivity of hepatocellular carcinoma cells to molecular targeted agents. Cell Death & Disease, 10, 784.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to Participate
All patients signed a written informed consent form.
Consent to Publish
Patients signed informed consent regarding publishing their data.
Ethical Approval
The study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Y., Zhang, L., Wu, Q. et al. Long Noncoding RNA CRNDE Functions as an Oncogene to Facilitate Aggressive Behaviors of Nasopharyngeal Carcinoma Cells by Modulating miR-3163/TWIST1 Axis. Mol Biotechnol 64, 463–471 (2022). https://doi.org/10.1007/s12033-021-00425-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00425-5