Abstract
Lactate dehydrogenase B (LDHB) is widely expressed in adult somatic tissue and is one of the two subunits of lactate dehydrogenase, which is the key glycolytic enzyme and catalyzes the interconversion of pyruvate and lactate. However, the roles of LDHB in glycolysis and tumor progression were obscure in different types of cancer. Here, we determined the roles of LDHB in pancreatic cancer development and progression. We found suppressed expression of LDHB in pancreatic cancer which was due to promoter hypermethylation and deceased expression of LDHB led to glycolytic transition. Functional analysis revealed that suppressed expression of LDHB promoted pancreatic cancer cells proliferation, invasion, and migration in hypoxia. Thus, LDHB might function as a suppressor of glycolysis and suppressed pancreatic cancer progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the past decades, a great lot of researches have proved that development and progression of tumors persistently depends on glycolysis even under normal oxygen concentrations and that deregulating cellular metabolism has been one of the hallmarks of cancer [1]. The glycolytic phenotype, which is also known as the “Warburg effect,” might be the result of stable genetic or epigenetic changes in oncogenes, as many glycolytic enzymes are upregulated by oncogenes such as Ras, Myc, etc. [2–4]. Lactate dehydrogenase (LDH), which catalyzes the interconversion of pyruvate and lactate, is the key glycolytic enzyme. LDH is a tetrameric enzyme which composes two types of subunits LDHA (muscle-type, M subunit) and LDHB (heart-type, H subunit), and results in five isozymes: M4 (LDH5), M3H1 (LDH4), M2H2 (LDH3), M1H3 (LDH2), and H4 (LDH1) [5]. It has been reported that LDHA is overexpressed in a series of cancer cells, including breast cancer, colorectal cancer, and non-small cell lung cancer, could be transcriptionally upregulated by c-Myc and hypoxia-inducible factor (HIF), and plays critical roles in cancer development and progression [6–10]. Suppressed expression of LDHB has been reported in many kinds of cancer due to promoter hypermethylation, whereas the significance of LDHB in tumor development is elusive [11, 12]. It has been reported that high LDHB expression marked the most glycolytic breast cancer cells and LDHB also was a metabolic marker of response to neoadjuvant chemotherapy in breast cancer [13, 14]. Furthermore, the elevation of serum LDHB correlated with the clinical stage in lung cancer and was also required for the growth of KRAS-dependent lung adenocarcinomas. However, other studies revealed that LDHA favored catalysis of pyruvate to lactate, whereas LDHB converted lactate to pyruvate and the shift from LDH1 to LDH5 promoted cancer progression [5, 8]. Furthermore, in liver cancer, decreased expression of LDHB enhanced cancer cell invasiveness via mitochondrial defects [15]. Thus, further studies which focus on the roles and mechanism of LDHB in the development and progression of different types of tumors are needed.
Pancreatic ductal adenocarcinoma, often called pancreatic cancer, is the seventh leading cause of cancer-related deaths globally, and its incidence is increasing annually [16]. Recently, surgery, radiotherapy, and chemotherapy for pancreatic cancer have improved, but the overall 5-year survival rate still did not break through 5 % [17]. Thus, investigation of the molecular mechanisms underlying pancreatic cancer development and progression is urgently needed [17]. Our previous study showed that LDHA was transcriptionally regulated by forkhead box protein M1 (FOXM1) and Kruppel-like factor 4 (KLF4) and promoted pancreatic cancer progression. And the expression of LDHB was also suppressed in many cases of pancreatic cancer; however, the mechanism of the decreased expression and the significance of LDHB in pancreatic cancer development and progression have not been demonstrated [18, 19].
In the present study, we sought to determine the roles and mechanism of LDHB in pancreatic cancer development and progression. We found that suppressed expression of LDHB in pancreatic cancer was also due to hypermethylation and restored expression of LDHB led to decreased LDH activity and lactate production but increased intracellular ATP concentration. Furthermore, LDHB decreased pancreatic cancer cells proliferation, invasion, and migration in hypoxia. Our findings provide a new insight into the sophisticated roles and mechanisms of LDHB in pancreatic cancer development and progression.
Materials and methods
Cell lines and culture conditions
The human pancreatic cancer cell lines PANC-1, MiaPaca-2, AsPC-1, BxPC-3, Hs766T, CaPan-1, and PATU8902 were purchased from the American Type Culture Collection (ATCC). The pancreatic cancer cell lines MDA Panc-28 (MDA28), FG, COLO357, L3.3, and L3.7 were described previously [18]. All of these cell lines were maintained in plastic flasks as adherent monolayers in Eagle’s minimal essential medium supplemented with 10 % fetal bovine serum, sodium pyruvate, non-essential amino acids, l-glutamine, and a vitamin solution (Flow Laboratories).
Human tissue specimens and immunohistochemical analysis
The tissue microarray (TMA) construction and immunohistochemical analysis were conducted as described previously [18]. Briefly, the TMA contained 154 patients with pancreatic ductal adenocarcinoma and was from the Pancreatic Cancer Tissue Bank at Shanghai Jiaotong University Affiliated First People’s Hospital (Shanghai, PR China). Among the 154 primary tumors, 34 tissues contain matched tumor adjacent tissue, and 22 among the 34 tissues include normal tissues. The TMA was prepared and processed for immunostaining by using anti-LDHB (Epitomics) antibodies. The staining results were scored by two investigators blinded to the clinical data as described previously [20].
Plasmids and small interfering RNAs
The plasmids pCMV6-XL5-LDHB (pLDHB) were purchased from Origene (SC319772). Small interfering RNAs (siRNAs) for LDHB (#1, 5′-ACCGCCAAUUCUAAGAUUGTT-3′ and #2, GGAUAUACCAACUGGGCUATT-3′), EZH2 (#1, 5′-GCUGAAGCCUCAAUGUUUAdTdT-3′, #2, 5′-GAAUGGAAACAGCGAAGGAdTdT-3′), and Bmi1 (#1, 5′-CCAGACCACUACUGAAUAUTT-3′, #2, 5′-GACAUUGCAUCUGAUCUGU-3′) were previously reported and synthesized by Life Technologies [15, 21, 22]. Negative control siRNA (Invitrogen, Grand Island, NY, US) and pCMV6-XL5 vector were used as control. Transfection of plasmids and siRNAs into pancreatic cancer cells was performed using Lipofectamine LTX and Lipofectamine 2000 CD (Invitrogen) transfection reagents, respectively. For transient transfection, cells were transfected with plasmids or siRNA at different doses as indicated for 48 h before the performance of functional assays. Pancreatic cancer cells treated with transfection reagent alone were included as mock controls.
Treatment with 5-aza-2′-deoxycytidine (5-azaC)
Pancreatic cancer cell lines were treated with 5-azaC at 5 μM for 4 and 5 days, and total RNA and proteins were extracted and further analyzed by quantitative real-time PCR and Western blot.
Quantitative real-time PCR
Quantitative real-time PCR analysis of the expression of LDHB was performed using total RNA and the SYBR Green reagent with an ABI Prism 7000HT sequence detection system [23]. The sequences of the PCR primers were as follows: LDHB, 5′-GGGAAAGTCTCTGGCTGATGAA-3′ (forward) and 5′-GGGAAAGTCTCTGGCTGATGAA-3′ (reverse); and β-actin, 5′-AGAGGGAAATCGTGCGTGAC-3′ (forward) and 5′-CAATAGTGATGACCTGGCCGT-3′ (reverse).
Western blot analysis
Standard Western blotting was carried out using primary antibodies against anti-LDHB (Epitomics), anti-LDHA (Santa Cruz), anti-EZH2 (Abcam), and anti-Bmi1 (Cell Signaling Technology). Equal protein-sample loading was monitored using an anti-α-tublin antibody (Oncogene). Secondary antibodies were anti-mouse IgG (Cell Signaling) and anti-rabbit IgG (Santa Cruz).
LDH activity, lactate production, glucose utilization, and intracellular ATP concentration assays
Pancreatic cancer cells (1 × 106) were transfected with plasmids or siRNAs for 48 h and prepared for LDH activity, lactate production, and intracellular ATP concentration assays using an LDH activity assay kit, lactate assay kit, and ATP bioluminescent somatic cell assay kit (Sigma) according to the manufacturer’s protocol. Glucose utilization assay was conducted as previously described [18]. Briefly, cancer cells were transfected with plasmids or siRNAs for 24 h. Then, the media were replaced with phenol red-free RPMI with 1 % FBS and continuously cultured for 48 h. Glucose concentrations in the media were measured using a colorimetric glucose assay kit (BioVision) and normalized according to cell number.
Cell growth assay
Cell growth was determined by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazoliumbromide; Sigma). Transfected cells were grown in complete medium under hypoxic (1 % oxygen) conditions. Each group was repeated in three wells for three times. After incubation for 72 h, MTT (5 mg/ml in phosphate-buffered saline, PBS) was added to each well. After 2 h of incubation with MTT, the plates were centrifuged at 500 g for 5 min. After careful removal of the solution, 0.1 ml of DMSO was added to each well and plates were shaken. The absorbance was recorded on a microplate reader at the wavelength of 540 nm.
Cell scratch-wound assay
Cell scratch-wound assay was performed to examine the capacity of cell migration. We altered the expression of LDHB in AsPC-1 and FG cells by transfecting with pLDHB or LDHB siRNA. Specifically, after the cells grew in 90–95 % confluence in six-well plates, the wound was generated by scratching the surface of the plates with a 10-μl pipette tip. Cells will be washed with serum-free DMEM under hypoxic (1 % oxygen) conditions and photographed at 0 and 12 or 24 h. The in vitro wound filling was determined by measurement of the cell-free areas in multiple fields using a service provided by Wimasis, which permits users to upload their images online and allows their images to be analyzed and results uploaded back to the researcher’s server [24].
Tumor cell invasion/migration assay
AsPC-1 and FG cells were transfected for 6 h with different reagents respective to different groups (mock, control, pLDHB or mock, control, siLDHB). Cells in each group were trypsinized, and 2 × 104 to 5 × 104 cells in a 300-μl volume of serum-free medium in each group were placed in the upper parts of modified Boyden chambers with a Matrigel-coated/uncoated membrane (Millipore). For both cell lines, 500 μl of DMEM with 10 % fetal bovine serum was used as chemoattractant and added to the lower chamber. After 24 h of incubation under hypoxic (1 % oxygen) conditions, invasive/migrated cells were fixed, stained, and counted under a microscope in five randomly selected fields at a magnification of 200×.
Statistical analysis
The two-tailed Chi-squared test or Fisher’s exact test was used to determine the significance of the difference among the covariates of patient specimens. The significance of the data was determined using the Student t test (two-tailed) or one-way ANOVA. In all of the tests, P values less than 0.05 were considered statistically significant. The SPSS software program (version 13.0; SPSS Inc, New York, USA) was used for statistical analysis.
Results
Expression of LDHB in pancreatic cancer tissues and cell lines
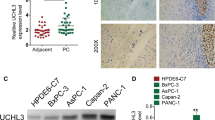
To investigate the roles of LDHB on pancreatic cancer development and progression, we first used IHC to analyze the expression of LDHB in the TMA which contained 154 primary pancreatic tumor tissues, 34 tumor adjacent tissues, and 22 normal pancreas tissues, and the clinicopathologic characteristics of TMA were described previously [18]. We found that LDHB staining was localized predominantly in the cytoplasm and most adjacent normal and normal pancreatic tissues were strong positive LDHB staining, whereas the expression of LDHB was suppressed and even lost in pancreatic cancer tissues (Fig. 1A). The expression levels of LDHB in non-malignant tissues were much higher than in cancer tissues (Fig. 1B left panel). We then further investigated the relationship between LDHB expression and clinicopathologic parameters of the pancreatic tumor. The results showed that the expression of LDHB did not significantly associate with gender, age, regional lymph node metastasis, and differentiation of pancreatic cancer (P > 0.05, Table 1), but negatively correlated with pT stages and TNM stages (P < 0.001, Fig. 1B middle and right panels). The expression pattern of LDHB was different from LDHA, as LDHA was much higher expressed in tumor tissues as compared to adjacent tissues and normal pancreas tissues and was positively associated with T stages, TNM stages, regional lymph node metastasis, and tumor differentiation [18]. We then used Western blot to further confirm the protein levels of LDHB and LDHA of cancer and associated normal tissues in the cases with LDHB suppressed and silent expression. The protein levels of LDHB in normal tissues were much higher than in cancer tissues, whereas LDHA was much higher in cancer tissues (Fig. 1C), which was consistent with the IHC results. Furthermore, we investigated the expression levels of LDHB and LDHA in human pancreatic cancer cell lines. Expression of LDHA was elevated in all cell lines, whereas the expression of LDHB was high in some cell lines but lost in AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells (Fig. 1D).
Expression of LDHB in pancreatic cancer tissues and cell lines. Immunostaining of LDHB in pancreatic tumor tissue microarray (TMA) with a specific anti-LDHB antibody. A (a) Non-malignant pancreatic tissues showing strong LDHB staining, (b) pancreatic cancer tissue showing complete absence of LDHB, (c) pancreatic cancer tissue showing strong LDHB staining. B The expression levels of LDHB were significantly suppressed in tumor tissues (TT) than in non-malignant pancreatic tissues, whereas there was no difference in LDHB expression between adjacent normal tissue (TN) and normal tissue (NN). The expression of LDHB was negatively correlated with pT and TNM stages. C Verification of expression of LDHB and LDHA in the paired normal pancreatic tissues (strong LDHB staining tissues) and pancreatic cancer tissues (negative or weak LDHB staining tissues) by Western blot. D Expression of LDHB and LDHA in pancreatic cancer cell lines
Promoter hypermethylation silenced LDHB in pancreatic cancer
Studies have revealed that promoter hypermethylation contributed to the silent expression of LDHB in prostate cancer, gastric cancer, and colorectal cancer [11, 12]. However, whether promoter hypermethylation silences LDHB in pancreatic cancer has not been demonstrated. Indeed, the promoter region of LDHB gene contains typical CpG islands (Fig. 2A). We incubated AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells, which lost the expression of LDHB, with whole medium or medium containing the demethylating agent 5-azaC. We found that the expression of LDHB in both mRNA and protein levels were restored in the 5-azaC-treated groups (Fig. 2B, C). These data demonstrated that promoter hypermethylation also led to silent expression of LDHB in pancreatic cancer. We then anticipated whether enhancer of zeste homolog 2 (EZH2) and B cell-specific Moloney murine leukemia virus integration site 1 (Bmi1) mediated the DNA methylation of LDHB promoter. However, the results showed that knockdown of EZH2 or Bmi1 by siRNAs in LDHB expressing or non-expressing cells did not change the expression levels of LDHB (Fig. 2D, E).
Regulation of LDHB expression by promoter hypermethylation in pancreatic cancer. A Diagram of CpG islands in the promoter region of LDHB. Reactivation of LDHB expression. AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells were treated with 5-azaC (5 μM) for 4 and 5 days. Quantitative real-time PCR (B) and Western blot (C) were used to analyze the mRNA and protein levels of LDHB, respectively. D, E Knock down the expression of EZH2 and Bmi1 and analyze their effect on LDHB expression by Western blot
Suppressed LDHB expression promoted the glycolytic phenotype of pancreatic cancer cells
LDHB is a major subunit of LDH, which is the key glycolytic enzyme and catalyzes the interconversion between pyruvate and lactate. However, the roles of LDHB in glycolytic phenotype of cancer have remained obscure. Thus, we knock down or restored expression of LDHB and analyzed the effect of LDHB on LDH activity, lactate production, glucose utilization, and intracellular ATP production in pancreatic cancer cells. The pCMV6-XL5-LDHB vector effectively expressed LDHB. Both siRNA#1 and siRNA#2 could knock down LDHB, and siRNA#1 was more effective (Fig. 3A). We then used pCMV6-XL5-LDHB (pLDHB) and siRNA#1 (siLDHB) for further experiments.
Influence of LDHB on pancreatic cancer glycolytic phenotype. A Transient transfection of pCMV6-XL5-LDHB effectively overexpressed the expression of LDHB in AsPC-1 and MiaPaca-2 cells, and siLDHB#1 and siLDHB#2 effectively knocked down the expression of LDHB in FG and PANC-1 cells. AsPC-1 was transfected with pCMV6-XL5-LDHB or control vector (B), FG cells was transfected with LDHB siRNA or control siRNA (C). LDH activity assay kit, lactate assay kit, ATP bioluminescent somatic cell assay kit and colorimetric glucose assay kit were used to analyze the LDH activity, lactate production, intracellular ATP concentration, and glucose utilization. D AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells transfected with pLDHB (D1) and FG, BxPC-3, PANC-1 and MDA28 cells transfected with LDHB siRNA (D2) were cultured under hypoxic (1 % oxygen) condition for 72 h. The proliferation of the cells was measured by MTT
The results showed that overexpression of LDHB in AsPC-1 cells led to reduced LDH activity and lactate production but elevated intracellular ATP level, whereas knockdown of LDHB in FG cells increased LDH activity and lactate production but decreased intracellular ATP concentration (Fig. 3B, C). However, the utilization of glucose did not change significantly according to the altered expression of LDHB (Fig. 3B, C). These results indicated that suppressed expression of LDHB promoted the glycolytic phenotype of pancreatic cancer and restored expression of LDHB could increase mitochondrial respiratory activity.
LDHB suppressed pancreatic cancer cells proliferation, migration, and invasion under hypoxia
Pancreatic cancer is rich in stroma and lack of oxygen. We then analyze the effect of LDHB on pancreatic cancer growth and metastasis under hypoxia. We ectopically expressed LDHB in AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells and knockdown of LDHB in FG, BxPC-3, PANC-1, and MDA28 cells. MTT assay was used to measure the proliferation of the cells and found that restored expression of LDHB inhibited cell proliferation and knockdown of LDHB increased cell proliferation under hypoxia (Fig. 3D1, D2, *, P < 0.05). To assess the impact of LDHB expression on pancreatic cancer migration and invasion under hypoxia, the transfected AsPC-1 and FG cells were wounded by scratching and the results showed that ectopic expression of LDHB decreased the flattening and spreading of AsPC-1 cells (Fig. 4A1, *, P < 0.05), whereas knockdown of LDHB promoted flattening and spreading of FG cells (Fig. 4B1, *, P < 0.05). Furthermore, the results of migration and invasion assay also revealed that restored expression of LDHB repressed migration and invasion of AsPC-1 cells (Fig. 4A2, A3, *, P < 0.05), whereas knockdown of LDHB promoted the migration and invasion of FG cells (Fig. 4B2, B3, *, P < 0.05). These data revealed that LDHB could suppress pancreatic cancer growth, migration, and invasion, which was consistent with the effect of LDHB on hepatoma cells [15].
Influence of LDHB expression on migration and invasion of pancreatic cancer cells. For cell scratch-wound assay, AsPC-1 (A1) and FG cells (B1) were transfected with pLDHB and siLDHB for 48 h under hypoxic condition, respectively. The cultures were then wounded by scratching and maintained for additional 24 or 12 h under hypoxic condition. Cell cultures were photographed, and migration was measured by the cell-free areas in multiple fields (inserted number represented the percentage of gap areas ± SD). The migration and invasion of AsPC-1 (A2 and A3) and FG (B2 and B3) cells were analyzed by the cell migration and invasion assays as described in “Materials and methods.” Representative cancer cell migrated or invaded was photographed; data represent mean ± SD of triplicates. Note *P<0.05 (one-way ANOVA) in a comparison of the pLDHB- or siLDHB-treated groups with the mock and control groups
Discussion
LDHA and LDHB are the predominant LDH subunits that are widely expressed in adult somatic tissue [25]. LDHA has reported to play critical roles in promoting the development and progression of many types of cancer, including pancreatic cancer, whereas the roles of LDHB are depending on different types of tumors. In our previous study, we found that the expression of LDHB was absent in many cases of pancreatic cancer tissues and cell lines [18, 19]. However, the expression pattern, mechanism, and effect of suppressed expression of LDHB have not been fully revealed in pancreatic cancer. It has been reported that promoter hypermethylation led to decreased expression of LDHB in prostate cancer, gastric cancer, and colorectal cancer [11, 12]. In the current study, we demonstrated that suppressed and even lost expression of LDHB in pancreatic cancer was also due to promoter hypermethylation, as treated AsPC-1, MiaPaca-2, CaPan-1, and PATU8902 cells with the demethylating agent 5-azaC resulted in restored expression of LDHB. Furthermore, we showed that the expression of LDHB was absent in about a quarter of pancreatic cancer tissues (38/154 cases), which was lower than in prostate cancer tissues (45 %) but much higher than in gastric cancer tissues (7 %) [11, 12]. Researches have demonstrated that polycomb group (PcG) proteins interacted with DNA methyltransferases (DNMTs) and led to DNA methylation of certain gene promoters [26, 27]. PcG of proteins contain two multimeric protein complexes: polycomb complex 1 (PRC1) and polycomb complex 2 (PRC2) [28]. Bmi1 and EZH2 are the key subunits of PRC1 and PRC2, respectively. In this study, we further determined whether the absent expression of LDHB in pancreatic cancer was due to Bmi1- and EZH2-mediated promoter DNA hypermethylation. The results demonstrated that PRC1 and PRC2 might not mediate the promoter hypermethylation of LDHB, as knockdown of Bmi1 and EZH2 did not change the expression states of LDHB.
Loss of LDHB in pancreatic cancer cells led to elimination of LDH1-4 and leaved only LDH5 which was expected to be the highest efficiency isoenzyme to catalyze the conversion of pyruvate to lactate [11, 29]. However, the roles of LDHB in glycolysis were obscure in different types of cancer [11, 13–15]. Here, we showed that loss of LDHB led to glycolytic transition and respiratory dysfunction, as knockdown of LDHB resulted in increased LDH activity and lactate production but decreased intracellular ATP concentration.
To determine the roles of LDHB in pancreatic cancer biology, the effect of LDHB on pancreatic cancer growth and metastasis was assessed. We measured cells proliferation in different LDHB expression groups under hypoxia and found that suppressed expression of LDHB promoted cells proliferation under hypoxia. Furthermore, we found that ectopic expression of LDHB decreased migration and invasion of AsPC-1 cells, whereas knockdown of LDHB promoted the migration and invasion of FG cells. It has been reported that suppressed expression of LDHB increased invasion of hepatoma cells via inducing claudin-1 expression, but claudin-1 inhibited pancreatic cancer metastasis [15]. However, suppressed expression of LDHB might promote pancreatic cancer cell migration and invasion by enhancing glycolytic phenotype which was closely correlated with increased metastasis in cancers [18, 30].
In summary, this study for the first time identified the suppressor role of LDHB in pancreatic cancer development and progression by inhibiting glycolysis. Four lines of both experimental and clinical evidence were provided. Firstly, we found that the expression of LDHB was suppressed and even lost in pancreatic cancer tissues and was negatively correlated with disease pT classification and TNM stages. Secondly, decreased expression of LDHB in pancreatic cancer was due to promoter hypermethylation. Thirdly, restored expression of LDHB reduced LDH activity and lactate production but elevated intracellular ATP concentration. Fourthly, restored expression of LDHB repressed pancreatic cancer cell proliferation, invasion, and migration. Collectively, these findings not only further demonstrated the molecular mechanism underlying pancreatic cancer development and progression, but also indicated that combination of LDHA inhibitors and demethylating agents might be a promising new therapeutic strategy for pancreatic cancer, which requires further studies.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70.
Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800.
Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72.
Drent M, Cobben NA, Henderson RF, Wouters EF, van Dieijen-Visser M. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur Respir J. 1996;9:1736–42.
Koukourakis MI, Kontomanolis E, Giatromanolaki A, Sivridis E, Liberis V. Serum and tissue LDH levels in patients with breast/gynaecological cancer and benign diseases. Gynecol Obstet Invest. 2009;67:162–8.
Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, Harris AL. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003;89:877–85.
Koukourakis MI, Giatromanolaki A, Simopoulos C, Polychronidis A, Sivridis E. Lactate dehydrogenase 5 (LDH5) relates to up-regulated hypoxia inducible factor pathway and metastasis in colorectal cancer. Clin Exp Metastas. 2005;22:25–30.
Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–63.
Lin Q, Cong X, Yun Z. Differential hypoxic regulation of hypoxia-inducible factors 1alpha and 2alpha. Mol Cancer Res. 2011;9:757–65.
Leiblich A, Cross SS, Catto JW, Phillips JT, Leung HY, Hamdy FC, Rehman I. Lactate dehydrogenase-B is silenced by promoter hypermethylation in human prostate cancer. Oncogene. 2006;25:2953–60.
Maekawa M, Taniguchi T, Ishikawa J, Sugimura H, Sugano K, Kanno T. Promoter hypermethylation in cancer silences LDHB, eliminating lactate dehydrogenase isoenzymes 1-4. Clin Chem. 2003;49:1518–20.
Dennison JB, Molina JR, Mitra S, Gonzalez-Angulo AM, Balko JM, Kuba MG, Sanders ME, Pinto JA, Gomez HL, Arteaga CL, et al. Lactate dehydrogenase B: a metabolic marker of response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2013;19:3703–13.
McCleland ML, Adler AS, Shang Y, Hunsaker T, Truong T, Peterson D, Torres E, Li L, Haley B, Stephan JP, et al. An integrated genomic screen identifies LDHB as an essential gene for triple-negative breast cancer. Cancer Res. 2012;72:5812–23.
Kim JH, Kim EL, Lee YK, Park CB, Kim BW, Wang HJ, Yoon CH, Lee SJ, Yoon G. Decreased lactate dehydrogenase B expression enhances claudin 1-mediated hepatoma cell invasiveness via mitochondrial defects. Exp Cell Res. 2011;317:1108–18.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17.
Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng S, Gao Y, Huang S, Xie K. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin Cancer Res. 2014;20:2595–606.
Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J, Zhu Z, Gao Y, Xie K. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin Cancer Res. 2014;20:4370–80.
Wang L, Wei D, Huang S, Peng Z, Le X, Wu TT, Yao J, Ajani J, Xie K. Transcription factor Sp1 expression is a significant predictor of survival in human gastric cancer. Clin Cancer Res. 2003;9:6371–80.
Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–82.
Chen S, Sheng C, Liu D, Yao C, Gao S, Song L, Jiang W, Li J, Huang W. Enhancer of zeste homolog 2 is a negative regulator of mitochondria-mediated innate immune responses. J Immunol. 2013;191:2614–23.
Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, Feng XH, Sawaya R, Medema RH, Hung MC, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. J Clin Invest. 2014;124:564–79.
Khoo CP, Micklem K, Watt SM. A comparison of methods for quantifying angiogenesis in the Matrigel assay in vitro. Tissue Eng Part C Methods. 2011;17:895–906.
Draoui N, Feron O. Lactate shuttles at a glance: from physiological paradigms to anti-cancer treatments. Dis Model Mech. 2011;4:727–32.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4.
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6.
Raaphorst FM. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum Mol Genet. 2005;14:R93–100.
Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL. Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia. 2005;7:1–6.
Walenta S, Wetterling M, Lehrke M, Schwickert G, Sundfor K, Rofstad EK, Mueller-Klieser W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–21.
Acknowledgments
Supported by Shanghai Jiao Tong University Oversea Training Scholarship (to J. Cui and M. Quan); Grants 81171887 and 91229117 from the National Natural Science Foundation of China (to L. Wang).
Conflict of interest
No potential conflicts of interest were disclosed.
Ethical standard
The use of the tissue microarray was approved by the medical ethics committee.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jiujie Cui and Ming Quan have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Cui, J., Quan, M., Jiang, W. et al. Suppressed expression of LDHB promotes pancreatic cancer progression via inducing glycolytic phenotype. Med Oncol 32, 143 (2015). https://doi.org/10.1007/s12032-015-0589-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-015-0589-8