Abstract
The study was aimed to explore the underlying mechanisms and identify the potential target genes and key miRNAs for acute myeloid leukemia (AML) treatment by bioinformatics analysis. The microarray data of GSE9476 were downloaded from Gene Expression Omnibus database. A total of 64 samples, including 26 AML and 38 normal samples, were used to identify differentially expressed genes (DEGs) between AML and normal samples. The functional enrichment analysis was performed, and protein–protein interaction (PPI) network of the DEGs was constructed by Cytoscape software. Besides, the target miRNAs for DEGs were identified. Totally, 323 DEGs were identified, including 87 up-regulated and 236 down-regulated genes. Not only up-regulated genes but also down-regulated genes were related to hematopoietic-related functions. Besides, down-regulated genes were also enriched in primary immunodeficiency pathway. Tumor necrosis factor (TNF), interleukin 7 receptor (IL7R), lymphocyte-specific protein tyrosine kinase (LCK), CD79a molecule and immunoglobulin-associated alpha (CD79A) were identified in these functions. TNF and LCK were hub nodes in PPI networks. miR-124 and miR-181 were important miRNAs in this study. The hematopoietic-related functions and primary immunodeficiency pathway may be associated with AML development. Genes, such as TNF, IL7R, LCK and CD79A, may be potential therapeutic target genes for AML, and miR-124 and miR-181 may be key miRNAs in AML development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) is one of the most common hematopoietic malignancies [1]. It is a heterogeneous clonal disorder of hematopoietic progenitor cells, characterized by uncontrolled proliferation of white blood cells that gradually replace normal hematopoiesis in the bone marrow. Approximately 6500 new cases (almost children and adolescents) in the USA of AML are diagnosed annually [2]. Its incidence increases progressively with age [3]. The 5-year survival rate of AML is less than 15 % [1]. Therefore, an improved understanding mechanism on the pathogenesis of AML would supply new insights for the diagnosis and treatment of AML.
Numerous studies in molecular biology have been done to decipher the pathogenesis of AML. The carcinogenesis of AML is a process of accumulation of genetic changes in hematopoietic cells growth, proliferation differentiation and apoptosis [3, 4]. For example, over-expression of KIAA1524 (CIP2A) is related to active cell proliferation and arrest cell differentiation [4]. It has been reported that the phosphoinositide 3-kinase (PI3-kinase) signaling pathway plays an important role in the regulation of tumor cell survival and proliferation [5]. The work of Grandage et al. [6] found that blocking PI3-kinase had direct antileukemic effect and that it was useful in the treatment of AML. Besides, miR-424 and miR-155 deregulation was involved in the pathogenesis of AML with nucleophosmin-1 and fms-related tyrosine kinase 3 mutations, respectively [7]. Although many factors have been detected in AML, the pathogenic mechanisms of AML are still not clearly demonstrated. It is lack of potential target genes for AML treatment.
The microarray data (GSE9476) were analyzed with microarray analysis to identify the differentially expressed genes (DEGs) between AML and normal samples by Stirewalt et al. [8]. In this study, we downloaded these data and used different methods to identify the DEGs. The functional analysis was performed, and protein–protein interaction (PPI) network was constructed. What is more, the target miRNAs for DEGs were identified. The purpose of this study was to explore the underlying mechanisms and identify the potential target genes and key miRNAs in the AML development.
Data and methods
Affymetrix microarray data
The gene expression profile data of GSE9476 based on the platform of GPL96 (Affymetrix Human Genome U133A Array) (Affymetrix Inc., Santa Clara, CA, USA) were downloaded from Gene Expression Omnibus (GEO) database in National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/geo/), which was deposited by Stirewalt et al. [8]. The datasets available in this analysis contained 64 samples, including leukemic blasts from 26 AML patients and normal hematopoietic cells from 38 healthy donors.
Data preprocessing and differential expression analysis
The probe IDs were converted into corresponding gene names based on the annotation information on the platform. When multiple probes corresponded to a same gene, the average expression value was calculated to represent the gene expression level. The raw microarray data were normalized with the PreprocessCore package (http://master.bioconductor.org/packages/release/bioc/html/preprocessCore.html) [9] in R. The limma package (http://master.bioconductor.org/packages/release/bioc/html/limma.html) [10] in R was used to identify DEGs between AML and normal samples. Then, log2-fold change (log2FC) was calculated to identify genes with expression-level differences. Only DEGs with |log2FC| > 1.0 and adjusted P value <0.05 were selected.
Functional enrichment analysis
Gene Ontology (GO) database (http://geneontology.org/) [11] is a set of a large number of gene annotation terms. Kyoto Encyclopedia of Genes and Genomes (KEGG) knowledge database (http://www.kegg.jp/) [12] is applied to identify the significantly enriched metabolic or signal pathways for target genes. Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) [13] is a tool that provides a comprehensive set of functional annotation for large list of genes. GO categories and KEGG pathway enrichment analyses were performed for DEGs using DAVID, respectively. P value <0.05 was the cutoff criterion for the functional enrichment analysis.
Protein–protein interaction (PPI) network construction
Search Tool for the Retrieval of Interacting Genes (STRING, http://www.string-db.org/) [14] is an online database, which collects comprehensive information of proteins. The STRING online tool was applied to analyze the interactions of protein pairs, and only the interaction with confidence score > 0.4 was selected as significant. PPI network was constructed with Cytoscape software (http://cytoscape.org/) [15].
Prediction of target miRNAs for DEGs
Web-based Gene Set Analysis Toolkit (WebGestalt, http://genereg.ornl.gov/webgestalt/) [16] is an integrated system for exploring large sets of gene. WebGestalt was used to identify miRNAs corresponding to DEGs. MiRNAs with DEGs number > 2 and raw P value <0.05 were identified as target miRNAs. The gene-miRNA network was visualized with Cytoscape software.
Results
Identification of DEGs
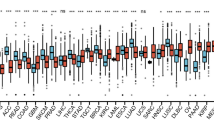
As shown in Fig. 1, the raw expression data were normalized after preprocessed. A total of 323 DEGs were identified between AML and normal samples, including 87 up-regulated genes and 236 down-regulated genes.
Functional enrichment analysis
The top five GO terms enriched by up- and down-regulated genes are shown in Table 1, respectively. The up-regulated genes were mainly enriched in GO biological process (BP) terms of defense response, hemopoiesis and inflammatory response. The most enriched GO terms of down-regulated genes were relevant with BP terms of hemopoiesis, hemopoietic or lymphoid organ development and immune system development.
The KEGG pathways of up- and down-regulated genes are shown in Table 2, respectively. The up-regulated genes were mainly enriched in hematopoietic cell lineage, systemic lupus erythematosus and AML. DEGs including tumor necrosis factor (TNF) and fms-related tyrosine kinase 3 (FLT3) were identified in the significantly function of hematopoietic cell lineage. Additionally, down-regulated genes were related to primary immunodeficiency, hematopoietic cell lineage and porphyrin and chlorophyll metabolism. The down-regulated genes including lymphocyte-specific protein tyrosine kinase (LCK) and CD79a molecule, immunoglobulin-associated alpha (CD79A) were enriched in primary immunodeficiency pathway. Down-expression of interleukin 7 receptor (IL7R) was identified in hematopoietic cell lineage pathway.
PPI network construction
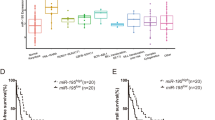
The PPI networks of up- and down-regulated genes are shown in Fig. 2, respectively. The up-regulated network was constructed with 31 nodes and 28 edges (A). The proteins TNF (degree = 6), TIMP metallopeptidase inhibitor 1 (TIMP1, degree = 6) and cathepsin D (CTSD, degree = 6) were hub nodes in this network. The down-regulated PPI network was constructed with 129 nodes and 316 edges (B). LCK (degree = 26), granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) (GZMB, degree = 17), CD79A (degree = 17), chemokine (C–C motif) receptor 7 (CCR7, degree = 15) and CD27 molecule (CD27, degree = 15) were hub proteins in this network.
PPI networks of DEGs. a PPI network of up-regulated genes. b PPI network of down-regulated genes. Nodes stand for proteins, and edges represent interactions between two proteins. The thickness of lines represents degree of interaction between nodes. Thicker line indicates stronger interaction. The shades of color represent expression value difference of DEGs between AML and normal samples. The deeper the color, the greater the different expression value
Prediction of target miRNAs for DEGs
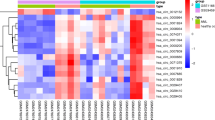
The target miRNAs for up- and down-regulated genes are presented in Fig. 3, respectively. In the up-regulated gene-miRNA network, the miR-124 (n = 6) and miR-499 (n = 4) regulated the most up-regulated genes (A). Polypyrimidine tract-binding protein 2 (PTBP2) was regulated by 4 miRNAs, such as miR-124A and miR-499. Besides, miR-181 (n = 13) and miR-23 (n = 12) families regulated the most DEGs in down-regulated gene-miRNA network (B). Gene transmembrane and coiled-coil domain family 1 (TMCC1) was the hub node and was regulated by 8 miRNAs, including miR-181 family and miR-221.
Discussion
AML is one of the most common and deadly forms of hematopoietic malignancies [1]. Understanding the molecular mechanism of AML is of critical importance for management policy. In this study, the gene expression profile data of GSE9476 were downloaded from GEO database to identify DEGs between AML and normal samples using bioinformatics analysis. Totally, 323 DEGs including 87 up- and 236 down-regulated genes were selected. The functional enrichment analysis results showed that not only up-regulated genes but also down-regulated genes were related to hemopoiesis and hematopoietic cell lineage functions. Genes TNF and IL7R were identified in these functions. Besides, down-regulated genes, LCK and CD79A, were enriched in primary immunodeficiency pathway. TNF and LCK were hub nodes in PPI networks. miR-124 and miR-181 were important miRNAs in this study, and TMCC1 was regulated by miR-181 family. These DEGs, their related functions and miRNAs may be involved in AML progress.
The hematopoietic-related functions (hemopoiesis and hematopoietic cell lineage pathway) were the significant functions in this study. TNF and IL7R were identified in these functions. TNF is a monocyte-derived cytotoxin that has been implicated in tumor regression and cell apoptosis [17]. It has been reported that TNF induces potent growth inhibition of myeloid progenitor cells, and it is a potential growth stimulator of human multipotent hematopoietic cells [18]. TNF exerts proliferative effects on human AML blasts, and the effects are mediated by both p55 and p75 TNF-receptor (TNFR) in AML cells [19]. In this study, TNF was up-regulated gene in AML samples, which was consistent with a previous study that Cimino et al. [20] reported that serum TNF level was significantly increased in AML patients. It was hub node with high degree of 6 in the up-regulated PPI network, and TNF was enriched in hematopoietic functions, suggesting that hematopoietic-related gene TNF may play an important role in AML development. Additionally, IL7R was also identified in hematopoietic functions. IL7R is a receptor for IL7 and is a protein found on the surface of cells. IL7 belongs to interleukin family. IL11 (another member of the interleukin family) is a stromal factor that has a number of hematopoietic-related effects cooperating with IL3 and stem cell factor [21]. It has been reported that IL7R mutations may contribute to the pathogenesis of acute leukemias [22]. In our study, IL7R was down-regulated gene in AML samples, suggesting that it may play an important role in AML carcinogenesis. Our results showed that TNF and IL7R may be associated with AML progress and that hematopoietic-related functions may play a key role in the pathogenesis of AML.
A primary immunodeficiency pathway was identified as an important function associated with the progression of AML. LCK and CD79A were identified in this pathway. LCK is involved in primary T cell immunodeficiency [23], and it is a member of the Src family of tyrosine kinases. Members of the Src family of tyrosine kinases are signaling intermediates that can regulate cell metabolism, proliferation, differentiation and migration in many liquid cancers including AML [24]. The work of Dos Santos et al. [25] indicated that silencing Lyn (a member of the Src family of tyrosine kinases) expression by small interfering RNA in AML cells strongly inhibited proliferation. However, the evidence concerning the impact of LCK in AML is rare. In this study, down-expressed LCK was enriched in the primary immunodeficiency pathway and was hub node in PPI network, suggesting that LCK may play an important role in AML progression by regulating primary immunodeficiency pathway. In addition, CD79A is a B lymphocyte antigen receptor. It is associated with B cell-mediated immunodeficiency [26]. CD79A is a marker for B cell neoplasms including AML in routinely processed tissue samples [27]. Cruse et al. [28] reported that the aberrant expression of CD79A portended a poor prognosis in AML. Down-expressed CD79A was hub node in PPI network in this study. It suggested that primary immunodeficiency pathway may affect the progress of AML by inhibiting the expression of LCK and CD79A.
Apart from DEGs and their functions, miRNAs such as miR-124 and miR-181 family may be important for the development of AML. It has been reported that miR-124 reduces the retinoblastoma protein phosphorylation [29], as well as its role as a tumor suppressor. The work of Vázquez et al. [30] found that the expression of MDS1 and EVI1 complex locus (EVI1) in AML patients was associated with decreased expression of miR-124. The EVI1 gene codes for a transcription factor involved in the development and progression of high-risk AML [31]. Furthermore, miR-181 was reported that its expression was used for molecular risk assessment in AML patients [32]. In this study, TMCC1 was the target gene of members of the miR-181 family. TMCC1 was down-expressed in AML samples, and its expression was inversely correlated with the expression level of miR-181. Therefore, it indicated that miR-181 was over-expressed in AML. It has been reported that high expression of miR-181 may reduce the aggressiveness of AML [32]. Taking these factors into account, we speculated that miR-124 and miR-181 may be key regulators in AML development.
In conclusion, our study shows that hematopoietic-related functions and primary immunodeficiency pathway may be closely associated with AML development. Genes such as TNF, IL7R, LCK and CD79A may be potential therapeutic target genes for AML. miR-124 and miR-181 may be key miRNAs in AML development. However, further studies are still required to confirm our results.
References
Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999;341(14):1051–62.
Ries LG, Smith MA, Gurney J, Linet M, Tamra T, Young J et al. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995; 1999.
Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24(1):35–63.
Wang J, Li W, Li L, Yu X, Jia J, Chen C. CIP2A is over-expressed in acute myeloid leukaemia and associated with HL60 cells proliferation and differentiation. Int J Lab Hematol. 2011;33(3):290–8.
Vivanco I, Sawyers CL. The phosphatidylinositol 3-kinase–AKT pathway in human cancer. Nat Rev Cancer. 2002;2(7):489–501.
Grandage V, Gale R, Linch D, Khwaja A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kB, MAPkinase and p53 pathways. Leukemia. 2005;19(4):586–94.
Faraoni I, Laterza S, Ardiri D, Ciardi C, Fazi F, Lo-Coco F. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5(1):26.
Stirewalt DL, Meshinchi S, Kopecky KJ, Fan W, Pogosova-Agadjanyan EL, Engel JH, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosom Cancer. 2008;47(1):8–20.
Bolstad B. preprocessCore: a collection of pre-processing functions. R package version. 2013;1(0).
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):1–25.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9.
Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6(1):60.
Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(suppl 2):W169–75.
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(suppl 1):D561–8.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Suppl 2):W741–8.
Fransen L, Mūller R, Marmenout A, Tavernier J, Van der Heyden J, Kawashima E, et al. Molecular cloning of mouse tumour necrosis factor cDNA and its eukaryotic expression. Nucleic Acids Res. 1985;13(12):4417–29.
Bettaieb A, Record M, Come M, Bras A-C, Chap H, Laurent G, et al. Opposite effects of tumor necrosis factor alpha on the sphingomyelin-ceramide pathway in two myeloid leukemia cell lines: role of transverse sphingomyelin distribution in the plasma membrane. Blood. 1996;88(4):1465–72.
Carter A, Haddad N, Draxler I, Israeli E, Raz B, Rowe JM. Expression and role in growth regulation of tumour necrosis factor receptors p55 and p75 in acute myeloblastic leukaemia cells. Br J Haematol. 1996;92(1):116–26.
Cimino G, Amadori S, Cava M, De Sanctis V, Petti M, Di Gregorio A, et al. Serum interleukin-2 (IL-2), soluble IL-2 receptors and tumor necrosis factor-alfa levels are significantly increased in acute myeloid leukemia patients. Leukemia. 1991;5(1):32–5.
Ozaki K, Leonard WJ. Cytokine and cytokine receptor pleiotropy and redundancy. J Biol Chem. 2002;277(33):29355–8.
Kim MS, Chung NG, Kim MS, Yoo NJ, Lee SH. Somatic mutation of IL7R exon 6 in acute leukemias and solid cancers. Hum Pathol. 2013;44(4):551–5.
Hauck F, Randriamampita C, Martin E, Gerart S, Lambert N, Lim A et al. Primary T-cell immunodeficiency with immunodysregulation caused by autosomal recessive LCK deficiency. J Allergy Clin Immunol 2012;130(5):1144–52. e11.
Frame MC. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta (BBA) Rev Cancer. 2002;1602(2):114–30.
Dos Santos C, Demur C, Bardet V, Prade-Houdellier N, Payrastre B, Récher C. A critical role for Lyn in acute myeloid leukemia. Blood. 2008;111(4):2269–79.
Mackay IR, Rosen FS, Buckley RH. Primary immunodeficiency diseases due to defects in lymphocytes. N Engl J Med. 2000;343(18):1313–24.
Mason DY, Cordell J, Brown M, Borst J, Jones M, Pulford K, et al. CD79a: a novel marker for B-cell neoplasms in routinely processed tissue samples. Blood. 1995;86(4):1453–9.
Cruse JM, Lewis RE, Pierce S, Lam J, Tadros Y. Aberrant expression of CD7, CD56, and CD79a antigens in acute myeloid leukemias. Exp Mol Pathol. 2005;79(1):39–41.
Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–9.
Vázquez I, Maicas M, Marcotegui N, Conchillo A, Guruceaga E, Roman-Gomez J, et al. Silencing of hsa-miR-124 by EVI1 in cell lines and patients with acute myeloid leukemia. Proc Natl Acad Sci. 2010;107(44):E167–8.
Wieser R. The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene. 2007;396(2):346–57.
Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117(4):1121–9.
Acknowledgments
This study was supported by Natural Science Foundation of Heilongjiang Province of China (D201252).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, Y., Zhang, X., Zhao, Y. et al. Identification of potential therapeutic target genes, key miRNAs and mechanisms in acute myeloid leukemia based on bioinformatics analysis. Med Oncol 32, 152 (2015). https://doi.org/10.1007/s12032-015-0572-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-015-0572-4