Abstract
Interleukin-18 (IL-18) is a pleiotropic, pro-inflammatory cytokine with dual effects on tumor development and progression. The -607(C/A) and -137(G/C) polymorphisms in IL-18 gene region have been implicated in cancer risk; however, data from published studies with individually low statistical power are conflicting. To clarify the role of IL-18 -607(C/A) and -137(G/C) genotype in global cancer, we examined all the available published studies through a pooled analysis approach. Overall, IL-18 -607A allele was associated with increased total cancer risk when compared with -607C allele (OR = 1.14, 95% CI = 1.01–1.28, P = 0.010), as well as in the heterozygote comparison (OR = 1.10, 95% CI = 1.04–1.15, P = 0.256) and the dominant model (OR = 1.07, 95% CI = 1.03–1.11, P = 0.124). Furthermore, IL-18 -137(G/C) polymorphism was associated with increased nasopharyngeal carcinoma risk. In the stratified analysis for -607(C/A) polymorphism, a significantly increased cancer risk in Asian population was found, as well as subgroup in source of control. Similar results were found in the stratified analysis for -137(G/C) polymorphism. Our pooled analysis supported that IL-18 is a good candidate for large-scale epidemiological case–control studies that may be a low-penetrance susceptibility biomarker for cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, a considerable number of factors involved in angiogenesis, inflammation, and thrombosis have been associated with increased risk for several types of cancers. Interleukin-18 (IL-18) is one such factor, previously correlated to angiogenesis, inflammation, coronary artery disease and cancer [1–3]. IL-18, formerly called interferon-γ-inducing factor, is a novel 18.3-KDa cytokine produced by activated macrophages, keratinocytes, Kupffer cells, intestinal epithelial cells and osteoblasts [4]. IL-18 can promote tumor cells, as evidenced by the elevated expression of IL-18 in tumor cells [5, 6]. IL-18 expression was also elevated in the tumor versus nontumor area in patients with gastric cancer and was related to distant metastasis [7]. Higher IL-18 levels were found in serum from breast cancer patients with or without metastasis and were proposed as an important marker of breast cancer progression [8]. It also enhances immune escape by promoting the expression of Fas ligand in tumor cells and downregulating major histocompatibility complex class I expression [9, 10].
IL-18 gene locus is located on chromosome 11q22.2-q23.3 [11]. Two single nucleotide polymorphisms (SNPs) in the promoter of IL-18 gene at the position -607(C/A)[rs1946518] and -137(G/C)[rs187238] have been identified. A change from C to A at position -607 disrupts a potential cAMP-responsive element binding protein binding site. A change at position -137 from G to C changes the H4TF-1 nuclear factor binding site to a binding site for an unknown factor found in the GM-CSF promoter. The functional significance of these two SNPs has been shown that the C allele at position -607 and the G allele at position -137 were attributed to the IL-18 higher transcription and protein production [11, 12]. These two SNPs have been investigated in several types of cancer such as renal cell carcinoma [13], ovarian cancer [14, 28], breast cancer [15], nasopharyngeal carcinoma [16, 20, 22], cervical cancer [17, 29], esophageal squamous cell carcinoma [18], prostate cancer [19], lung cancer [21], stomach cancer [23], colorectal cancer [23, 25], head and neck squamous cell carcinoma [24], oral cancer [26] and lymphomas [27].
To clarify the effect of variation within the role of IL-18 in cancer, hence, we performed a pooled analysis of all eligible case–control studies to derive a more precise estimation of the association of -607(C/A) and -137(G/C) polymorphisms in IL-18 and cancer risk.
Materials and methods
Publication search
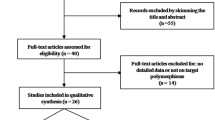
We conducted searches on the PubMed database (http://www.ncbi.nlm.nih.gov/), last search updated on April 2010, with the keywords ‘IL-18’, ‘polymorphism’ and ‘cancer’. Using these terms, a total of 35 articles were retrieved, of which 17 [13–29] articles according to the inclusion criteria reported on studies examining the association between IL-18 -607(C/A) or -137(G/C) polymorphism and cancer risk.
Inclusion criteria
Studies testing the association between IL-18 -607(C/A) and/or -137(G/C) gene polymorphisms and cancer were considered if all the following inclusion criteria were found: (a) the study assessed the correlation between global cancer and at least one of the polymorphism cited above; (b) case–control studies; (c) control subjects matched with case patients for age and gender and (d) only full-text manuscripts were included.
Exclusion criteria
Major exclusion criteria were (a) no control population; (b) no available genotype frequency; (c) duplication of the previous publications and (d) manuscripts with a clear bias of accrual.
Data extraction
Two of the authors reviewed results of each of the database searches to make sure that published papers are not missed. Data were collected on the first author’s last name, year of publication, country of origin, ethnicity, cancer type, sample size (cases/controls), source of control and Hardy–Weinberg equilibrium of controls. Ethnicity was categorized as European, Asian, and African.
Statistical analysis
Odds ratios (ORs) with 95% confidence intervals (CIs) were used to measure the strength of the association between IL-18 -607(C/A) or -137(G/C) polymorphism and cancer risk based on the genotype frequencies in cases and controls. We explored the association between allele A of IL-18 -607(C/A) or allele C of -137(G/C) and cancer risk, as well as the heterozygote comparison, the homozygote comparison and the risk of cancers in dominant and recessive models, respectively. Stratified analyses were also performed by cancer type (if one cancer type contained only one individual study, it was combined into the ‘other cancers’ group), ethnicity and source of control.
A pooled analysis is not a simple average of the effects in all studies. Rather, it is a more precise study. To take into account the possibility of heterogeneity across the studies, a statistical test for heterogeneity was carried out based on the Q statistic, in which a P-value greater than 0.05 suggested a lack of heterogeneity [30]. Random effects model (DerSimonian and Laird [30]) was used when P value for heterogeneity test < 0.05; otherwise, fixed effects model (Mantel–Haenszel [31]) was used.
Hardy–Weinberg equilibrium (HWE) in the controls group was tested using the Pearson chi-square test for goodness of fit; P < 0.05 was considered significant.
The presence of publication bias was examined by visual inspection of funnel plots, and formally evaluated with Begg’s adjusted rank correlation test and Egger’s regression asymmetry test [32], P < 0.05 was considered statistically significant.
All statistical analyses were undertaken using the program STATA version 10.0 (Stata Corporation, TX).
Results
Eligible studies
Overall, we identified 17 articles (30 studies; 17 different first authors) to evaluate the association of IL-18 -607(C/A) or -137(G/C) polymorphism with risk for cancer. Characteristics of studies of -607(C/A) or -137(G/C) polymorphism are summarized in Tables 1 and 2, respectively. Therefore, there were 15 studies with 2,137 cases and 3,117 controls concerning the -607(C/A) polymorphism and 15 studies with 2,372 cases and 3,476 controls concerning the -137(G/C) polymorphism. For -607(C/A) polymorphism, there were 3 studies of nasopharyngeal carcinoma, two studies of colorectal cancer and ten studies of other cancers; in the subgroup of ethnicity, 4 were carried out in European population, ten were in Asian population and one was in African population. Hospital-based controls were used in seven studies. For -137(G/C) polymorphism, there were 3 studies of nasopharyngeal carcinoma, two studies of ovarian cancer and ten studies of other cancers; -137(G/C) polymorphism was similar to -607(C/A) polymorphism in the subgroup of ethnicity. Hospital-based controls were used in eight studies. The distribution of genotypes in the controls was consistent with the Hardy–Weinberg equilibrium in all studies.
IL-18 -607(C/A) polymorphism
In the overall analysis, significant association could be observed between cancer risk and the variant genotypes of IL-18 -607(C/A) in different genetic models: in the contrast of A allele vs. C allele (OR = 1.14, 95% CI = 1.01–1.28, P heterogeneity = 0.010), the heterozygote comparison AC vs. CC (OR = 1.10, 95% CI = 1.04–1.15, P heterogeneity = 0.256) and the dominant models (AA+AC) vs. CC (OR = 1.07, 95% CI = 1.03–1.11, P heterogeneity = 0.124). In the stratified analysis by cancer type, significant association between IL-18 -607(C/A) polymorphism and nasopharyngeal carcinoma was found: in the heterozygote comparison AC vs. CC (OR = 1.12, 95% CI = 1.02–1.24, P heterogeneity = 0.703; Table 3 and Fig. 1) and the dominant models (OR = 1.09, 95% CI = 1.01–1.17, P heterogeneity = 0.928). Moreover, there was also association between IL-18 -607(C/A) polymorphism and Asian population in the contrast of A allele vs. C allele (OR = 1.19, 95% CI = 1.02–1.40, P heterogeneity = 0.006), the heterozygote comparison (OR = 1.10, 95% CI = 1.04–1.17, P heterogeneity = 0.570) and the dominant models (OR = 1.08, 95% CI = 1.03–1.12, P heterogeneity = 0.186). In the subgroup of source of control, also significant association was found (data not show).
IL-18 -137(G/C) polymorphism
In the overall analysis, no association could be observed between cancer risk and the variant genotypes of IL-18 -137(G/C) in different genetic models (data not show). In the stratified analysis by cancer type, significant association between IL-18 -137(G/C) polymorphism and nasopharyngeal carcinoma was found: in the contrast of C allele vs. G allele (OR = 1.30, 95% CI = 1.12–1.50, P heterogeneity = 0.126), in the heterozygote comparison (OR = 1.23, 95% CI = 1.05–1.43, P heterogeneity = 0.285), the homozygote comparison (OR = 1.83, 95% CI = 1.18–2.84, P heterogeneity = 0.351; Table 4 and Fig. 2), the dominant models (OR = 1.25, 95% CI = 1.09–1.43, P heterogeneity = 0.156) and the recessive models (OR = 1.67, 95% CI = 1.07–2.63, P heterogeneity = 0.439). Moreover, there was also association between IL-18 -137(G/C) polymorphism and Asian population in the heterozygote comparison (OR = 1.30, 95% CI = 1.05–1.60, P heterogeneity = 0.015). Finally, there was also significant association between IL-18 -137(G/C) polymorphism and hospital-based controls in the recessive models (OR = 1.35, 95% CI = 1.02–1.78, P heterogeneity = 0.074).
Sensitivity analysis
We used sensitivity analysis to determine whether modification of the inclusion criteria of the pooled analysis affected the final results. These were carried out by limiting the pooled analysis to the studies conforming to HWE and altering corresponding statistic variables and analysis models. Moreover, no other single study influenced the summary OR qualitatively as indicated by sensitivity analysis.
Bias diagnosis
The Begg’s funnel plot and Egger’s test were performed to access the publication bias of literatures. The shape of the funnel plots did not reveal any evidence of obvious asymmetry in the heterozygote comparison of IL-18 -607(C/A) and -137(G/C) polymorphism. Then, Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not suggest any evidence of publication bias (AC vs. CC, t = 1.66, P = 0.121; CC vs. GG, t = −0.44, P = 0.668. Figs. 3, 4).
Discussion
The overall goal of pooled analysis is to combine the results of previous studies to arrive at summary conclusions about a body of research. It is most useful in summarizing prior research when individual studies are small, and when they are individually too small to yield a valid conclusion. Taking into consideration that chronic inflammation promotes the development of cancer, a number of polymorphisms of genes encoding for pro-inflammatory cytokines have been studied and a correlation with cancer risk has been found.
To the best of our knowledge, this is the first pooled analysis to explore the association between IL-18 gene polymorphism and overall cancer risk, involving about 2,137 cases and 3,117 controls of -607(C/A) polymorphism and 2,372 cases and 3,476 controls of -137(G/C) polymorphism. We found that both IL-18 -607(C/A) and -137(G/C) polymorphisms were associated with significant increase in cancer risk. Given the potential roles of IL-18 in the immune system against tumor cells, angiogenesis, metastasis, proliferation and immune escape, the IL-18 polymorphisms may modulate the risk of cancer. Giedraitis et al. [11] and Bai et al. [12] found that the -607(C/A) variant disrupts a potential cAMP-responsive element binding protein binding site and the -137(G/C) variant changes the H4TF-1 nuclear factor binding site, which can be attributed to the IL-18 higher transcription and protein production leading to impaired IL-18 function. In our pooled analysis, we found that subjects carrying the -607A or -137C allele were associated with higher risk of cancer than those with the wild-type allele, which confirmed the hypothesis described earlier.
Our results suggested that there was an association between the -607(C/A) polymorphism and cancer risk among Asians but not among Europeans and Africans (all genetic models), as well as among nasopharyngeal carcinoma and other cancers but not among colorectal cancer. Moreover, the -137(G/C) polymorphism was associated with increased cancer risk among Asians but not among Europeans and Africans, as well as among nasopharyngeal carcinoma but not among ovarian cancer and other cancers in all genetic models. Some factors can influence these results. First, it is now widely accepted that differences in the distribution of various ethnicities between cases and controls may be a source of confounding when pooling studies. Second, cancer is a multifactorial disease that results from complex interactions between many genetic and environmental factors. This means that there will not be a single gene or single environmental factor that has large effects on cancer susceptibility [33]. Environmental factors (e.g. tobacco smoke, dietary factors, infectious agents, and radiation) add to the carcinogenic load to which humans are exposed, but exact numbers for added risk are generally less well established. Third, studies with ‘negative’ results take longer time to be published in time-lag bias, whereas enthusiastic results are published much more quickly [34]. Fourth, small studies with ‘negative’ results are never published in publication bias, whereas equally small studies with similar quality but ‘positive’ results would appear in the literature [35]. We examined these possibilities and found that ‘positive’ studies were reported more in -607A and -137C allele carriers’ studies (especially in the European and Asian studies).
In this study, we also found an association between the -607(C/A) polymorphism and cancer risk among studies using hospital-based and population-based controls, the same as the -137(G/C) polymorphism.
Some limitations of this pooled analysis should be mentioned. First of all, the numbers of published studies included in our pooled analysis were not sufficiently large for a comprehensive analysis, particularly for any given cancer (especially colorectal cancer and ovarian cancer) and ethnicity (especially African) site. Second, publication bias might have occurred and our Egger’s test results may have a substantial risk of being affected by such bias. Third, the interactions between gene–gene, gene–environment and even different polymorphic loci of the same gene may modulate cancer risk. Fourth, in some IL-18 polymorphism studies, a small number of cases and controls were included. Fifth, our pooled analysis was based on unadjusted estimates. A more precise analysis should be conducted if individual information including other covariates such as age, sex and cancer stage becomes available. In spite of these, our pooled analysis also had two advantages. First, substantial number of cases and controls were pooled from different studies, which significantly increased statistical power of the analysis. Second, the quality of case–control studies included in the current pooled analysis was satisfactory based on our selection criteria.
Conclusions
Overall, in the present our pooled analysis showed the evidence that IL-18 -607(C/A) and -137(G/C) polymorphisms were associated with increased cancer risk in Asian population and nasopharyngeal carcinoma, which supported the hypothesis that these polymorphisms may be low-penetrance susceptibility cancer biomarkers. Therefore, further well-designed large studies, particularly referring to gene–gene and gene–environment interactions are warranted. These future studies should lead to better and comprehensive understanding of the association between the IL-18 -607(C/A) and -137(G/C) polymorphisms and cancer risk.
References
Mallat Z, Henry P, Fressonnet R, Alouani S, Scoazec A, Beaufils P, et al. Increased plasma concentrations of interleukin-18 in acute coronary syndromes. Heart. 2002;88(5):467–9.
Dinarello CA. Interleukin-18 and the pathogenesis of inflammatory diseases. Semin Nephrol. 2007;27(1):98–114.
Nilkaeo A, Bhuvanath S. Role of interleukin-18 in modulation of oral carcinoma cell proliferation. Mediators Inflamm. 2006;2006(3):67120.
Okamura H, Tsutsi H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378(6552):88–91.
Park H, Byun D, Kim TS, Kim YI, Kang JS, Hahm ES, et al. Enhanced IL-18 expression in common skin tumors. Immunol Lett. 2001;79(3):215–9.
Udagawa N, Horwood NJ, Elliott J, Mackay A, Owens J, Okamura H, et al. Interleukin-18 (interferon-gamma-inducing factor) is produced by osteoblasts and acts via granulocyte/macrophage colony-stimulating factor and not via interferon-gamma to inhibit osteoclast formation. J Exp Med. 1997;185(6):1005–12.
Ye ZB, Ma T, Li H, Jin XL, Xu HM. Expression and significance of intratumoral interleukin-12 and interleukin-18 in human gastric carcinoma. World J Gastroenterol. 2007;13(11):1747–51.
Eissa SA, Zaki SA, El-Maghraby SM, Kadry DY. Importance of serum IL-18 and RANTES as markers for breast carcinoma progression. J Egypt Natl Canc Inst. 2005;17(1):51–5.
Cho D, Song H, Kim YM, Houh D, Hur DY, Park H, et al. Endogenous interleukin-18 modulates immune escape of murine melanoma cells by regulating the expression of Fas ligand and reactive oxygen intermediates. Cancer Res. 2000;60(10):2703–9.
Bubeník J. Tumour MHC class I downregulation and immunotherapy (Review). Oncol Rep. 2003;10(6):2005–8.
Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112(1–2):146–52.
Bai J, Zhang Y, Lin M, Zeng X, Wang Z, Shen J. Interleukin-18 gene polymorphisms and haplotypes in patients with oral lichen planus: a study in an ethnic Chinese cohort. Tissue Antigens. 2007;70(5):390–7.
Sáenz-López P, Carretero R, Vazquez F, Martin J, Sánchez E, Tallada M, et al. Impact of interleukin-18 polymorphisms-607 and -137 on clinical characteristics of renal cell carcinoma patients. Hum Immunol. 2010;71(3):309–13.
Samsami Dehaghani A, Shahriary K, Kashef MA, Naeimi S, Fattahi MJ, Mojtahedi Z, et al. Interleukin-18 gene promoter and serum level in women with ovarian cancer. Mol Biol Rep. 2009;36(8):2393–7.
Khalili-Azad T, Razmkhah M, Ghiam AF, Doroudchi M, Talei AR, Mojtahedi Z, et al. Association of interleukin-18 gene promoter polymorphisms with breast cancer. Neoplasma. 2009;56(1):22–5.
Farhat K, Hassen E, Bouzgarrou N, Gabbouj S, Bouaouina N, Chouchane L. Functional IL-18 promoter gene polymorphisms in Tunisian nasopharyngeal carcinoma patients. Cytokine. 2008;43(2):132–7.
Qi T, Wang Q, Zheng L, Yang HL, Bao J. Correlation of serum IL-18 level and IL-18 gene promoter polymorphisms to the risk of cervical cancer. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28(5):754–7.
Wei YS, Lan Y, Liu YG, Tang H, Tang RG, Wang JC. Interleukin-18 gene promoter polymorphisms and the risk of esophageal squamous cell carcinoma. Acta Oncol. 2007;46(8):1090–6.
Liu Y, Lin N, Huang L, Xu Q, Pang G. Genetic polymorphisms of the interleukin-18 gene and risk of prostate cancer. DNA Cell Biol. 2007;26(8):613–8.
Pratesi C, Bortolin MT, Bidoli E, Tedeschi R, Vaccher E, Dolcetti R, et al. Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother. 2006;55(1):23–30.
Farjadfar A, Mojtahedi Z, Ghayumi MA, Erfani N, Haghshenas MR, Ghaderi A. Interleukin-18 promoter polymorphism is associated with lung cancer: a case-control study. Acta Oncol. 2009;48(7):971–6.
Nong LG, Luo B, Zhang L, Nong HB. Interleukin-18 gene promoter polymorphism and the risk of nasopharyngeal carcinoma in a Chinese population. DNA Cell Biol. 2009;28(10):507–13.
Haghshenas MR, Hosseini SV, Mahmoudi M, Saberi-Firozi M, Farjadian S, Ghaderi A. IL-18 serum level and IL-18 promoter gene polymorphism in Iranian patients with gastrointestinal cancers. J Gastroenterol Hepatol. 2009;24(6):1119–22.
Asefi V, Mojtahedi Z, Khademi B, Naeimi S, Ghaderi A. Head and neck squamous cell carcinoma is not associated with interleukin-18 promoter gene polymorphisms: a case-control study. J Laryngol Otol. 2009;123(4):444–8.
Nikiteas N, Yannopoulos A, Chatzitheofylaktou A, Tsigris C. Heterozygosity for interleukin-18–607 A/C polymorphism is associated with risk for colorectal cancer. Anticancer Res. 2007;27(6B):3849–53.
Vairaktaris E, Serefoglou ZC, Yapijakis C, Agapi C, Vassiliou S, Nkenke E, et al. The interleukin-18–607A/C polymorphism is not associated with risk for oral cancer. Anticancer Res. 2007;27(6B):4011–4.
Andrie E, Michos A, Kalampoki V, Pourtsidis A, Moschovi M, Polychronopoulou S. Genetic variants in immunoregulatory genes and risk for childhood lymphomas. Eur J Haematol. 2009;83(4):334–42.
Bushley AW, Ferrell R, McDuffie K, Terada KY, Carney ME, Thompson PJ, et al. Polymorphisms of interleukin (IL)-1alpha, IL-1beta, IL-6, IL-10, and IL-18 and the risk of ovarian cancer. Gynecol Oncol. 2004;95(3):672–9.
Sobti RC, Shekari M, Tamandani DM, Malekzadeh K, Suri V. Association of interleukin-18 gene promoter polymorphism on the risk of cervix carcinogenesis in north Indian population. Oncol Res. 2008;17(4):159–66.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in metaanalysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–60.
Ioannidis JP. Effect of the statistical significance of results on the time to completion and publication of randomized efficacy trials. JAMA. 1998;279(4):281–6.
Dickersin K, Min YI, Meinert CL. Factors influencing publication of research results. Follow-up of applications submitted to two institutional review boards. JAMA. 1992;267(3):374–8.
Acknowledgments
This study was supported by the program of key medical department of Jiangsu Province. Department of urology of Jiangsu Province Hospital (XK17 200904).
Author information
Authors and Affiliations
Corresponding authors
Additional information
The authors Yuan-Yuan Mi, Qian-Qian Yu, and Meng-Lei Yu contributed equally to this work and each is considered as first author.
Rights and permissions
About this article
Cite this article
Mi, YY., Yu, QQ., Yu, ML. et al. Review and pooled analysis of studies on -607(C/A) and -137(G/C) polymorphisms in IL-18 and cancer risk. Med Oncol 28, 1107–1115 (2011). https://doi.org/10.1007/s12032-010-9569-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12032-010-9569-1