Abstract
Aim
To evaluate the possible relevance of the IL-18-137 G>C (rs187238), IL-18-607 C>A (rs1946518) and IL-4-590 C>T (rs2243250) polymorphisms to the genetic susceptibility of head and neck cancer.
Methods
Data were retrieved from PubMed, EMBASE, Web of Science and CNKI databases, and the results were independently analysed by two reviewers using Stata 14.0 software.
Results
After searching for and assessing the literature, a total of thirteen studies involving 2,959 patients newly diagnosed as head and neck cancer and 3,622 controls from healthy donors were analysed. The results suggested that a strong relationship between patients and healthy controls was observed in the IL-18-137 G>C polymorphism in consistence with the result (CC vs. GG + GC: OR = 1.63, P = 0.004; CC vs. GG: OR = 1.82, P = 0.001). When stratified by cancer type, ethnicity and the source of control samples, significant and elevated risks were obtained in the genetic susceptibility to Asian patients with NPC in all genetic models and in those studies using the PCR-RFLP test method. In addition, comparable results were obtained for the IL-18-607 C>A polymorphism, especially for Asian patients with NPC.
Conclusions
It should be a potential association between IL-18 variants and nasopharyngeal carcinoma. Furthermore, IL-18 gene variants might be considered as a critical role in predicting the occurrence of nasopharyngeal carcinoma in Asian population. However, the IL-4-590 C>T polymorphism does not influence the development of head and neck cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the sixth most common malignancy in the world, head and neck cancer (HNC) occurs in epithelial tissues of various origins consisting of the oral and maxillofacial cavities, pharynx and larynx [1, 2]. Among the different types of HNC, oral cancer and nasopharyngeal carcinoma (NPC) are the most common [3, 4]. More than 90% of these malignancies are squamous cell carcinoma (SCC) [5]. HNC has become a serious global health problem, with estimated more than 550,000 new cases and 300,000 deaths every year [6]. In 2015, there were 106,540 newly diagnosed cases of HNC and 11,710 expected related deaths in the USA alone [7]. Epidemiological evidence indicates that the overall 5-year survival of HNC patients has not significantly improved in the past decade [8]. Pathogenetic mechanisms of head and neck organs are involved into a multifactorial process arising from genetic alterations in oncogenes and tumour suppressor genes as well as due to the interaction of environmental factors such as smoking, alcohol intake and betel quid chewing, as well as lack of oral hygiene. According to previous studies, HPV infection has also emerged as a risk factor for the transformation of normal tissues into malignant lesions [9,10,11]. Recently, chronic inflammation stimulated by exposure to chemical, bacterial and viral agents has been proved that it has an important effect on the development and progression of cancer. Immune dysfunction induced by certain autoimmune reactions, also enhances the occurrence of various cancers, including head and neck cancer [12,13,14,15,16]. Cytokines are low-molecular-weight regulatory polypeptides of the immune system, which accumulate in the immune microenvironment. Functional single nucleotide polymorphisms (SNPs) on cytokine-encoding genes can strongly induce the proliferation of malignant cell by immune system disorders and enhance the capability of malignant transformation and tumour growth [12,13,14,15,16]. SNPs on the gene encoding the pro-inflammatory interleukins IL-18 are located on chromosome 11q22 [17], which induce alterations in its promoter region (-607 C>A, -137 G>C), and these genetic polymorphisms are responsible for the progression of some malignancies, such as esophageal squamous cell carcinoma [18]. The anti-inflammatory cytokines IL-4 is associated with humoral immune responses and induces a potent cytotoxic response against tumors, which is secreted by Th2 cells [19, 20]. It is also an autocrine growth factor, responsible for the induction of IgE production by B cell, and it can antagonize the function of IFN-γ and inhibit the activation of macrophages [21]. A key SNP on the promoter region of the IL-4 gene (-590 C>T), referred to as rs2243250 [22], is relevant to cancer. On the other hand, increased serum levels of IL-18 have been tested in cancers of various origins (colon, gastrointestinal tract and breast carcinomas) [21, 23,24,25]. To date, there have been a few studies investigating the association of IL-18-137 G>C, IL-18-607 C>A and IL-4-590 C>T polymorphisms with HNC risk, but the results have been inconsistent. Until now, there has been no specific meta-analysis or systematic review on the risk of HNC in relation to polymorphisms in IL-18 nor IL-4 polymorphism. Given that, we propose to perform a meta-analysis to better understand the relationship between HNC risk and SNPs on the IL-18 and IL-4 genes.
Methods
Search strategy
A search strategy based on combinations of the key words such as: “IL-18” or “interleukin-18”; “IL-4”, or “interleukin-4”; “polymorphism” or “variant”; “head and neck cancer” and “oral cancer”, “nasopharyngeal cancer”, “pharynx cancer”, or “larynx cancer”, was applied on all studies selected before June 2017, using PubMed, EMBASE, Web of Science and CNKI databases without language restriction. Combined phrases with information on specific genes or SNPs from studies were also used. References cited in selected original studies and previous meta-analyses and review articles were scanned to avoid missing to be included in.
Inclusion criteria
The inclusion criteria were as follows: (a) case–control study design, (b) relevance of polymorphisms in IL-18 and IL-4 to head and neck cancer risk/susceptibility, and (c) sufficient genotype data or data allowed to be calculated.
Exclusion criteria
The exclusion criteria were as follows: (a) review articles, (b) absence of the proposed SNPs or completed data on genotypes, (c) repeated publications by the same author or team, (d) studies based on animals or cell lines, and (e) unreliable and unreasonable materials and methods.
Data extraction
The extracted data included: the first author’s name (first name for record), year of publication, country, ethnicity (Asian or Caucasian), sources of controls, cancer types, genotyping methods, allele counts in HNC cases and controls, p for the Hardy–Weinberg equilibrium (HWE) using Fisher’s exact test, and Newcastle-Ottawa Scale (NOS) scores. Importantly, some details about patients, such as stage classification of cancer were also obtained. All the information was collected by two investigators independently. A third reviewer was required (Li) when the results were inconsistent.
Quality assessment
All the information was collected by two investigators independently. A third reviewer was required (Li) when the results were inconsistent. NOS scores were used to assess the quality of individual case–control studies. In brief, assessment scores ranged from 0 points (worst) to 9 points (best). A final score > 6 was considered as high quality.
Statistical analysis
The potential association of the three functional SNPs with head and neck cancer risk was assessed by odds ratio (OR) with 95% confidence interval (95% CI). P < 0.05 was defined significant. For rs1946518 polymorphism, the pooled ORs were calculated for the allele model (A vs. C), dominant model (CA + AA vs. CC), recessive model (AA vs. CC + CA) and codominant model (CA vs. CC, AA vs. CC). The same genetic models were also applied in rs187238 and rs2243250 polymorphisms. Necessarily, subgroup analyses of ethnicity, cancer types, source of controls and genotyping methods were also performed and statistically evaluated. Cochran’s Q statistic and I 2 method was applied in heterogeneity [26]. I 2 = 0–50% meant no heterogeneity and I 2 = 50–75% meant moderate heterogeneity, and I 2 > 75% suggested high heterogeneity. The fixed-effect model (also called the Mantel–Haenszel method) was chosen for meta-analysis only when the heterogeneity tests yielded significant results (p > 0.1 or I 2 < 40%) [27]. Otherwise, the random-effects model (also called the DerSimonian and Laird method) [28] was selected. All analyses were conducted using Stata 14.0 (Stata Corporation, College Station, TX, USA).
Results
Studies characteristics
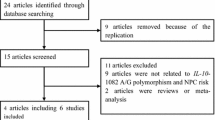
The flowchart of the search process is presented in Fig. 1 and followed the method described in previous publications [29, 30]. A total of 204 studies were retrieved through databases searches using several combinations of keywords. After removing repeated records, an additional 130 studies were excluded for irrelevant title. Then, two papers of meta-analyses, one of case-only study and one lacking of specific data, were excluded. The remaining 14 full-text articles got assessed further for eligibility. Among the excluded studies, the data derived from two different studies had been pooled into one manuscript. Finally, thirteen studies satisfied the inclusion criteria of this analysis, being the basis of three independent study panels (Table 1). Of those, eight studies on IL-18-137 G>C polymorphism [29,30,31,32,33,34,35,36], nine studies on IL-18-607 C>A polymorphism [29,30,31,32,33,34,35,36,37], and four studies on IL-4-590 C>T polymorphism [38,39,40,41], respectively. Nine studies involved Asian populations [29,30,31, 33, 35, 36, 38, 39, 41], and four studies involved Caucasian populations [32, 34, 37, 40]. Regarding cancer type, five studies reported on nasopharyngeal carcinoma [29, 30, 32,33,34], seven reports on oral cancer/ oral squamous cell carcinoma (OSCC) [35,36,37,38,39,40,41], and one reported on HNC [31] and pharyngeal squamous cell carcinoma (PSCC) [41]. In terms of the genotyping method, seven studies adopted polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [23, 29, 30, 32, 33, 37, 38, 40], two studies adopted the TaqMan assay (Applied Biosystems) [36, 41], two studies used PCR [35, 39], and two studies used alleles specific-PCR (AS-PCR) [31, 34]. Only three studies deviated from HWE in the three SNPs, respectively.
Meta-analysis
After reviewing the literature, a total of 2959 patients of HNC and 3,622 controls from healthy population were incorporated into this meta-analysis. The pooled counts of three genotypes of the proposed SNP variants are shown in Table 2.
Association between rs187238 polymorphism and HNC risk
With regard to rs187238 polymorphism, eight studies with a total of 1,792 patients and 1,900 cancer-free controls were analysed. Significant results were found in the following genetic models: CC vs. GG + GC: OR = 1.63, P = 0.004; CC vs. GG: OR = 1.82, P = 0.001, without heterogeneity (Table 2). However, none of other three models yielded significant association (all P > 0.05). And heterogeneities existed in other three genotype models, after they were analysed based on subgroups, the differences in ethnicity and cancer type may be responsible for these heterogeneities. In addition, significant and elevated risks were obtained in the genetic susceptibility to Asian patients with NPC in all genetic models and in those studies using the PCR-RFLP test method, but not in Caucasian patients with any types of HNC (Dominant model: OR = 1.89, P = 0.000; Recessive model: OR = 2.07, P = 0.004; Allele model: OR = 1.84, P = 0.000; GC vs. GG: OR = 1.79, P = 0.000; CC vs. GG: OR = 2.50, P = 0.000) (Table 2; Fig. 2).
Association of rs1946518 polymorphism with head and neck cancer risks
The results of the analysis of rs1946518 polymorphism presented as forest plots provided obvious evidence of association between the SNP and head and neck cancer susceptibility between cases and controls (CA + AA vs. CC: OR 1.18, 95% CI 1.02–1.36, P = 0.022; A vs. C: OR 1.10, 95% CI 1.00–1.20, P = 0.043 and CA vs. CC: OR 1.18, 95% CI 1.01–1.37, P = 0.032) (Table 2). Overall, the data from 1941 cases and 1989 controls were analysed. No heterogeneity was observed among the studies in any of the genetic models (Table 2). The pooled ORs were used in a fixed effects model. After stratified analysis, IL-18-607 C>A was found to be relevant to increased risk in only Asian populations with NPC for three models (CA + AA vs. CC: OR 1.36, P = 0.021; AA vs. CC: OR 1.39, P = 0.045 and CA vs. CC: OR 1.36, P = 0.033), but without any replication in Caucasian patients nor oral cancer (Table 2; Fig. 3).
No difference between rs2243250 polymorphism and head and neck cancer risk
For rs2243250 polymorphism, five studies with 1018 patients and 1633 healthy donors were included. Heterogeneities existed in all genotype models. ORs were calculated using a random effects model. No relevance was found in IL-4-590 C>T susceptibility to HNC in any genotypes. In terms of the analyses based on subgroups, the differences in genotyping method may be helpful to explain these heterogeneities, and there was no any association obtained under any genetic model (all P > 0.05) (Table 2).
Sensitivity analysis
The estimated pooled OR change slightly showed in sensitivity analysis, which indicated that our results were statistically stable, by removing one study at a time from the analysis (Figs. 4, 5).
Publication bias
Begg’s test and Egger’s test (figures not shown) were performed to assess the publication bias among overall selected studies. As a result, there was no obvious evidence of publication bias in IL-18 and IL-4-590 gene polymorphisms under the dominant model (P > 0.05) (Figs. 6, 7).
Discussion
IL-18 and IL-4 levels have been found to be significantly increased in HNSCC cell lines [19, 42]. Recent reports confer that IL-4 may achieve the ability in inducing the growth of HNSCC cell lines through a paracrine mechanism [43]. Similar results have also been found in ovarian carcinomas, prostate cancer, melanoma and various types of lymphomas [22, 43,44,45,46]. The number of T cells expression by IL-4 in oral cancer patients is significantly higher than that in normal controls [23, 47]. Evidence has been shown that IL-4 could directly increase the proliferation rate of HNSCC cell lines [19]. There was a slight increase observed in the frequency of the T allele in oral cancer patients, compared to controls in a study on Chinese populations. Increased levels of IL-18 have been observed in many cancers, such as esophageal, colon, and skin carcinoma [48, 49]. A previous study has suggested that the overexpression of IL-18 could downregulate cyclin D1 expression and affect the caspase-dependent cell death, which decreased cell viability and induced apoptosis, respectively, in human tongue SCC cells [50]. However, the mechanisms of how IL-18-137 G>C, IL-18-607 C>A and IL-4 -590 C>T mutations influence the risk of HNC and the extent to which the variants are responsible for HNC risk are still unclear. Previous studies have shown that the specific associations between IL-18-137 G>C, IL-18-607 C>A and IL-4-590 C>T polymorphisms and HNC risk are controversial. Therefore, it is worth conducting a meta-analysis from all available studies to figure out these specific associations. In the present meta-analysis, thirteen case–control studies were involved, consisting of 2959 patients and 3622 healthy donors. There was a slight association between IL-18-137 G>C and HNC risk in some genetic models. Interestingly, in the stratified analyses, a significant increase of risk was found in Asian patients diagnosed with NPC and assessed with the PCR-RFLP method in overall genetic models. Furthermore, for the IL-18-607 C>A polymorphism, the results indicated that a high frequency of the A variant was found in HNC patients, indicating that the genetic variant could have an effect on the progression of head and neck cancer by influencing the maturation of IL-18 or by altering the interaction of IL-18-607 with its corresponding genes or targets. Moreover, the subgroup analyses showed a significant increase in the risk to Asian patients with nasopharyngeal carcinoma. In general, we found strong associations between SNPs on the IL-18 gene and head and neck cancer, especially for nasopharyngeal carcinoma in Asian population. In contrast, there was no any significant association of HNC risks with the IL-4 -590 C>T polymorphism.
To the best of our knowledge, this is the most comprehensive quantitative assessment of polymorphisms in these two key cytokines, related to inflammation that is specifically focused on their functions in the pathogenetic process of head and neck cancer. It is noteworthy that there are certain limitations of our analyses. First, heterogeneities exist in some genetic models. It is known that heterogeneity in a systematic review refers to the variability among studies [51]. In our study, differences in factors such as ethnicity, cancer type, cancer stage, age and gender among the selected studies could have resulted in heterogeneity. Meanwhile, the lack of unified methods to collect and analyze samples could change these results. Second, small-study effects are unavoidable in such a meta-analysis with a small sample size and a limited number of papers. Third, we could not determine the interaction between the environmental factors and genetic mutation and distribution or disease stage, age and gender. Nevertheless, we believe that these quantitative results can provide some evidence on how these key variants in cytokines play roles in the pathogenesis and progression of head and neck cancer.
In conclusion, the current data reveal that rs187238 and rs1946518 polymorphisms are more likely to represent potentially valuable genetic biomarkers that are related to indicate the susceptibility to nasopharyngeal carcinoma. Furthermore, IL-18 gene variants may serve as important biomarkers in predicting the occurrence of nasopharyngeal carcinoma in Asian population. However, the IL-4-590 C>T (rs2243250) polymorphism does not influence the development of head and neck cancer. Nevertheless, more high-quality studies with completed data, including multiple populations from different ethnic backgrounds and more efficient, strict and unified genotyping methods are required for further evaluation.
References
Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet 371:1695–1709
Warnakulasuriya S (2009) Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 45:309–316
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65:87–108
Pai SI, Westra WH (2009) Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu Rev Pathol 4:49–70
Saba NF, Goodman M, Ward K, Flowers C, Ramalingam S, Owonikoko T et al (2011) Gender and ethnic disparities in incidence and survival of squamous cell carcinoma of the oral tongue, base of tongue, and tonsils: a surveillance, epidemiology and end results program-based analysis. Oncology 81:12–20
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67:7–30
Bell RB, Kademani D, Homer L, Dierks EJ, Potter BE (2007) Tongue cancer: Is there a difference in survival compared with other subsites in the oral cavity? J Oral Maxillofac Surg 65:229–236
Alnuaimi AD, Wiesenfeld D, O’Brien-Simpson NM, Reynolds EC, McCullough MJ (2015) Oral Candida colonization in oral cancer patients and its relationship with traditional risk factors of oral cancer: a matched case-control study. Oral Oncol 51:139–145
Gupta B, Johnson NW (2014) Systematic review and meta-analysis of association of smokeless tobacco and of betel quid without tobacco with incidence of oral cancer in South Asia and the Pacific. PLoS One 9:e113385
Young D, Xiao CC, Murphy B, Moore M, Fakhry C, Day TA (2015) Increase in head and neck cancer in younger patients due to human papillomavirus (HPV). Oral Oncol 51:727–730
Jewett A, Head C, Cacalano NA (2006) Emerging mechanisms of immunosuppression in oral cancers. J Dent Res 85:1061–1073
Lalla RV, Tanzer ML, Kreutzer DL (2003) Identification of a region of the fibrin molecule involved in upregulation of interleukin-8 expression from human oral squamous cell carcinoma cells. Arch Oral Biol 48:263–271
Min R, Zun Z, Siyi L, Wenjun Y, Lizheng W, Chenping Z (2011) Increased expression of Toll-like receptor-9 has close relation with tumour cell proliferation in oral squamous cell carcinoma. Arch Oral Biol 56:877–884
Wei H, Hongya P, Linlin J, Mujiang A, Kuijie W, Duohong Z et al (2011) IFN-gamma enhances the anti-tumour immune response of dendritic cells against oral squamous cell carcinoma. Arch Oral Biol 56:891–898
Young MR, Wright MA, Lozano Y, Matthews JP, Benefield J, Prechel MM (1996) Mechanisms of immune suppression in patients with head and neck cancer: influence on the immune infiltrate of the cancer. Int J Cancer 67:333–338
Giedraitis V, He B, Huang WX, Hillert J (2001) Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol 112:146–152
Wei YS, Lan Y, Liu YG, Tang H, Tang RG, Wang JC (2007) Interleukin-18 gene promoter polymorphisms and the risk of esophageal squamous cell carcinoma. Acta Oncol 46:1090–1096
Myers JN, Yasumura S, Suminami Y, Hirabayashi H, Lin W, Johnson JT et al (1996) Growth stimulation of human head and neck squamous cell carcinoma cell lines by interleukin 4. Clin Cancer Res 2:127–135
Olver S, Apte S, Baz A, Kienzle N (2007) The duplicitous effects of interleukin 4 on tumour immunity: how can the same cytokine improve or impair control of tumour growth? Tissue Antigens 69:293–298
Toi M, Bicknell R, Harris AL (1992) Inhibition of colon and breast carcinoma cell growth by interleukin-4. Cancer Res 52:275–279
Le Beau MM, Lemons RS, Espinosa R 3rd, Larson RA, Arai N, Rowley JD (1989) Interleukin-4 and interleukin-5 map to human chromosome 5 in a region encoding growth factors and receptors and are deleted in myeloid leukemias with a del(5q). Blood 73:647–650
Agarwal A, Rani M, Saha GK, Valarmathi TM, Bahadur S, Mohanti BK et al (2003) Disregulated expression of the Th2 cytokine gene in patients with intraoral squamous cell carcinoma. Immunol Invest 32:17–30
Lissoni P, Brivio F, Rovelli F, Fumagalli G, Malugani F, Vaghi M et al (2000) Serum concentrations of interleukin-18 in early and advanced cancer patients: enhanced secretion in metastatic disease. J Biol Regul Homeost Agents 14:275–277
Ye ZB, Ma T, Li H, Jin XL, Xu HM (2007) Expression and significance of intratumoral interleukin-12 and interleukin-18 in human gastric carcinoma. World J Gastroenterol 13:1747–1751
Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J (2006) Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 11:193–206
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
Curtin F (2017) Meta-analysis combining parallel and cross-over trials with random effects. Res Synth Methods 8:263–274
DuBing ZJ, WeiYE-Sheng (2012) Interleukin-18 gene genetic polymorphisms and risk of nasopharyngeal carcinomar in Han population from Sichuan China. Med J West China 24(9)
Nong LG, Luo B, Zhang L, Nong HB. Interleukin-18 gene promoter polymorphism and the risk of nasopharyngeal carcinoma in a Chinese population
Asefi V, Mojtahedi Z, Khademi B, Naeimi S, Ghaderi A (2009) Head and neck squamous cell carcinoma is not associated with interleukin-18 promoter gene polymorphisms: a case-control study. J Laryngol Otol 123:444–448
Farhat K, Hassen E, Bouzgarrou N, Gabbouj S, Bouaouina N, Chouchane L (2008) Functional IL-18 promoter gene polymorphisms in Tunisian nasopharyngeal carcinoma patients. Cytokine 43:132–137
Lina PGLBTYL (2013) Research on interleukin-18 gene promoter polymorphisms and genetic susceptibility of nasopharyngeal carcinoma. Laboratory Medicine, June 2013, vol 28, no 6
Pratesi C, Bortolin MT, Bidoli E, Tedeschi R, Vaccher E, Dolcetti R et al (2006) Interleukin-10 and interleukin-18 promoter polymorphisms in an Italian cohort of patients with undifferentiated carcinoma of nasopharyngeal type. Cancer Immunol Immunother 55:23–30
Singh PK, Ahmad MK, Kumar V, Hussain SR, Gupta R, Jain A et al (2014) Effects of interleukin-18 promoter (C607A and G137C) gene polymorphisms and their association with oral squamous cell carcinoma (OSCC) in northern India. Tumour Biol 35:12275–12284
Tsai HT, Hsin CH, Hsieh YH, Tang CH, Yang SF, Lin CW et al (2013) Impact of interleukin-18 polymorphisms -607A/C and -137G/C on oral cancer occurrence and clinical progression. PLoS One 8:e83572
Vairaktaris E, Serefoglou ZC, Yapijakis C, Agapi C, Vassiliou S, Nkenke E et al (2007) The interleukin-18-607A/C polymorphism is not associated with risk for oral cancer. Anticancer Res 27:4011–4014
Gaur P, Mittal M, Mohanti B, Das S (2011) Functional variants of IL4 and IL6 genes and risk of tobacco-related oral carcinoma in high-risk Asian Indians. Oral Dis 17:720–726
Tsai MH, Chen WC, Tsai CH, Hang LW, Tsai FJ (2005) Interleukin-4 gene, but not the interleukin-1 beta gene polymorphism, is associated with oral cancer. J Clin Lab Anal 19:93–98
Vairaktaris E, Yapijakis C, Serefoglou Z, Avgoustidis D, Critselis E, Spyridonidou S et al (2008) Gene expression polymorphisms of interleukins-1 beta, -4, -6, -8, -10, and tumor necrosis factors-alpha, -beta: regression analysis of their effect upon oral squamous cell carcinoma. J Cancer Res Clin Oncol 134:821–832
Yang CM, Chen HC, Hou YY, Lee MC, Liou HH, Huang SJ et al (2014) A high IL-4 production diplotype is associated with an increased risk but better prognosis of oral and pharyngeal carcinomas. Arch Oral Biol 59:35–46
Riedel F, Adam S, Feick P, Haas S, Gotte K, Hormann K et al (2004) Expression of IL-18 in patients with head and neck squamous cell carcinoma. Int J Mol Med 13:267–272
Obiri NI, Siegel JP, Varricchio F, Puri RK (1994) Expression of high-affinity IL-4 receptors on human melanoma, ovarian and breast carcinoma cells. Clin Exp Immunol 95:148–155
Lan Q, Zheng T, Rothman N, Zhang Y, Wang SS, Shen M et al (2006) Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood 107:4101–4108
Mainou-Fowler T, Proctor SJ, Taylor PR (2004) Interleukin 4 production by peripheral blood lymphocytes in patients with classical Hodgkin lymphoma. Leuk Res 28:159–166
Takeshi U, Sadar MD, Suzuki H, Akakura K, Sakamoto S, Shimbo M et al (2005) Interleukin-4 in patients with prostate cancer. Anticancer Res 25:4595–4598
Manchanda P, Sharma SC, Das SN (2006) Differential regulation of IL-2 and IL-4 in patients with tobacco-related oral squamous cell carcinoma. Oral Dis 12:455–462
Diakowska D, Markocka-Maczka K, Grabowski K, Lewandowski A (2006) Serum interleukin-12 and interleukin-18 levels in patients with oesophageal squamous cell carcinoma. Exp Oncol 28:319–322
Wen Z, Ouyang Q, Chen D, Su X (2003) [Interleukin 18 expression in colon cancer and adenoma]. Sichuan Da Xue Xue Bao Yi Xue Ban 34:262–264
Liu W, Han B, Sun B, Gao Y, Huang Y, Hu M (2012) Overexpression of interleukin-18 induces growth inhibition, apoptosis and gene expression changes in a human tongue squamous cell carcinoma cell line. J Int Med Res 40:537–544
Moreno SG, Sutton AJ, Thompson JR, Ades AE, Abrams KR, Cooper NJ (2012) A generalized weighting regression-derived meta-analysis estimator robust to small-study effects and heterogeneity. Stat Med 31:1407–1417
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by the Chongqing Municipal Key Laboratory of Oral Biomedical Engineering of Higher Education and the Program for Innovation Team Building at Institutions of Higher Education in Chongqing in 2016, NO CXTDG201602006.
Conflict of interest
We declare that we have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Xiao, TT., Li, X., Xu, Y. et al. Significant association of the cytokine variants with head and neck cancer risk: evidence from meta-analysis. Eur Arch Otorhinolaryngol 275, 483–496 (2018). https://doi.org/10.1007/s00405-017-4820-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-017-4820-4