Abstract
Glioblastoma multiforme (GBM) is one of the most lethal malignancies of the central nervous system characterized by high mortality rate. The complexity of GBM pathogenesis, progression, and prognosis is not fully understood yet. GBM-derived extracellular vesicles (EVs) carry several oncogenic elements that facilitate GBM progression. The purpose of this study was to identify systems level molecular signatures from GBM-derived EVs using integrative analysis of publicly available transcriptomic data generated from plasma and serum samples. The dataset contained 19 samples in total, of which 15 samples were from plasma (11 GBM patients and 4 healthy samples) and 4 samples were from serum (2 GBM and 2 healthy samples). We carried out statistical analysis to identify differentially expressed genes (DEGs), functional enrichment analysis of the DEGs, protein–protein interaction networks, module analysis, transcription factors and target gene regulatory networks analysis, and identification of hub genes. The differential expression of the identified hub genes were validated with the independent TCGA-GBM dataset. We have identified a few crucial genes and pathways associated with GBM prognosis and therapy resistance. The DEGs identified from plasma were associated with inflammatory processes and viral infection. On the other hand, the hub genes identified from the serum samples were significantly associated with protein ubiquitinylation processes and cytokine signaling regulation. The findings indicate that GBM-derived plasma and serum DEGs may be associated with distinct cellular processes and pathways which facilitate GBM progression. The findings will provide better understanding of the molecular mechanisms of GBM pathogenesis and progression. These results can further be utilized for developing and validating minimally invasive diagnostic and therapeutic molecular biomarkers for GBM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is a lethal disease and one of the most frequent malignancies of the central nervous system in adults. GBM is associated with poor prognosis and high mortality rate with the majority of the patients dying within 1 year of diagnosis (Ohgaki and Kleihues 2007; LOBAMRDI and ASSEM 2017). The overall median survival of GBM patients is approximately 14 months even after the aggressive surgical resection followed by standard regimens of chemotherapy and radiotherapy. The recent clinical practices for molecular characterization of disease status in GBM patients are feasible with tissue specimens obtained from biopsy and open surgery. However, both the approaches mentioned are invasive measures to assess the underlying pathogenesis in GBM. The standard management of GBM patients is also limited due to the lack of efficient monitoring system and therapeutic approaches such as MRI which cannot detect small tumors. Therefore, there is an urgent need to develop standard diagnostic approaches that would impart timely evaluation of disease status and therapy response in GBM patients (Santiago-Dieppa et al. 2014; Osti et al. 2019).
The extracellular vesicles (EVs) play an important role in clinical research purpose as diagnostic or prognostic biomarker in GBM progression and clinical treatment. EVs are mainly involved in the mechanism of intracellular communication and secrete DNA, mRNA, proteins, and membrane receptors to the target cells. Recent studies revealed that GBM cells release EVs that contains tumor-specific RNAs and proteins which can be detected in the systemic blood circulation of GBM patients. EVs have been suggested to function in normal development of nervous system and neuron growth (Raposo and Stoorvogel 2013; Xu et al. 2018). EVs are known to be involved in wide array of pathological processes that are principle to cancer development such as angiogenesis, tumor invasion, cell proliferation, and chemoresistance. There is increasing evidences to show differential cargo content between glioma-derived EVs and EVs that are secreted by normal glial cells. The glioma-derived EVs contain several oncogenic factors which they release to neighboring cells through signal transduction and facilitate glioma progression (Basu and Ghosh 2019; Osti et al. 2019). Recent study reported that GBM patient tends to show increased concentration of EVs compared to that of healthy controls. This indicated that blood-based EVs can act as a potential biomarker for GBM diagnosis, identifying molecular markers and evaluating therapy resistance (Lane et al. 2019). The EVs from serum, plasma, and cerebrospinal fluid (CSF) provide important diagnostic and prognostic information. Cancer cell–specific mRNA and miRNA can be detected from EVs derived from blood and CSF (Whitehead et al. 2020). CSF has direct connection with the brain tumor cells. Patients suffering from GBM and other neurological disorders show increased concentration of CSF proteins and other tumor-specific cells. Sequence analysis of CSF based EVs can be used to detect genomic alteration in GBM. However, the isolation of CSF is a complex and painful procedure compared to the isolation of serum and plasma from blood (Saugstad et al. 2017; Duan et al. 2020). Hence, transcriptomic analysis of GBM-derived EVs from serum and plasma bear potential to identify non-invasive diagnostic and prognostic biomarkers for GBM.
In this study, the GBM-derived EVs (plasma and serum) RNA-seq dataset were selected for integrative analysis. We performed functional enrichment, protein–protein interaction network analysis and module analysis of these DEGs for identification of hub genes in plasma and serum samples. Subsequently, the expression levels of the hub genes were validated against the independent TCGA-GBM dataset.
Materials and Methodology
Data Retrieval
The experimental design of the selected dataset (GSE106804) involves GBM derived extracellular vesicles isolated from both serum and plasma. The sequencing of GBM-derived EVs was carried out at the Broad Institute of Harvard and MIT, and the gene expression profiles were generated using Illumina Hiseq2000 platform (Reátegui et al. 2018). The selected dataset was retrieved and downloaded from the NCBI-GEO (https://www.ncbi.nlm.nih.gov/gds) in SRA format. The dataset included 19 samples in total, of which 15 samples were from plasma (11 GBM patients and 4 healthy samples) and 4 samples were from serum (2 GBM and 2 healthy samples).
Analysis of the RNA-Seq Data
The retrieved dataset for GBM-derived EVs-RNA from both plasma and serum samples were converted to fastq format using fastq-dump. The raw reads in FASTQ format were preprocessed through in-house RNA seq analysis pipeline. The preprocessed high-quality reads after quality control were mapped using Hisat2 against the ensemble reference human genome Homo sapiens (GRCh38) (Kim et al. 2015). The mapped reads were then quantified at the feature (gene) level to obtain the count data for each gene. We have used DESeq2 package (Love et al. 2014) for identification of DEGs. The dataset for both serum and plasma were transformed separately in DESeq2 using variance stabilizing transformation. Finally, the list of significant DEGs for both plasma and serum samples were selected by considering p-adjusted value < 0.05.
Functional Annotation and Pathway Enrichment Analysis
For better understanding of the biological role of significant DEGs in GBM-derived plasma and serum samples, functional annotation and pathway enrichment analysis were performed using DAVID tool (Dennis et al. 2003). DAVID provides statistically significant functional annotations for a given set of gene lists. For our study, we carried out gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis using DAVID online tool. A p- value < 0.05 was considered as statistically significant.
Construction of PPI Networks and Identification of Hub Genes
Protein–protein interaction (PPI) networks were constructed using the STRING database with DEGs identified from the both plasma and serum (Szklarczyk et al. 2019). Cytoscape version 3.7.2 was used to analyze the networks (Paul Shannon et al. 1971). The Molecular Complex Detection (MCODE) algorithm implemented within cytoscape software was used to detect tightly connected module (Pruitt et al. 2001). MCODE scores greater than or equal to 4 and the number of nodes > 4 were set as cutoff criteria with the default parameters (degree cutoff ≥ 2, node score cutoff ≥ 2, K-core ≥ 2, and max depth = 100). CytoHubba, another cytoscape plugin (Chin et al. 2014) was used to detect hub genes in the PPI network topology. The CytoHubba uses an ensemble approach by employing five calculation methods: MNC (maximum neighborhood component), degree, edge percolated component (EPC), eccentricity (EcC), and betweeness. The top ranked genes identified using these five algorithms were further intersected to identify consensus hub genes from plasma and serum networks.
TFs-hub Gene Regulatory Networks
We have carried out the transcription factors (TFs) and hub gene network analysis to identify the TFs that were associated with the hub gene from plasma and serum PPI networks. To perform the TFs to hub gene interaction analysis, we have used the JASPAR database (https://jaspar.genereg.net/) which stores manually curated and non-redundant TF-binding profiles as position frequency matrices (PFMs). The hub TFs were identified from the JASPAR database, and regulatory network visualizations were done using NeworkAnalyst tool (Zhou et al. 2019).
Independent Validation and Clinical Significance of the Identified Hub Genes
GEPIA database was used to validate the expression of hub genes identified from PPI network and module analysis. The Gene Expression Profiling Interactive Analysis (GEPIA) database (Tang et al. 2017) is a Web-based tool that provides rapid and customizable functionalities based on The Cancer Genome Atlas (TCGA) and GTEx data. We considered p < 0.01 and fold change > 2 as a threshold. Additionally, to verify the genetic alterations associated with these hub genes, cBioPortal (https://www.cbioportal.org/) was used to summarize possible transcriptional (mRNA) change and mutational alterations. The results were reported as an OncoPrint. To further confirm the prognostic value of hub genes, we have evaluated the survival curve of the eleven hub genes identified from PPIs network analysis. The overall survival (OS) curve of each hub gene was determined by log rank test. A p value < 0.05 was considered as statistically significant.
Results
Identification of DEGs in GBM-Derived Plasma and Serum Samples
The analysis of the RNA-Seq data identified 636 and 269 significant DEGs from GBM-derived serum samples and plasma samples, respectively. Among the total significant DEGs, the number of upregulated DEGs in serum samples was more than the downregulated DEGs. In GBM-derived plasma samples, we have observed that all the 269 DEGs were upregulated compared to that of normal plasma samples (supplementary Fig. 1a). Overexpression of some of these critical genes might play important role during GBM development. In total, 24 DEGs were found to be common between serum and plasma samples (supplementary Fig. 1b). All the common DEGs between plasma and serum samples were upregulated. Hierarchical clustering of GBM-derived plasma and serum DEGs showed distinct patterns in GBM samples compared to the healthy donors (HD) (supplementary Fig. 1c).These findings indicated that GBM-derived plasma and serum samples might carry distinct group of genes which could potentially reflect the molecular signatures of GBM progression.
PPI network and module analysis of plasma and serum DEGs. a, b The PPI network with 269 plasma DEGs and 636 serum DEG constructed using string database. The DEGs with highest degree are represented with large circular nodes. c, d Module analysis of PPI network identified significant module with 14 nodes and 38 edges from PPI network of plasma. A significant module with 21 nodes and 210 edges was identified from PPI network of serum
Functional Annotations and Pathway Enrichment Analysis Revealed Distinct Biological Processes Associated with GBM-Derived Plasma and Serum DEGs
After identification of significant DEGs, the functional GO terms and pathways enrichment analysis were carried out with significant DEGs of plasma and serum using DAVID tool. The significance of the functional biological process and pathways were selected by considering a threshold of p value less than 0.05. The functional enrichment analysis shows that the plasma and serum DEGs are involved with distinct biological processes and pathways. The top five significant functional annotations with GO terms and KEGG pathways for plasma and serum are listed in Tables 1 and 2, respectively.
The Hub Genes Identified Through PPI Analysis
PPI networks were constructed with plasma and serum DEGs to determine the potential interaction between the DEGs. The PPI networks were constructed using the STRING database with confidence score > 0.4, and only the query protein has been considered and visualized by Cytoscape software (Fig. 1a, b). The module analysis revealed that the all the plasma hub genes except CREB1 were present in a module containing 14 nodes and 38 edges (Fig. 1c). These genes were associated with innate immune responses and ion transport. The hub genes from serum samples were present in a module containing 21 nodes and 210 edges (Fig. 1d). This module was enriched in protein ubiquitinylation processes and interleukin-mediated signaling.
The cytoscape plugin, Cytohubba, was used to screen the top ranked 25 hub genes from plasma and serum PPIs network by five topological scoring measures which include MNC, degree, EPC, EcC, and betweeness (Table 3). The predicted hub genes from each of the algorithm were further intersected for the identification of consensus significant hub genes in plasma and serum (supplementary Fig. 2). The hub genes identified from plasma were CASP3, CREB1, NFKBIA, JAK2, TLR1, GRN, and GDI2. Similarly, the hub genes identified from serum were SOCS3, UBE2S, KLHL5, and FZR1.
Identification of Key TFs Associated with the Predicted Hub Genes
The JASPAR database was used to identify the transcriptional regulators of the hub genes identified from plasma and serum. The TFs for the hub were predicted to construct the network (Fig. 2). In total 37 nodes and 50 interactions were identified from plasma hub genes and TFs. Similarly, an interaction network of 33 nodes and 45 was obtained from the serum hub genes. The hub genes showed association with various TFs (Table 4). The JAK2 gene identified from plasma had the highest degree of TFs. On the other hand, in serum, FZR1 is connected with maximum number of TFs. However, no TF was found to be associated with CASP3. From the hub gene and TF networks, it was observed that the family of FOX TFs such as FOXC1 and FOXL1 were the highly connected TFs associated with both plasma and serum hub genes. The FOX genes belong to superfamily of TFs which are associated with several important biological and cellular processes. Therefore, any genetic alteration of FOX can lead to aberrant of cells and may result in carcinogenesis. The FOXC class such as FOXC1 is known to play a crucial role in cancer formation. However, the mechanism of action of FOXC1 TF is still unclear. The expression of FOXL1 is reported to be associated with overall survival and worst clinical outcome in high-grade glioma patients. Hence FOXL1 might serve as candidate marker for clinical outcome of GBM patients (Yang et al. 2017; Chen et al. 2019).
Validation of the Expression and Genetic Alteration of the Identified Hub Genes
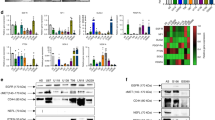
The differential expressions of the predicted hub genes were validated using the GEPIA database. The box plots of the expression levels showed that hub genes identified from both plasma and serum were significantly overexpressed in GBM samples from TCGA-GBM dataset compared with normal tissues (p value < 0.01 and log fold change of 1). This was in agreement with our results (Fig. 3a).
Validation of the mRNA expression level and associated genetic alteration of hub genes identified from GBM-derived plasma and serum. a The gene expression level of ten hub genes was validated between TCGA-GBM dataset and normal brain tissues (red: tumor samples; green: normal samples). All the ten hub genes were upregulated in GBM as compared to controls. The red * is considered as p value < 0.01. b The oncoprint from cBioportal shows that hub genes are genetically dysregulated in 32% (44/136) of the GBM cases
The cBioPortal was used to analyze the genomic alteration of hub genes with a study from GBM (TCGA, Firehouse Legacy dataset) which showed that 32% (44/136) of GBM cases undergo genetic alteration which includes amplification, deletion, mRNA upregulation, mRNA downregulation, and several mutations (Fig. 3b).
The prognostic significance of the eleven hub genes (Table 3) identified from GBM-derived plasma and serum samples were analyzed using the GEPIA survival plot to determine whether the hub gene expression is correlated with the survival of GBM patients from TCGA dataset. According to the median group cutoff, the patients were grouped into high and low risk group. The survival analysis plot showed that four genes (CASP3, GRN, GDI2, and SOCS3) was significantly associated with poor overall survival of GBM patients using p value < 0.05; at confidence interval (CI) = 0.95 (Fig. 4).
Discussions
In this study, we have performed integrative transcriptomic analysis of plasma- and serum-derived EVs from GBM samples and identified 269 and 636 significant DEGs in plasma and serum, respectively. The plasma and serum samples showed more upregulated DEGs than downregulated DEGs. To further elucidate the role of these DEGs in GBM-derived plasma and serum samples, we have carried out functional enrichment analysis and PPIs analysis for identification of hub genes. TFs associated with the hub genes were predicted, and finally, the expression levels of each of the hub genes were validated against TCGA-GBM dataset.
The hub genes identified from plasma were mainly linked to viral carcinogenesis and innate immune response regulation. Several studies reported link between viral carcinogenesis and glioblastoma pathogenesis. Viruses such as herpes virus, papilloma virus, and polygamous virus are reported to be detected in GBM tissues and cell lines. The role of viral infection and its mechanism in GBM is still obscure (Alibek et al. 2013; Akhtar et al. 2018; Foster and Cobbs 2019). Studies reported that most of the human cancers occur due to pathogen-related infections are associated with inflammation. There are numerous oncogenic viral pathogens such as HPVs, EBVs, and hepatitis B and C viruses which create an inflammatory environment that favors tumor development. Moreover, the class type of inflammatory or immune response such as redness, swelling and pain in tumors is ascertained by the characteristics pathogenic feature involved with tumor generation. There are reports which state that most of the human cancers are associated with chronic inflammation likely to be caused by viral infections, autoimmunity, and dysregulation in immune response. There are oncogenic viral pathogens such as human papillomaviruses (HPVs), Epstein-Barr viruses (EBVs) and hepatitis B and C viruses which create an inflammatory environment that favors tumor development (Goldszmid et al. 2014; Liggett 2014; Gonzalez et al. 2018). It was reported that viruses such as human cytomegalovirus (HCMV) were associated with GBM development or gliomagenesis (McFaline-Figueroa and Wen 2017). A recent study demonstrated that a significant percentage of GBM patients were infected by HCMV, and the tumor cells expressed several HCMV gene products. Moreover, inflammation or immunosuppresion can lead to reactivation of HCMV which interferes with various cellular processes involved in oncogenesis (Cobbs et al. 2002). Studies revealed that GBM patients treated with antiviral drug valganciclovir along with standard therapy showed high survival rates which suggests that valganciclovir may inhibit the viral oncogenic processes that can lead to GBM pathogenesis (Fornara et al. 2016).
The hub genes JAK2 and NFKBIA are known to be pro-inflammatory mediators that are involved in the multiple cellular pathways associated with cancer development. JAK2 was also reported to be involved in HPV-related cervical cancer. This indicates that JAK2 might play critical role in malignant tumor related to virus through immunomodulation (Kundu and Surh 2008; Abdolmaleki and Sohrabi 2018). The NF-kB signaling plays a significant role in GBM and provides a negative prognosis in GBM patients. The activation of NF-kB is one of the hallmarks of inflammation-induced cancer. In GBM, inflammation can be detected through infiltration of macrophages/microglia and lymphocytes, production of inflammatory cytokines, and NF-kB pathway activation (Puliyappadamba et al. 2014). The activity of NF-kB is found to be significantly higher in gliomas as compared to that of normal brain tissues. However, recent studies reported that NFKBIA is deleted in 25% of glioblastoma patients (Kinker et al. 2016). In our study, the expression of NFKBIA was significantly upregulated. TLR1 belongs to the class of Toll-like receptor family (TLRs) that plays an important role in both innate and adaptive immune system regulation. It binds to the microbial repertoire and initiates a series of signaling cascade and activates numerous inflammatory genes. There are evidences that some viruses encode specific proteins that inactivate the TLR-mediated signaling and contribute to virulence (Noreen and Arshad 2015). Studies revealed that TLR2 and TLR1 makes a heterodimer complex and acts as a functional sensor for HCMV (Cobbs et al. 2002). CASP3 is an important member of the cysteine protease family which plays a key role in apoptotic regulation. CASP3 along with CASP9 were reported to be overexpressed in malignant brain tumor including GBM (Yamabe et al. 1999; Ray et al. 2002). CREB1 acts as an oncogene and was reported to be involved in the development of wide array of cancers through transcriptional activation of many genes (Chen et al. 2017). There are several reports which suggested that CREB1 acts as an important gene in the progression of GBM pathogenesis (Mantamadiotis et al. 2012; Tan et al. 2012). Further studies will be needed to understand the mechanism of CREB1 behind GBM progression. The GRN gene encodes a protein called progranulin which is involved in proliferation of active cells that are dividing rapidly such as fibroblasts, immune cells, and certain brain cells. The GRN gene was reported to be associated with tumor progression in astrocytoma and can serve as a prognostic marker in GBM. A recent study showed that the survival rate of granulin-positive tumors has lower survival rate and higher recurrence than granulin-negative tumors (Ryu et al. 2017; Vachher et al. 2020). GDI2 belongs to the class of Rho GDP dissociation inhibitor family which regulates several important cellular processes. GDI2 is mostly expressed by hematopoetic cells and were reported to be highly dysregulated in ovarian adenocarcinoma (Lee et al. 2010). GDI2 was found to be highly overexpressed in anaplastic thyroid cancer (ATC) and are also involved in the development of liver metastasis in colorectal cancer (Onda et al. 2004; Yamasaki et al. 2007). A study on differential ceramide distribution and nitric oxide (NO) exposure in glioma cell lines reported GDI2 as one of the potential marker associated with tumor aggressiveness in high grade glioma cell lines (Leone et al. 2015). However, the role of GDI2 in GBM pathogenesis or progression has not been reported yet.
The hub genes identified from PPIs network of serum are significantly associated with ubiquitin mediated proteolysis and response to interleukin-6. Ubiquitins are highly conserved and abundant 9-kD protein that regulates several cellular processes, such as cell proliferation and apoptosis, and are also known to function in inflammatory processes such as cancer. Studies reported that dysregulation in ubiquitin mediated proteolysis of important cell cycle related genes or house-keeping genes may promote cancer. A high percentage of GBM patients possess EGFR mutation which is likely to be caused by CBL-mediated ubiquitination (Mani and Gelmann 2005; Hoeller et al. 2006), which indicated that protein ubiquitinylation might involve in GBM pathogenesis. The UBE2S (ubiquitin enzyme E2 S) gene was reported to be highly expressed in the majority of patients suffering from glioma III–IV grades. The overexpression of UBE2S is associated with reduced sensitivity towards chemotherapy (Hu et al. 2017). The KLHL5 (Kelch-like family member 5) is a recently identified novel protein coding gene related to action binding protein which plays important role in various cellular processes such as cell cycle regulation, apoptosis, metabolic processes, etc. In cancer, KLHL5 decreases the anticancer drug sensitivity as they are involved in degradation of cell cycles genes related to chemotherapeutics (Xu et al. 2003; Schleifer et al. 2018). However, no studies related to KLHL5 have been reported in GBM. FZR1 (fizzy and cell division cycle 20 related 1) is an activator for anaphase promoting complex/cyclosome (APC/C) that functions in the mitosis process and also functions in the regulation of G1 phase of the cell cycle. Most of the FZR1 substrate such as mitotic and S-phase cyclins, DNA replication factors are highly overexpressed in wide array of human cancers (Sigi et al. 2009; Wan et al. 2017). The role of FZR1 gene in GBM is still poorly understood. SOCS3 (suppressor of cytokine signaling) is a regulator of cytokine signaling pathway which inactivate cytokine signaling in wide range of cell types that mainly includes immune cells and cells of the CNS. The expression of SOCS3 was found to be dysregulated in various solid tumors including GBM which facilitates tumor progression through increased signal transduction and lead to radiotherapy resistance in GBM (Zhou et al. 2007; Baker et al. 2009; Ventero et al. 2019).
Using systems level integrative analysis of transcriptomic data, our study identified significant hub genes from GBM derived EVs in plasma and serum samples, which were CASP3, CREB1, NFKBIA, JAK2, TLR1, GRN, GDI2, SOCS3 UBE2S, KLHL5, and FZR1. Among the 11 hub genes, CASP3, GRN, GDI2, and SOCS3 were associated with poor clinical outcome in GBM and might act as potential biomarkers for the prognosis of GBM patients. The results from the present study will provide the basis for conducting targeted experiments for further clinical validation.
Conclusion
The present study generated a system level molecular understanding of DEGs from GBM-derived EVs in plasma and serum. The identified hub genes can further be validated to develop clinically usable blood-based diagnostic and prognostic biomarkers for GBM.
Abbreviations
- GBM:
-
Glioblastoma multiforme
- EVs:
-
Extracellular vesicles
- DEGs:
-
Differentially expressed genes
- PPIs:
-
Protein–protein interactions
- GO:
-
Gene ontology
- TFs:
-
Transcription factors
- OS:
-
Overall survival
References
Abdolmaleki M, Sohrabi A (2018) Characterization of JAK2 V617F (1849 G > T) mutation in cervical cancer related to human papillomavirus and sexually transmitted infections. J Cancer Prev 23:82–86. https://doi.org/10.15430/jcp.2018.23.2.82
Akhtar S, Vranic S, Cyprian FS, Al MAE (2018) Epstein-barr virus in gliomas: Cause, association, or artifact? Front Oncol 8:1–10. https://doi.org/10.3389/fonc.2018.00123
Alibek K, Kakpenova A, Baiken Y (2013) Role of infectious agents in the carcinogenesis of brain and head and neck cancers. Infect Agent Cancer 8:1. https://doi.org/10.1186/1750-9378-8-7
Baker BJ, Akhtar LN, Benveniste EN (2009) SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol 30:392–400. https://doi.org/10.1016/j.it.2009.07.001
Basu B, Ghosh MK (2019) Extracellular vesicles in glioma: From diagnosis to therapy. BioEssays 41:1–9. https://doi.org/10.1002/bies.201800245
Chen A, Zhong L, Lv J (2019) FOXL1 overexpression is associated with poor outcome in patients with glioma. OncolLett 18:751–757. https://doi.org/10.3892/ol.2019.10351
Chen J, Zhang C, Mi Y et al (2017) CREB1 regulates glucose transport of glioma cell line U87 by targeting GLUT1. Mol Cell Biochem 436:79–86. https://doi.org/10.1007/s11010-017-3080-3
Chin CH, Chen SH, Wu HH et al (2014) cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC SystBiol 8:S11. https://doi.org/10.1186/1752-0509-8-S4-S11
Cobbs CS, Harkins L, Samanta M et al (2002) Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res 62:3347–3350
Dennis G, Sherman BT, Hosack DA, et al (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:. https://doi.org/10.1186/gb-2003-4-9-r60
Duan H, Hu JL, Chen ZH et al (2020) Assessment of circulating tumor DNA in cerebrospinal fluid by whole exome sequencing to detect genomic alterations of glioblastoma. Chin Med J (Engl) 133:1415–1421. https://doi.org/10.1097/CM9.0000000000000843
Fornara O, Bartek J, Rahbar A et al (2016) Cytomegalovirus infection induces a stem cell phenotype in human primary glioblastoma cells: Prognostic significance and biological impact. Cell Death Differ 23:261–269. https://doi.org/10.1038/cdd.2015.91
Foster H, Cobbs CS (2019) Viruses and Glioblastoma: Affliction or Opportunity? 67–86. https://doi.org/https://doi.org/10.1007/978-3-030-04155-7_4
Goldszmid RS, Dzutsev A, Trinchieri G (2014) Host immune response to infection and cancer: Unexpected commonalities. Cell Host Microbe 15:295–305. https://doi.org/10.1016/j.chom.2014.02.003
Gonzalez H, Hagerling C, Werb Z (2018) Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev 32:1267–1284. https://doi.org/10.1101/GAD.314617.118
Hoeller D, Hecker CM, Dikic I (2006) Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer 6:776–788. https://doi.org/10.1038/nrc1994
Hu L, Li X, Liu Q et al (2017) UBE2S, a novel substrate of Akt1, associates with Ku70 and regulates DNA repair and glioblastomamultiforme resistance to chemotherapy. Oncogene 36:1145–1156. https://doi.org/10.1038/onc.2016.281
Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Kinker GS, Thomas AM, Carvalho VJ et al (2016) Deletion and low expression of NFKBIA are associated with poor prognosis in lower-grade glioma patients. Sci Rep 6:1–9. https://doi.org/10.1038/srep24160
Kundu JK, Surh YJ (2008) Inflammation: Gearing the journey to cancer. Mutat Res - Rev Mutat Res 659:15–30. https://doi.org/10.1016/j.mrrev.2008.03.002
Lane R, Simon T, Vintu M, et al (2019) Cell-derived extracellular vesicles can be used as a biomarker reservoir for glioblastoma tumor subtyping. Commun Biol 2:. https://doi.org/10.1038/s42003-019-0560-x
Lee DH, Chung K, Song JA et al (2010) Proteomic identification of paclitaxel-resistance associated hnRNP A2 and GDI 2 proteins in human ovarian cancer cells. J Proteome Res 9:5668–5676. https://doi.org/10.1021/pr100478u
Leone R, Giussani P, De Palma S et al (2015) Proteomic analysis of human glioblastoma cell lines differently resistant to a nitric oxide releasing agent. MolBiosyst 11:1612–1621. https://doi.org/10.1039/c4mb00725e
Liggett (2014) 基因的改变NIH Public Access. Bone 23:1–7. https://doi.org/10.1038/jid.2014.371
Lombardi MY, Assem M (2017) Glioblastoma Genomics: A Very Complicated Story. Glioblastoma 3–25. https://doi.org/10.15586/codon.glioblastoma.2017.ch1
Love MI, Anders S, Huber W (2014) Differential analysis of count data - the DESeq2 package
Mani A, Gelmann EP (2005) The ubiquitin-proteasome pathway and its role in cancer. J ClinOncol 23:4776–4789. https://doi.org/10.1200/JCO.2005.05.081
Mantamadiotis T, Papalexis N, Dworkin S (2012) CREB signalling in neural stem/progenitor cells: Recent developments and the implications for brain tumour biology. BioEssays 34:293–300. https://doi.org/10.1002/bies.201100133
McFaline-Figueroa JR, Wen PY (2017) The viral connection to glioblastoma. Curr Infect Dis Rep 19:2–6. https://doi.org/10.1007/s11908-017-0563-z
Noreen M, Arshad M (2015) Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunol Res 62:234–252. https://doi.org/10.1007/s12026-015-8640-6
Ohgaki H, Kleihues P (2007) Genetic pathways to primary and secondary glioblastoma. Am J Pathol 170:1445–1453. https://doi.org/10.2353/ajpath.2007.070011
Onda M, Emi M, Yoshida A et al (2004) Comprehensive gene expression profiling of anaplastic thyroid cancers with cDNA microarray of 25 344 genes. EndocrRelat Cancer 11:843–854. https://doi.org/10.1677/erc.1.00818
Osti D, Del BM, Rappa G et al (2019) Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res 25:266–276. https://doi.org/10.1158/1078-0432.CCR-18-1941
Paul Shannon 1, Andrew Markiel 1, Owen Ozier, 2 Nitin S. Baliga, 1 Jonathan T. Wang, 2 Daniel Ramage 2, et al (1971) Cytoscape: A Software Environment for Integrated Models. Genome Res 13:426. https://doi.org/10.1101/gr.1239303.metabolite
Pruitt KD, Hogue CW, Groll M et al (2001) Mcode. Nucleic Acids Res 29:137–140. https://doi.org/10.1093/nar/29.1.137
Puliyappadamba VT, Hatanpaa KJ, Chakraborty S, Habib AA (2014) The role of NF-κB in the pathogenesis of glioma. Mol Cell Oncol 1:. https://doi.org/10.4161/23723548.2014.963478
Raposo G, Stoorvogel W (2013) Extracellular vesicles exosomesmicrovesicles and friends. J Cell Biol 200:373–383. https://doi.org/10.1083/jcb.201211138
Ray SK, Patel SJ, Welsh CT et al (2002) Molecular evidence of apoptotic death in malignant brain tumors including glioblastomamultiforme: Upregulation of calpain and caspase-3. J Neurosci Res 69:197–206. https://doi.org/10.1002/jnr.10265
Reátegui E, Van Der Vos KE, Lai CP, et al (2018) Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat Commun 9:. https://doi.org/10.1038/s41467-017-02261-1
Ryu JI, Han MH, Cheong JH et al (2017) Expression of the growth factor granulin in patients with brain tumors: Relevance to prognosis. Biomed Res 28:5583–5588
Santiago-Dieppa DR, Steinberg J, Gonda D et al (2014) Extracellular vesicles as a platform for “liquid biopsy” in glioblastoma patients. Expert Rev MolDiagn 14:819–825. https://doi.org/10.1586/14737159.2014.943193
Saugstad JA, Lusardi TA, Van Keuren-Jensen KR, et al (2017) Analysis of extracellular RNA in cerebrospinal fluid. J Extracell Vesicles 6:. https://doi.org/10.1080/20013078.2017.1317577
Schleifer RJ, Li S, Nechtman W et al (2018) KLHL5 knockdown increases cellular sensitivity to anticancer drugs. Oncotarget 9:37429–37438. https://doi.org/10.18632/oncotarget.26462
Sigi R, Wandkee C, Rauch V et al (2009) Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J Cell Sci 122:4208–4217. https://doi.org/10.1242/jcs.054197
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Tan X, Wang S, Zhu L et al (2012) cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. ProcNatlAcadSci U S A 109:15805–15810. https://doi.org/10.1073/pnas.1207787109
Tang Z, Li C, Kang B et al (2017) GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102. https://doi.org/10.1093/nar/gkx247
Vachher M, Arora K, Burman A, Kumar B (2020) NAMPT, GRN, and SERPINE1 signature as predictor of disease progression and survival in gliomas. J Cell Biochem 121:3010–3023. https://doi.org/10.1002/jcb.29560
Ventero MP, Fuentes-Baile M, Quereda C et al (2019) Radiotherapy resistance acquisition in glioblastoma. Role of SOCS1 and SOCS3. PLoS ONE 14:1–21. https://doi.org/10.1371/journal.pone.0212581
Wan L, Chen M, Cao J et al (2017) The APC/C E3 ligase complex activator fzr1 restricts braf oncogenic function. Cancer Discov 7:424–441. https://doi.org/10.1158/2159-8290.CD-16-0647
Whitehead CA, Kaye AH, Drummond KJ et al (2020) Extracellular vesicles and their role in glioblastoma. Crit Rev Clin Lab Sci 57:227–252. https://doi.org/10.1080/10408363.2019.1700208
Xu J, Gu S, Wang S et al (2003) Characterization of a novel splicing variant of KLHL5*, a member of the kelch protein family. MolBiol Rep 30:239–242. https://doi.org/10.1023/A:1026372901766
Xu R, Rai A, Chen M et al (2018) Extracellular vesicles in cancer — implications for future improvements in cancer care. Nat Rev ClinOncol 15:617–638. https://doi.org/10.1038/s41571-018-0036-9
Yamabe K, Shimizu S, Ito T et al (1999) Cancer gene therapy using a pro-apoptotic gene, caspase-3. Gene Ther 6:1952–1959. https://doi.org/10.1038/sj.gt.3301041
Yamasaki M, Takemasa I, Komori T et al (2007) The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int J Oncol 30:129–138. https://doi.org/10.3892/ijo.30.1.129
Yang Z, Jiang S, Cheng Y et al (2017) FOXC1 in cancer development and therapy: Deciphering its emerging and divergent roles. TherAdv Med Oncol 9:797–816. https://doi.org/10.1177/1758834017742576
Zhou G, Soufan O, Ewald J et al (2019) NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47:W234–W241. https://doi.org/10.1093/nar/gkz240
Zhou H, Miki R, Eeva M et al (2007) Reciprocal regulation of SOCS1 and SOCS3 enhances resistance to ionizing radiation in glioblastomamultiforme. Clin Cancer Res 13:2344–2353. https://doi.org/10.1158/1078-0432.CCR-06-2303
Acknowledgments
NR acknowledges Institutional Fellowship from Tezpur University for pursuing her PhD. PB acknowledges the infrastructural support provided from the Department of Molecular Biology and Biotechnology at the Tezpur University.
Funding
This work was supported by the Ramalingaswami Re-entry Fellowship Grant to Dr. Pankaj Barah by the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Contributions
PB developed the concept. NR and MG analyzed the data. NR, MG, DKB, and PB interpreted the results and written the manuscript. NR and MG contributed equally to this work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Publicly available gene expression data has been used for this analysis.
Consent for publication
All authors have seen and approved the manuscript for submission.
Competing interests
The authors of this manuscript declare no competing interest.
Availability of data and materials
The dataset used for this study is available at NCBI-GEO database (GEO Accession No. GSE106804).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roy, N., Gaikwad, M., Bhattacharrya, D.K. et al. Identification of Systems Level Molecular Signatures from Glioblastoma Multiforme Derived Extracellular Vesicles. J Mol Neurosci 71, 1156–1167 (2021). https://doi.org/10.1007/s12031-020-01738-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-020-01738-x