Abstract
Extensive experimental studies indicate that autologous bone marrow mesenchymal stem cells (BMSCs) are able to ameliorate experimental autoimmune encephalomyelitis (EAE) and potentially multiple sclerosis. However, the impact that the inflammatory environment present in EAE may have on the biological properties of BMSCs expanded in vitro for transplantation is yet to be clarified. It was investigated whether BMSCs isolated from EAE-induced C57bl6/J mice and expanded in vitro preserve the properties of BMSCs isolated from healthy donors (BMSCs-control). The mesenchymal origin, the differentiation potential, and the transcriptional expression profile of six histone-modifying genes were studied in both groups of BMSCs. BMSCs-EAE exhibited distinct morphology and larger size compared to BMSCs-control, higher degree of proliferation and apoptosis, differences in the adipogenesis and the osteogenesis induction, and differential expression of stromal markers and markers of progenitor and mature neuronal/glial cells. Moreover, BMSCs-EAE exhibited different expression patterns on a number of histone-modifying genes compared to controls. We recorded manifold differences, both phenotypical and functional, of in vitro expanded BMSCs-EAE in comparison to their healthy donor-derived counterparts that may be attributed to the inflammatory environment they originated from. Whether our findings may be of any clinical relevance needs to be clarified in future studies, in vivo.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is a chronic inflammatory multifocal demyelinating disease of the central nervous system (CNS) affecting mainly young adults. Immunosuppressive and immunomodulatory drugs at least partly delay the progress of the disease (Grigoriadis 2002). In addition, cell-based therapies are being developed and expected to influence disease outcome via a number of identified mechanisms of actions (Martino et al. 2010). Among the several cellular sources considered for transplantation in MS are the bone marrow mesenchymal stem cells (BMSCs).
BMSCs comprise a distinct nonhematopoietic cell population residing in the bone marrow (BM) (representing 0.01–0.001 % of total bone marrow cells). Since they were first described (Friedenstein et al. 1974), BMSCs have attracted scientific attention due to their stem-like characteristics. In addition to differentiation into cells of mesenchymal origin such as bone, cartilage, and muscle (Baksh et al. 2004), these cells have initially been reported to transdifferentiate into almost any cell type of the three lineages (Black and Woodbury 2001; Jiang et al. 2002; Li et al. 2007; Rizvi et al. 2006; Sanchez-Ramos et al. 2000; Schwartz et al. 2002; Tseng et al. 2007; Woodbury et al. 2000). BMSCs’ strong candidacy as therapeutic agent arises from the fact that not only can they be relatively easily isolated from the adult BM and expanded in vitro for several passages, but they are also considered poorly immunogenic and able to possess immunomodulatory properties (Kassis et al. 2008; Uccelli et al. 2006).
The therapeutic potential of murine BMSCs in autoimmune demyelination has been examined following autologous cell transplantation (Kassis et al. 2008; Zappia et al. 2005; Gerdoni et al. 2007) or with cells originating from other species such as human (Gordon et al. 2010) in experimental autoimmune encephalomyelitis (EAE), the animal model of MS. It has already been shown that BMSCs have a beneficial effect in the clinical course of EAE by means of immunomodulation and/or engraftment in the CNS (Gerdoni et al. 2007; Gordon et al. 2010; Kassis et al. 2008; Zappia et al. 2005). However, there is increasing evidence that long-term engraftment and the transdifferentiation of the cells may hardly be considered an important mechanism involved in the therapeutic effect of BMSCs (Uccelli et al. 2011a, b).
Autologous transplantation of BMSCs has been proposed as the method of choice for MS (Freedman et al. 2010). In this concept, BMSCs derived from the same patient destined to be transplanted require multiple passages to reach adequate numbers for transplantation. Moreover, small clinical trials where autologous BMSCs were either intrathecally (Karussis et al. 2010; Mohyeddin Bonab et al. 2007; Yamout et al. 2010) or intravenously (Karussis et al. 2010; Rice et al. 2010) administered in MS patients indicated the safety of these procedures. However, when experimental studies are considered, the donors of the cells were either naive inbred animals (Gerdoni et al. 2007; Kassis et al. 2008; Zappia et al. 2005) or non-MS patients (Gordon et al. 2010). Under this view, there still may be a missing point over the beneficial effect of BMSCs if autologous transplantation to patients with MS is considered.
Indeed, the impact that the inflammatory environment present in EAE may have on the biological properties of BMSCs destined to be transplanted is yet to be clarified. To address this question, we studied BMSCs isolated from EAE-induced and healthy C57bl6/J mice and expanded them for a period of four to five passages after which the cells are considered appropriate for transplantation (Kassis et al. 2008). Our findings indicated that there are some differences between EAE- and naïve animal-derived BMSCs namely with regard to their size and shape, proliferation rate, and differentiation capacities. Moreover, BMSCs originating from EAE-induced mice exhibited different expression patterns on a number of histone-modifying genes compared to controls, thereby indicating potential dysregulation of histone acetylation/deacetylation equilibrium in these cells.
Materials and Methods
Induction of EAE
Inbred female 6–8-week-old C57BL/6 mice (Hellenic Pasteur Institute, Athens, Greece) were housed in a pathogen-free animal facility of the B’ Department of Neurology, AHEPA University Hospital, in accordance to the European Union Guidelines (Official Journal of the European Communities/No L 374/11). Animal facilities and experimentation were subjected to The Animals Act 160/3.05.1991 approved by institutional Ethics Committee and the Veterinary Animal Experimentation Authority, Perfecture of Thessaloniki (license nr. 19EE07). All mice were fed a regular diet and given water without antibiotics ad libitum.
EAE was induced as previously described (Grigoriadis et al. 2011; Lourbopoulos et al. 2011). Briefly, animals were immunized with subcutaneous injection at the left para-lumbar region of 300 μg of MOG35–55 (Sigma) emulsified in 200 μl of a solution containing 100 μl complete Freund’s adjuvant (IFA, Sigma) supplemented with 4 mg/ml Mycobacterium tuberculosis H37RA (Difco) and 100 μl filtered phosphate-buffered saline (PBS) (day 0). In addition, on day 0 and on day 2 post-induction (DPI), 400 and 200 ng of pertussis toxin (Sigma) diluted in 0.5 ml of filtered PBS were administered IP, correspondingly. A booster of 300 μg of MOG35–55 in CFA was administered on 7 DPI. Animals were examined daily and evaluated for clinical signs of disease.
Isolation and Culture of BMSCs
BMSCs were isolated and cultured from EAE-induced mice at the acute phase of the disease (days 20–25 post-induction) and age-matched naïve controls, under standard protocol (Gerdoni et al. 2007; Grigoriadis et al. 2011; Kassis et al. 2008). The entire procedure was repeated three times, resulting in three control vs. EAE culture experiments (eight animals/cell group per experiment).
Briefly, the medium used consisted of α-MEM (Gibco) supplemented with 20 % heat-inactivated fetal bovine serum (Gibco), 1 % l-glutamine (Gibco), and 1 % antibiotics–antimycotics (Gibco). Mice were euthanized by total body anesthesia followed by cervical dislocation. Femurs and tibias were removed and cleared of connective, adipose tissue and muscles. The bones were placed in a plate with fresh medium (15 % FBS) on ice. The bone marrow was flushed out of each bone’s central canal with a 19 × g syringe containing fresh culture medium into a 50 ml falcon. The cellular solution was aspirated several times by 19 × g, 21 × g, and 23 × g needles successively to reach a single cell suspension, and then, it was centrifuged at 240 × g for 10 min at room temperature. The supernatant was removed and the pellet was resuspended to 10 ml fresh medium. Cells were counted with a hemocytometer.
Cells were seeded at a concentration of 4 × 106 cells/cm2 in T25 flasks (Nunclon) and maintained in an incubator at 37 °C, 5 % CO2, and 95 % air. At day 2, half of the medium was replaced with fresh 20 % FBS medium. At day 7, the whole medium was aspirated from the flasks and replaced with fresh 20 % FBS medium. Half of the total medium was refreshed every 3–4 days. The cell density and morphology were monitored under an inverted microscope. Attached cells developed into colonies within 3–7 days. When cells reached 70–80 % confluence, they were harvested as follows. First, the medium was aspirated from the flasks and the residual FBS was washed out with 2–3 ml PBS (Gibco). Then, 0.05 % trypsin–0.01 % EDTA (Sigma, St. Louis, MO, USA) was added and incubated at 37 °C for 5 min. Full fresh medium (20 % FBS) was added in order to inactivate the trypsin. The cell solution was collected using cell scrapers into a sterile 50-ml tube and centrifuged at 400 × g for 10 min at room temperature. The supernatant was removed and the cell pellet was resuspended in 1–2 ml of fresh medium. The cells were counted and subcultured in T75 flasks at a ratio of 1:3 for five consecutive passages. The area that each cell of the BMSC-EAE and the BMSC-control group at passage 5 covered in the culture flask was measured from 10 different visual fields using the software ImageJ (v.1.38).
Culture of Fibroblasts
Fibroblast-like L929 strain (ATCC) was cultured at a density of 1 × 106 cells in 10 ml of cell maintenance medium [α-MEM (Gibco) enriched with 10 % heat-inactivated FBS (Gibco), 1 % l-glutamine (200 mM, Gibco), and 1 % penicillin/streptomycin solution (Life Technologies)] in a T75 flask (Corning) and kept in an incubator at 37 °C, 5 % CO2, and 95 % air. When cells reached 70–80 % confluence, they were split as follows. The culture medium was aspirated from the flask and 10 ml of PBS was added to wash out the remaining medium. Then, 4 ml of trypsin–EDTA 0.5 % (Gibco) was added to the flask and incubated for 2 min. Trypsin was inactivated by 10 ml of fresh medium and the cells were harvested into a new flask at a density of 1.25 × 106 cells/10 ml of culture medium. The cells were incubated at 37 °C, 5 % CO2, and 95 % air. The medium was renewed every 3–4 days.
Differentiation Assays
BMSCs from both groups were induced toward the osteogenic and adipogenic lineages. Cells from the third passage were collected by trypsinization as described previously and plated at a density of 200,000 cells/well at a 12-well plate for osteogenic or adipogenic differentiation. Three wells were seeded per cell group for each differentiation condition along with two wells which served as blanks. After 2 to 3 days of incubation at 37 °C, 5 % CO2, and 95 % air where cells reached confluence, the medium was changed to osteogenic or adipogenic induction medium, respectively.
The osteogenic induction medium consisted of DMEM (Sigma), 10 % FBS, 10 mM β-glycerol-phosphate (Sigma), 50 μΜ l-ascorbic acid-2-phosphate (Sigma), and 10−7 M dexamethasone (Sigma). The medium was replaced every 2 days for approximately 21 days. The adipogenic induction medium consisted of α-ΜΕΜ, 10 % FBS, 10−7 M dexamethasone, and 10 μg/ml insulin (Sigma). The medium was replaced every 2 days for approximately 21 days.
In order to detect osteoblastic differentiation, the von Kossa staining was used which identifies the deposition of minerals excreted by bone-differentiated cells. The medium was aspirated from each well and the cells were fixed using paraformaldehyde (PFH-Panreac) 4 % for 20 min at room temperature. PFH was removed and two washes with distilled water followed. The cells were then stained with 1 % silver nitrate (Baker) under intense light for 1 h. Cells were then washed with distilled H2O and incubated with 5 % sodium thiosulfate (Riedel de Haen) for 5 min and washed with distilled H2O. Calcium salts were stained dark brown-black and cells were counterstained with 1 % Nuclear Fast Red (Sigma). Evaluation of osteogenesis was performed using light microscopy (Axioplan 2, Zeiss) under ×40 optical fields. A digital camera (Nikon, DS-5Mc) was used to capture images. Ten randomly chosen optical fields (images) were captured from each well (thus producing 30 images per cell group). Images were further analyzed using the ImageJ software and the “threshold” mode in order to quantify the dark brown-black mineral deposits in arbitrary units (AU). Data are from three independent experiments (average mean ± SEM). The adipogenic differentiation was evaluated by Oil Red O (Sigma) staining. The medium was aspirated and PBS was used to rinse each well. PFH 4 % was added and cells were incubated for 20 min at room temperature. PFH was removed and two washes with distilled water followed. Six percent isopropanol (Panreac) was added and poured off. Oil Red O solution (stock solution 0.5 g diluted in 100 ml isopropanol, working solution—60 ml stock in 40 ml distilled water) was added and left to incubate for 10 min at room temperature. Finally, the wells were washed with isopropanol and distilled water and the plates were counterstained with hematoxylin (Merck) and mounted with Aqueous Mounting medium (Dako). Evaluation of adipogenesis was performed using similar methodology to osteogenesis. However, since adipogenesis followed a pattern of “clustered” lipid formation, images were captured from adipocytic aggregates in randomly chosen optical fields (10 images per well). Images were further analyzed using the ImageJ software and the “threshold” mode; red adipose tissue was quantified in AU using a 100-μm × 100-μm square surface placed in the center of the adipocytic aggregates (data were expressed in mean ± SEM). Furthermore, the diameter of the adipocytic vacuoles (droplets) was studied in both groups and was expressed in micrometer ± SEM. Data are from three independent experiments (average mean ± SEM).
Fluorescence Activated Cell Sorting (FACS) Analysis
In order to characterize the cells (Dominici et al. 2006), we studied the expression of CD45, CD11b, CD11c, CD44, CD73, and Sca-1 using FACS analysis. We excluded through gating the population of cells expressing CD45, the pan-leukocyte marker, present on leucocytes and their hematopoietic progenitors. In the subpopulation of CD45− cells, we studied the expression of the marker CD11b expressed mainly on monocytes/macrophages and microglia and to milder extent on granulocytes, NK cells, and subsets of dendritic cells, the CD11c marker of dendritic cells but also of monocytes, macrophages, neutrophils, and a subpopulation of B cells. In addition, we identified the expression of CD73 marker which is expressed on subsets of B and T cells, endothelial cells, dendritic cells, fibroblasts as well as neurons and oligodendrocytes along with mesenchymal stromal cells in a specific time point of maturation. Finally, we tested our cell populations for the markers CD44, which are found natively on hematopoietic stem cells and play a role on cell to cell interactions, cell adhesion and migration, and Sca-1 marker of immature hematopoietic progenitor cells. Fibroblast-like cells L929 were used as cellular controls to BMSCs extracted from both control and EAE groups.
BMSCs (control and EAE group) (passage 4) and L929 cells were harvested by trypsinization, divided into polystyrene FACS tubes (2 × 105 cells/tube), and labeled with rat anti-mouse CD45 (Leukocyte Common Antigen, PE-Cy™5, BD Pharmingen), rat anti-mouse CD73 (Ecto-5-nucleotidase, PE, BD Pharmingen), rat anti-mouse CD11b (Integrin αM chain, FITC, BD Pharmingen), rat anti-mouse CD44− (PE, BD Pharmingen), hamster anti-mouse CD11c (Integrin alpha-X chain, PE-Cy™7, BD Pharmingen), and rat anti-mouse SCA-1 (Ly-6A/E, FITC, BD Pharmingen) antibodies for 45 min in the dark. Fluorescence data were collected from 10,000 cells and measurements and analysis were performed using a FACScan (BD Biosciences, Franklin Lakes, NJ, USA). In addition, isotypic control was used for each run.
MTT Cell Proliferation Assay
MTT (tetrazolium) assay was performed for each cell group at passage 5 using the TACSTM MTT Cell Proliferation Assay (R&D Systems). Briefly, 106 cells/ml were seeded in a 24-well plate. A week later, MTT reagent solution (100 μl/well) was added to all wells, and the plate was incubated at 37 °C for 4 h. MTT detergent (1 ml/well) was added to dissolve the dark blue crystals and the plate was read 2 h later on a Molecular Devices Elisa microplate reader at 540 nm using 650 nm as reference filter.
Self-differentiation Protocol
BMSCs of the EAE-induced and the control group were harvested by trypsinization at passage 4 and re-plated at six-well plates at a density of 150,000 cells/well with fresh culture medium α-MEM supplemented with 20 % fetal bovine serum, 1 % l-glutamine, and 1 % antibiotics–antimycotics and then heat-inactivated. BMSCs were cultured without any changes in the media for 3 weeks (starvation) according to the protocol followed by Li et al. (2007). These plates were subjected to immunofluorescence.
Immunofluorescence Microscopy
Cells from both non- and self-differentiating cultures were fixed in 4 % PFH in PBS, blocked in antibody diluent (Dako), and incubated with the first antibody for 1 h. After repetitive washes in order to remove any excess antibody, the cells were incubated with fluorescent secondary antibody and counterstained with DAPI (Invitrogen, Molecular Probes). A Zeiss Axioplan 2 microscope was used for observations and images were captured with Nikon digital light camera (DS-5Mc). The cells were stained for the expression of the differentiation markers: PSA-NCAM for the presence of immature neural-committed progenitors, NG2 for the presence of glia progenitors, CNPase for the presence of mature oligodendrocytes, GFAP for the presence of mature astrocytes, and NeuN for the presence of mature neurons.
The antibodies used were mouse anti-p53 (Dako), mouse anti-Ki67 (Dako), rabbit anti-GFAP (Dako), mouse anti-NeuN (Chemicon), mouse anti-PSA-NCAM (Chemicon), rabbit anti-NG2 (Chemicon), rabbit anti-caspase-3 (R&D Systems), mouse anti-CNPase (Chemicon), and rabbit anti-BDNF (Santa Cruz). Corresponding secondary antibodies used were 568 Alexa Fluor Rhodamine goat anti-mouse (Invitrogen, Molecular Probes), 488 Alexa Fluor FITC goat anti-rabbit (Invitrogen, Molecular Probes), 568 Alexa Fluor Rhodamine goat anti-rabbit (Invitrogen, Molecular Probes), 488 Alexa Fluor FITC goat anti-mouse (Invitrogen, Molecular Probes), and Rhodamine goat IgM anti-mouse (Jackson ImmunoResearch). Measurements were performed from 10 to 15 randomly selected visual fields for each staining and the immunofluorescence markers were quantified by measuring the percentage of positive cells out of the entire cell population.
RNA Extraction
Total RNA was extracted with the Mini RNA Isolation II kitTM (Zymo Research) from 106 L929 fibroblasts, BMSCs-control, and BMSCs-EAE according to the manufacturer’s instructions. Briefly, cells were harvested and pelleted by centrifugation for 5 min at 500 × g. The supernatant was completely removed and the cell pellet was suspended in 600 μl ZR RNA Buffer. After a brief vortex, each sample was transferred into a Zymo-Spin Column in a supplied collection tube and centrifuged at 10,470 × g for 1 min. The flow-through was discarded. Then, 350 μl of RNA wash buffer was added to the column and centrifuged at 10,470 × g for 1 min. The flow-through was discarded and the washing step was repeated. The column was transferred to an RNase-free tube. With the addition of 50 μl of DNase/RNase-free water directly onto the column membrane and the centrifugation at 7,269 × g for 1 minute, the total RNA was eluted. RNA was stored at −80 °C until further use.
cDNA Synthesis
The iScript™ cDNA Synthesis Kit (Bio-Rad) was used to reverse transcribe 1 μg of total RNA into its first strand complementary DNA, according to the manufacturer’s instructions. Briefly, the 20-μl reaction consisted of 4 μl of 5× iScript reaction mix (containing a blend of oligo(dT) and random primers, an MMLV RNase H+ reverse transcriptase and nuclease-free water), 1 μl of reverse transcriptase, 1 μg of template RNA, and up to 20 μl nuclease-free water. The reaction protocol included 5 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, and cooling at 4 °C.
Real-Time PCR
There are several genes that are considered useful in a routine test for BMSCs in tissue engineering and cell therapy (Igarashi et al. 2007). Among them are the TRIB2, KCTD12 (potassium channel tetramerization domain containing 12), PRG1 (proteoglycan 1, secretory granule), MGP1 (matrix G1a protein), IGF1 (insulin growth factor 1), IGFBP7 (insulin-like growth factor-binding protein 7), and LIF (leukemia inhibitory factor). Primer pairs for the above-mentioned genes as well as the histone acetyltransferases (CBP, P300, PCAF) and histone deacetylases (HDAC1, HDAC2, HDAC3) were designed for real-time PCR applications using Primer3 (Rozen and Skaletsky 2000), so that at least one of them targets to an exon–exon junction, and therefore, genomic DNA can be excluded as a template. The desired primer pairs were checked with BLAST for sequence homology with sequences originating from other species and their complete sequences are presented in supplementary Table 1.
All real-time PCR reactions were performed using the ICycler IQ5 Multicolor real-time PCR Detection System (Bio-Rad). Reactions were performed using the 2× Master Mix KAPA SYBR® FAST qPCR Kit (Kapa Biosystems). The reaction included 10 μl of the master mix, 0.4 μl of each primer (10 pmol), 1 μl of cDNA, and nuclease-free water up to 20 μl.
PCR conditions were 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and a final step of 72 °C for 5 min. Melting curve analysis was applied to all reactions to ensure specificity of the amplification product. The product size for each primer set was confirmed by agarose gel electrophoresis. Mus musculus beta actin mRNA was used in all reactions as a reference gene to normalize the transcript levels of target genes. The comparative threshold cycle, ΔΔCT method, was used for the relative quantification data analysis (REST software). All results were repeated in three independent experiments and performed in triplicate each time.
Karyotype
The cells from the BMSC-control and BMSC-EAE group of passage 6 were grown to T25 flasks up to the point where 60–70 % confluence was reached. Colcemide (Applichem) was added to the flasks and incubated for 16 h at 37 °C, 5 % CO2, and 95 % air.
Cells were trypsinized, using Trypsin-EDTA (Biochrom) and brought to a single cell suspension and resuspended into 7 ml KCl (0.56 % w/v) (Biochrom) and incubated for 30 min at 37 °C. The cells were centrifuged at 170 × g for 5 min and were fixed into ice-cold acetic acid–methanol mix, in a ratio1:3. Fixed cells, after repetitive washes, were placed on microscope slides and dried out overnight.
For the observation of metaphases, slides were pretreated with trypsin–EDTA, washed with FBS, and stained with Giemsa. The metaphases were captured using metaphor system on AxionPlan II (Zeiss) microscope and analyzed using the Ikaros software.
Statistical Analysis
The statistical analysis was performed using the SPSS 18.0 software. Normality of the data was tested using the Shapiro–Wilk and Kolmogorov–Smirnov tests. For parametric data, Student’s t test or one-way ANOVA with Dunnett’s and Bonferroni post hoc tests were used. For nonparametric data, the equivalent Mann–Whitney U test for comparison of two groups was used. Values of all scale data are taken from three independent experiments and are expressed as average mean ± SEM, unless noted. Statistical significance was set at p < 0.05.
Results
Morphology and Size
At each culture medium change, cells were observed using phase contrast microscopy in order to detect possible morphological differences between control and BMSCs-EAE. The culture of control cells resulted in a heterogeneous population which after repetitive re-culturing rendered a more homogenous population of spindle-shaped cells, which formed widely expanded networks or colonies, dispersed throughout the culture flask (Fig. 1a, e). BMSCs-EAE from passage 3 onwards exhibited a different morphology compared to controls. In particular, the population comprised of mixed cell types such as spindle cells with thin and long processes and sharp edges and multiradial cells with extensive spherical nuclei (Fig. 1b, f). Moreover, the size of the BMSCs-control was significantly lower compared to that of BMSCs-EAE (passage 5) (ImageJ) (1,079 ± 298.8 vs. 1,915 ± 482.5, respectively, p = 0.0002).
BMSCs-control (a, d), BMSCs-EAE (b, e), and L929 (c, f) after 24 h (a–c) and 48 h (d–f) of observation after their subculturing, under phase contrast microscopy. Morphological differences between cell lines were observed since BMSCs-control rendered a more homogenous population and BMSCs-EAE had different morphology such as spindle cells with long processes and multiradial cells with extensive spherical nuclei. g Proliferation MTT OD results of BMSCs-control, BMSCs-EAE, and L929 cells after their culturing for a week. BMSCs-EAE exhibited increased proliferation rate compared to control cells (p = 0.0005)
Proliferation
Interestingly enough, the time needed to reach confluence between passages was much shorter in the EAE compared to control BMSCs (data not shown). This resulted in significantly higher proliferation rate in BMSCs-EAE compared to BMSCs-control (Kruskal–Wallis test, p = 0.0005, Fig. 1g) after a week of culturing in passage 5. This proliferation rate exceeded even the rate of the L929 fibroblastic cells (Fig. 1c, d).
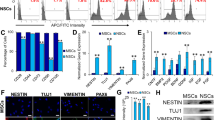
The proliferation rate was also studied using Ki-67/Mib-1 immunocytochemistry, which marks proliferating cells. Increased expression of the Mib-1-positive cells was observed in BMSCs-EAE compared to control cells (control 4.45 ± 1.43 % vs. EAE 26.16 ± 2.12 %, p = 0.0002) (Fig. 2a–c). In addition, when the cells were stained for cleaved caspase-3, an increased expression of this pro-apoptotic marker was identified in BMSCs-EAE compared to BMSCs-control (control 10.12 ± 0.76 % vs. EAE 15.23 ± 1.23 %, p = 0.0014) (Fig. 2c, d, f). Thus, increased proliferation along with increased apoptosis was evident for the BMSCs-EAE group.
Characterization of BMSCs
Fluorescence Activated Cell Sorting
FACS analysis indicated that both BMSCs-control and BMSCs-EAE stained positive for CD44 and Sca-1, whereas they were negative for the markers CD45, CD11c, CD11b, and CD73. No significant changes for any marker were identified between the two cell groups (data not shown). L929 cells served as control for the combination of the above markers. Representative data of the three independent experiments are summarized in Fig. 3.
Osteogenesis and Adipogenesis
Both BMSCs-EAE and BMSCs-control were able to differentiate into osteogenic lineage as identified with the von Kossa staining (Fig. 4a, c). However, the BMSCs-EAE exhibited significantly more intense osteogenesis with abundant mineral depositions (211,011.3 ± 10,271.7 AU) compared to BMSCs-control (15,344.3 ± 10,658 AU, p < 0.0001, Fig. 4e).
Osteogenic differentiation potential was assessed by culturing BMSCs-control and BMSC-EAE for 3 weeks with osteogenic mineralization medium and staining cells with von Kossa (a, c). The osteogenesis differentiation as evaluated with ImageJ (e). Both cell lines were able to differentiate into cells of the bone tissue with a higher prevalence of osteogenesis in BMSCs-EAE compared to controls (p < 0.0001). Adipogenic differentiation potential was assessed by culturing BMSCs-control and BMSC-EAE for 3 weeks with adipogenic induction medium and staining cells with Oil Red O (b, d). The adipogenesis differentiation as evaluated with ImageJ (f). A higher incidence of large lipid vacuoles was observed in BMSCs-EAE rather than in BMSCs-control (b, d; magnified fields)
In addition, as demonstrated by the Oil Red staining (Fig. 4b, d), both cell types were able to differentiate into the adipogenic lineage with BMSCs-EAE exhibiting a trend for more potent adipogenesis (72,948.5 ± 9,090.6 AU) compared to controls (49,777 ± 7,784.7 AU, p = 0.066, Fig. 4f). Interestingly enough, BMSCs-EAE exhibited large single or aggregated lipid vacuoles (lipid droplets) (diameter 16.3 ± 1.1 μm) which were absent from the corresponding BMSCs-control (diameter < 1 μm).
BMSCs-Related Gene Expression
BMSCs-EAE exhibited increased mRNA expression of TRIB2 (t test, p < 0.0001), IGF1 (t test, p = 0.0461), IGFBP7 (t test, p < 0.0001), and KCTD12 (t test, p = 0.0021) and a decreased PRG mRNA expression level (t test, p < 0.0001) compared to controls, whereas the expression level of MGP (t test, p = 0.0717) and LIF (t test, p = 0.0520) was not affected (Fig. 5).
mRNA relative expression of the genes TRIB2 (a), KCTD12 (b), MGP (c), PRG1 (d), IGFBP7 (e), IGF1 (f), and LIF (g) of the BMSCs-control and BMSCs-EAE using L929 as control. The expression of the above markers was used as an extra tool for the characterization of the mesenchymal cells. BMSCs-control and BMSCs-EAE although expressing the same genes, their levels of expression varied significantly in most cases (*p < 0.05, **p < 0.001, ***p < 0.0001)
Neuronal Cell Lineage Phenotypic Differentiation
The markers that exhibited variability between EAE and control BMSCs at passage 4 were PSA-NCAM (control 11.78 ± 5.9 vs. EAE 19.43 ± 5.7, p = 0.0021) and NeuN, which were markedly increased in BMSCs-EAE compared to controls (control 4.13 ± 0.72 vs. EAE 9.84 ± 0.51, p < 0.0001). On the contrary, NG2 was markedly increased in the control compared to BMSCs-EAE (9.21 ± 1.12 vs. 4.12 ± 0.84, correspondingly, p < 0.0001; Fig. 6). However, neither CNPase nor GFAP was detected in either studied groups (data not shown).
Neural differentiation factors (PSA-NCAM, NG2, and NeuN) immunocytochemistry (merged with DAPI) of undifferentiated BMSCs-control (a, c, e) and BMSCs-EAE (b, d, f). BMSCs-EAE exhibited increased expression of PSA-NCAM (p = 0.0021) and NeuN (p < 0.0001) and decreased expression of NG2 (p < 0.0001) compared to BMSCs-control (g–i)
The same staining procedure was performed following the self-differentiation protocol at the same passage (passage 4 following 3 weeks of starvation) of both EAE and control BMSCs. It was observed that the expression of mature glial markers GFAP and CNPase was similar between the groups (data not shown), whereas the mature neuronal marker NeuN was increased in the BMSCs-EAE group (control 0.82 ± 0.22 vs. EAE 13.46 ± 1.52, p < 0.0001). Furthermore, the expression of PSA-NCAM was increased in the BMSCs-EAE group after the self-differentiation protocol (control 7.24 ± 7.61 vs. EAE 17.17 ± 8.12, p = 0.0077), whereas the expression level of NG2 (control 4.05 ± 0.91vs. EAE 4.3 ± 1.02) was similar in both groups (Fig. 7).
Neural differentiation factors (PSA-NCAM, NG2, and NeuN immunocytochemistry, merged with DAPI) of spontaneously differentiated BMSCs-control (a, c, e) and BMSCs-EAE (b, d, f). BMSCs-EAE exhibited increased expression of the markers PSA-NCAM (p = 0.0077) and NeuN (p < 0.0001) compared to BMSCs-control (g, i). There was no statistically significant difference between BMSCs-control and BMSCs-EAE in the expression of NG2 (h)
The tumor suppressor protein p53 was detected neither in control nor in BMSCs-EAE, whereas the neurotrophic factor BDNF exhibited increased expression in undifferentiated control compared to BMSCs-EAE (control 7.28 ± 2.88 vs. EAE 1.35 ± 1.56, p < 0.0001). However, BDNF expression level was reversed after the spontaneous differentiation of the cells (control 0.49 ± 0.89 vs. EAE 8.74 ± 3.94, p < 0.0001) (Fig. 8).
Neurotrophic factor BDNF expression in BMSCs-control and BMSCs-EAE before (a, b) and after (c, d) spontaneous differentiation. BMSCs-EAE had lower expression of BDNF (p < 0.0001) compared to BMSCs-control before differentiation (e), whereas they exhibited higher expression (p < 0.0001) compared to BMSCs-control after self-differentiation (f)
Alterations in Histone-Modifying Genes Expression/Expression Pattern of Histone-Modifying Genes
We studied possible involvement of epigenetic mediators to the observed variability of cell characteristics. More specifically, we investigated the differences in the expression of HDAC1, HDAC2, HDAC3, PCAF, P300, and CBP between the two cell groups. There was increased expression of HDAC2, P300, and PCAF (p < 0.001), whereas a decrease in the expression of HDAC1 and CBP (p < 0.001) in the BMSCs-EAE compared to BMSCs-control was observed. However, the expression of HDAC3 was unaltered (p = 0.9889) in the two cell populations. In addition, no expression or minimum expression of the studied genes was noticed among the L929 cells (Fig. 9).
mRNA relative expression of histone deacetylases HDAC1 (a), HDAC2 (b), and HDAC3 (c) and histone acetyltransferases P300 (d), CBP (e), and PCAF (f) in BMSCs-control and BMSCs-EAE using L929 fibroblast-like cells as controls (***p < 0.0001). Differences in the expression pattern of the relevant epigenetic factors between the two cell lines are suggestive of a different mechanism of transcriptional regulation
Karyotype
Routine cytogenetic analysis of the BMSCs-control and the BMSCs-EAE, using G-band metaphase chromosomes, revealed normal phenotype with no apparent alterations in either group (40, XX) (Fig. 10).
Discussion
In the present study, we provided evidence for differences in the phenotypical and functional characteristics between EAE- and healthy-donor-derived BMSCs in vitro. Autologous BMSCs have been implied in several studies since the “immune privilege” of allogeneic BMSCs has been questioned (Eliopoulos et al. 2005; Nauta et al. 2006; Uccelli et al. 2007). Based on previous experimental studies (Gerdoni et al. 2007; Kassis et al. 2008; Zappia et al. 2005), autologous BMSC transplantation in MS is suggested and has already been performed in recent clinical studies (Karussis et al. 2010; Mohyeddin Bonab et al. 2007; Rice et al. 2010; Yamout et al. 2010) where the safety of the procedure has been identified. However, the experimental studies that have been performed may not clearly correspond to autologous BMSCs transplantation since the donors were healthy though not EAE-suffering inbred animals.
A number of comparison studies on the properties of BMSCs isolated from patients with neurodegenerative/autoimmune disorders and those derived from healthy donors have been reported. In particular, BMSCs derived from patients with Parkinson’s disease (Zhang et al. 2008), stroke, or brain injury (Zhang et al. 2007) have been reported to be similar to normal BMSCs. However, BMSCs derived from patients with the autoimmune disorder systemic sclerosis (SSc) exhibited increased expression of growth factors and chemokines associated with vascularization, a property which could be exploited for autologous cell therapy treatment in SSc (Cipriani et al. 2007). Other studies with patient- and healthy donor-derived BMSCs have reported no morphological changes whatsoever (Kastrinaki et al. 2008; Larghero et al. 2008).
In recent studies, BMSCs derived from MS patients have been examined in comparison to healthy controls. In 10 MS patients (five with relapsing–remitting and five with secondary progressive form of the disease), no differences with regard to proliferation, phenotype, or in vitro differentiation potential have been identified. However, difference in the interferon-inducible protein-10 (IP10) production was noticed in MS-BMSCs compared to control cells (Mazzanti et al. 2008). In a more recent study, BMSCs derived from five secondary progressive MS patients, plastic adherence, differentiation along adipogenic/osteogenic/chondrogenic lineages, proliferation, differentiation potential, and cell surface antigen expression were similar between MS patients and controls (Mallam et al. 2010).
Evidently, there is a controversy between our experimental data and those derived from recent human studies in MS (Mallam et al. 2010; Mazzanti et al. 2008). It may very well be that EAE is not MS or that murine BMSCs do not share completely the same properties with those of human origin. Alternatively, in human studies. BMSCs were probably derived from MS patients where the inflammatory process was not that pronounced at least at a level comparable to the one noticed in acute EAE. In addition, although most studies with MS patients have reported safety and preliminary evidence of efficacy of BMSCs, the number of participating patients was small. Larger clinical studies ought to be conducted to verify efficacy of this therapeutic approach (Uccelli et al. 2011a, b). Consequently, similar in vitro studies of BMSCs derived from selected patients with variable disease activity and concomitant inflammatory burden (progressive vs. relapsing forms of MS) would be of importance. Noteworthy, the rationale for phase I clinical trials with BMSC transplantation in MS was based on corresponding EAE studies where the animal model used was the same as the one performed in our study (Kassis et al. 2008; Gerdoni et al. 2007).
Cellular Morphology and Size
Our results indicated that BMSCs-EAE displayed a different morphology and size compared to controls. BMSCs, in general, comprise highly heterogeneous populations which tend to acquire a more homogenous picture as the culture passages progress. The dominant cell type after several passages is spindle-shaped with long processes. BMSCs-EAE apart from the typical cell type exhibited additional cell types such as multiradial cells with unusually long processes, large spherical nuclei, and generally occupied larger surface areas than BMSCs-control. These morphological inconsistencies were identifiable as soon as the cells entered the second passage onwards.
Mesenchymal Origin
Both cell groups complied with the generally accepted criteria of characterization of mesenchymal cells, in agreement with other previously published data with autologous patient- and healthy donor-derived BMSCs (Anjos-Afonso and Bonnet 2008). However, we identified a higher incidence of large lipid vacuoles in the BMSCs-EAE group and a prevalence of osteogenesis in the BMSCs-EAE cell group compared to controls. Moreover, on the basis that no general acceptance exists over the markers used in FACS analysis of nonhuman BMSCs (Anjos-Afonso and Bonnet 2008; Uccelli et al. 2006), we performed molecular analysis for the characterization of mesenchymal cells (Igarashi et al. 2007) where some significant differences were also evident between BMSCs-control and BMSCs-EAE. These markers are consistently expressed in high levels in the BMSCs and they are involved in the signal transduction via growth factors and/or cytokines (IGF1, LIF, IGFBP7) or in remodeling the extracellular matrix (MGP, PRG1) (Igarashi et al. 2007). Therefore, differences in their expression between the two groups of BMSCs may indicate potential changes in the correspondent pathways and need to be clarified in the future.
Proliferation Potential and Apoptosis
BMSCs-EAE were found to proliferate in higher rates than the controls by means of an MTT assay conducted after 1 week of expansion. Additionally, immunofluorescence for Mib1/Ki-67 exhibited elevated rates in comparison with the controls. These results are consistent with previously published data regarding the proliferation rate of BMSCs from patients with other autoimmune disorders, such as rheumatoid arthritis (Kastrinaki et al. 2008) and Crohn’s disease (Bernardo et al. 2009). The first survey of reference reported similar apoptosis rates between the cell groups under investigation, but that was not the case in the present study. Apoptosis rates measured by immunofluorescence for caspase-3 were significantly higher in BMSCs-EAE. High proliferation potential together with high apoptotic rates may be indicative of tumorigenic events (Gal et al. 2004; Snibson et al. 1999). However, in our set of experiments, the negative immunofluorescence for tumor suppressor protein p53 and the absence of karyotypic abnormalities for both cell groups indicated no potential DNA damages.
BMSCs Transdifferentiation Towards the Neuronal/Glial Lineage Phenotype
Another characteristic property of BMSCs is their ability to transdifferentiate into cells expressing neural phenotypes (Black and Woodbury 2001; Dezawa et al. 2004; Mahmood et al. 2003; Mezey et al. 2000; Sanchez-Ramos et al. 2000; Woodbury et al. 2000) either by special inductive media or spontaneously after prolonged culture (Li et al. 2007; Tseng et al. 2007). Interestingly enough, differences in the expression pattern of neural markers were also evident between the two groups of cells, thereby indicating another difference in the intrinsic characteristics of the BMSCs when exposed to the EAE environment. In particular, BMSCs-EAE were shown to express higher levels of neuronal phenotypes indicative for both neuronal progenitors and mature neurons, whereas BMSCs-control expressed higher levels of developing and adult oligodendrocyte precursor cells. However, it is questionable whether these changes (a) may also indicate a functional—not purely phenotypic—differentiation of BMSCs and (b) may be of any clinical relevance since the benefit following BMSC transplantation in EAE or in MS is not related to their transdifferentiation potential (Uccelli et al. 2006; Tyndall and Uccelli 2009).
Evidently, the EAE-inflammatory environment imposed changes in a number of phenotypic and functional characteristics of BMSCs. In order to further confirm the impact of the inflammatory environment in the overall biology of BMSCs, we investigated the HATs and HDACs gene expression. These histone-modifying genes are vulnerable to environmental factors (Khan and Khan 2010) and regulate gene expression in many ways, either by forming repressor/activator complexes (Ogryzko 2001) or by interacting/acting directly with transcription factors (Lin et al. 2006). They also interact with nonhistone proteins, thus playing a pivotal role in various mechanisms such as cell cycle, proliferation, apoptosis, and differentiation (Minucci and Pelicci 2006). The mRNA expression levels of three members of the HDAC (HDAC1, HDAC2, HDAC3) and the HAT family (CBP, P300, PCAF) were examined in BMSCs-EAE and BMSCs-control. Marked differences in the expression pattern displayed by BMSCs-EAE when compared to controls may be ascribed to the specific inflammatory environment from which they originated. Therefore, the possibility that histone acetylation changes may also be attributed to inflammatory environmental factors in EAE might not totally be excluded. Moreover, it is known that an inflammatory environment can itself be a trigger for epigenetic reprogramming (Backdahl et al. 2009). Similarly, epigenetic changes in CD4+ T cells from patients with active systemic lupus erythematosus have been identified (Hu et al. 2008). However, whether the HATs and HDACs gene expression changes noticed in the current study may or may not be part of an ongoing epigenetic mechanism needs to be clarified.
Overall, our in vitro study on the biological properties of BMSCs from EAE-suffering animals suggests a profound impact of the inflammatory environment of the cells compared to their healthy donor-derived counterparts despite the fact that BMSCs residing in the bone marrow are not directly implicated in the disease process. Distinct morphology, size, higher potency for adipogenesis and osteogenesis combined with higher proliferation/apoptosis rates and a tendency for neuronal phenotypes rather than oligodendrocyte precursor and mature cells’ phenotype were found for in vitro expanded BMSCs-EAE. In view of the ex vivo expansion and subsequent manipulation of the BMSCs as a necessary step prior to transplantation, a lot of parameters need to be taken into consideration. Interestingly enough, recent studies in primary BMSCs also suggest that distinct morphology and size combined with more potent adipogenesis might reflect the presence of various subsets of MSCs with differential functional properties or potency to a specific lineage (Rasini et al. 2013) thereby indicating intrinsic variability of the BMSC phenotype and/or environmental factors. In future studies, the properties of such defined MSC subpopulations (engraftment, differentiation potential, rates of proliferation and apoptosis and gene expression patterns of cytokines, growth factors, histone modifiers) from EAE-induced animals or MS patients will need to be explored in an in vivo setting to possibly exploit them in therapeutic interventions.
References
Anjos-Afonso F, Bonnet D (2008) Isolation, culture, and differentiation potential of mouse marrow stromal cells. Curr Protoc Stem Cell Biol. Chapter 2: Unit 2B.3
Backdahl L, Bushell A, Beck S (2009) Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int J Biochem Cell Biol 41(1):176–184
Baksh D, Song L, Tuan RS (2004) Adult mesenchymal stem cells: characterization, differentiation, and application in cell and gene therapy. J Cell Mol Med 8(3):301–316
Bernardo ME, Avanzini MA, Ciccocioppo R et al (2009) Phenotypical/functional characterization of in vitro-expanded mesenchymal stromal cells from patients with Crohn's disease. Cytotherapy 11(7):825–836
Black IB, Woodbury D (2001) Adult rat and human bone marrow stromal stem cells differentiate into neurons. Blood Cells Mol Dis 27(3):632–636
Cipriani P, Guiducci S, Miniati I et al (2007) Impairment of endothelial cell differentiation from bone marrow-derived mesenchymal stem cells: new insight into the pathogenesis of systemic sclerosis. Arthritis Rheum 56(6):1994–2004
Dezawa M, Kanno H, Hoshino M et al (2004) Specific induction of neuronal cells from bone marrow stromal cells and application for autologous transplantation. J Clin Invest 113(12):1701–1710
Dominici M, Le Blanc K, Mueller I et al (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8(4):315–317
Eliopoulos N, Stagg J, Lejeune L, Pommey S, Galipeau J (2005) Allogeneic marrow stromal cells are immune rejected by MHC class I- and class II-mismatched recipient mice. Blood 106(13):4057–4065
Freedman MS, Bar-Or A, Atkins HL et al (2010) The therapeutic potential of mesenchymal stem cell transplantation as a treatment for multiple sclerosis: consensus report of the International MSCT Study Group. Mult Scler 16(4):503–510
Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV (1974) Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation 17(4):331–340
Gal AA, Sheppard MN, Nolen JD, Cohen C (2004) p53, cellular proliferation, and apoptosis-related factors in thymic neuroendocrine tumors. Mod Pathol 17(1):33–39
Gerdoni E, Gallo B, Casazza S et al (2007) Mesenchymal stem cells effectively modulate pathogenic immune response in experimental autoimmune encephalomyelitis. Ann Neurol 61(3):219–227
Gordon D, Pavlovska G, Uney JB, Wraith DC, Scolding NJ (2010) Human mesenchymal stem cells infiltrate the spinal cord, reduce demyelination, and localize to white matter lesions in experimental autoimmune encephalomyelitis. J Neuropathol Exp Neurol 69(11):1087–1095
Grigoriadis N (2002) Interferon beta treatment in relapsing-remitting multiple sclerosis. A review. Clin Neurol Neurosurg 104(3):251–258
Grigoriadis N, Lourbopoulos A, Lagoudaki R et al (2011) Variable behavior and complications of autologous bone marrow mesenchymal stem cells transplanted in experimental autoimmune encephalomyelitis. Exp Neurol 230(1):78–89
Hu N, Qiu X, Luo Y et al (2008) Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol 35(5):804–810
Igarashi A, Segoshi K, Sakai Y et al (2007) Selection of common markers for bone marrow stromal cells from various bones using real-time RT-PCR: effects of passage number and donor age. Tissue Eng 13(10):2405–2417
Jiang Y, Jahagirdar BN, Reinhardt RL et al (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418(6893):41–49
Karussis D, Karageorgiou C, Vaknin-Dembinsky A et al (2010) Safety and immunological effects of mesenchymal stem cell transplantation in patients with multiple sclerosis and amyotrophic lateral sclerosis. Arch Neurol 67(10):1187–1194
Kassis I, Grigoriadis N, Gowda-Kurkalli B et al (2008) Neuroprotection and immunomodulation with mesenchymal stem cells in chronic experimental autoimmune encephalomyelitis. Arch Neurol 65(6):753–761
Kastrinaki MC, Sidiropoulos P, Roche S et al (2008) Functional, molecular and proteomic characterisation of bone marrow mesenchymal stem cells in rheumatoid arthritis. Ann Rheum Dis 67(6):741–749
Khan SN, Khan AU (2010) Role of histone acetylation in cell physiology and diseases. Clin Chim Acta 411:1401–1411
Larghero J, Farge D, Braccini A et al (2008) Phenotypical and functional characteristics of in vitro expanded bone marrow mesenchymal stem cells from patients with systemic sclerosis. Ann Rheum Dis 67(4):443–449
Li N, Yang H, Lu L, Duan C, Zhao C, Zhao H (2007) Spontaneous expression of neural phenotype and NGF, TrkA, TrkB genes in marrow stromal cells. Biochem Biophys Res Commun 356(3):561–568
Lin HY, Chen CS, Lin SP, Weng JR, Chen CS (2006) Targeting histone deacetylase in cancer therapy. Med Res Rev 26:397–413
Lourbopoulos A, Grigoriadis N, Lagoudaki R et al (2011) Administration of 2-arachidonoylglycerol ameliorates both acute and chronic experimental autoimmune encephalomyelitis. Brain Res 1390:126–141
Mahmood A, Lu D, Lu M, Chopp M (2003) Treatment of traumatic brain injury in adult rats with intravenous administration of human bone marrow stromal cells. Neurosurgery 53(3):697–702
Mallam E, Kemp K, Wilkins A, Rice C, Scolding N (2010) Characterization of in vitro expanded bone marrow-derived mesenchymal stem cells from patients with multiple sclerosis. Mult Scler 16(8):909–918
Martino G, Franklin RJ, Van Evercooren AB, Kerr DA (2010) Stem cell transplantation in multiple sclerosis: current status and future prospects. Nat Rev Neurol 6(5):247–255
Mazzanti B, Aldinucci A, Biagioli T et al (2008) Differences in mesenchymal stem cell cytokine profiles between MS patients and healthy donors: implication for assessment of disease activity and treatment. J Neuroimmunol 199(1–2):142–150
Mezey E, Chandross KJ, Harta G, Maki RA, McKercher SR (2000) Turning blood into brain: cells bearing neuronal antigens generated in vivo from bone marrow. Science 290(5497):1779–1782
Minucci S, Pelicci PG (2006) Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6:38–51
Mohyeddin Bonab M, Yazdanbakhsh S, Lotfi J et al (2007) Does mesenchymal stem cell therapy help multiple sclerosis patients? Report of a pilot study. Iran J Immunol 4(1):50–57
Nauta AJ, Westerhuis G, Kruisselbrink AB, Lurvink EG, Willemze R, Fibbe WE (2006) Donor-derived mesenchymal stem cells are immunogenic in an allogeneic host and stimulate donor graft rejection in a nonmyeloablative setting. Blood 108(6):2114–2120
Ogryzko VV (2001) Mammalian histone acetyltransferases and their complexes. Cell Mol Life Sci 58:683–692
Rasini V, Dominici M, Kluba T et al (2013) Mesenchymal stromal/stem cells markers in the human bone marrow. Cytotherapy. doi:10.1016/j.jcyt.2012.11.009
Rice CM, Mallam EA, Whone AL et al (2010) Safety and feasibility of autologous bone marrow cellular therapy in relapsing-progressive multiple sclerosis. Clin Pharmacol Ther 87(6):679–685
Rizvi AZ, Swain JR, Davies PS et al (2006) Bone marrow-derived cells fuse with normal and transformed intestinal stem cells. Proc Natl Acad Sci U S A 103(16):6321–6325
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana, Totowa, pp 365–386
Sanchez-Ramos J, Song S, Cardozo-Pelaez F et al (2000) Adult bone marrow stromal cells differentiate into neural cells in vitro. Exp Neurol 164(2):247–256
Schwartz RE, Reyes M, Koodie L et al (2002) Multipotent adult progenitor cells from bone marrow differentiate into functional hepatocyte-like cells. J Clin Invest 109(10):1291–1302
Snibson KJ, Bhathal PS, Hardy CL, Brandon MR, Adams TE (1999) High, persistent hepatocellular proliferation and apoptosis precede hepatocarcinogenesis in growth hormone transgenic mice. Liver 19(3):242–252
Tseng PY, Chen CJ, Sheu CC, Yu CW, Huang YS (2007) Spontaneous differentiation of adult rat marrow stromal cells in a long-term culture. J Vet Med Sci 69(2):95–102
Tyndall A, Uccelli A (2009) Multipotent mesenchymal stromal cells for autoimmune diseases: teaching new dogs old tricks. Bone Marrow Transplant 43(11):821–828
Uccelli A, Moretta L, Pistoia V (2006) Immunoregulatory function of mesenchymal stem cells. Eur J Immunol 36(10):2566–2573
Uccelli A, Pistoia V, Moretta L (2007) Mesenchymal stem cells: a new strategy for immunosuppression? Trends Immunol 28(5):219–226
Uccelli A, Morando S, Bonanno S, Bonanni I, Leonardi A, Mancardi G (2011a) Mesenchymal stem cells for multiple sclerosis: does neural differentiation really matter? Curr Stem Cell Res Ther 6(1):69–72
Uccelli A, Laroni A, Freedman M (2011b) Mesenchymal stem cells for the treatment of multiple sclerosis and other neurological diseases. Lancet Neurol 10:649–656
Woodbury D, Schwarz EJ, Prockop DJ, Black IB (2000) Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res 61(4):364–370
Yamout B, Hourani R, Salti H et al (2010) Bone marrow mesenchymal stem cell transplantation in patients with multiple sclerosis: a pilot study. J Neuroimmunol 227(1–2):185–189
Zappia E, Casazza S, Pedemonte E et al (2005) Mesenchymal stem cells ameliorate experimental autoimmune encephalomyelitis inducing T-cell anergy. Blood 106(5):1755–1761
Zhang ZX, Guan LX, Zhang K et al (2007) Cytogenetic analysis of human bone marrow-derived mesenchymal stem cells passaged in vitro. Cell Biol Int 31(6):645–648
Zhang Z, Wang X, Wang S (2008) Isolation and characterization of mesenchymal stem cells derived from bone marrow of patients with Parkinson's disease. Vitro Cell Dev Biol Anim 44(5–6):169–177
Acknowledgments
This work was supported by a Sanofi Aventis research grant. We thank Dr. T. Papadopoulos, Dr. T. Kelly, and Dr. E. Polyzoidou for their technical assistance and comments on various topics of the paper.
Conflict of Interest
No competing financial interests exist.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dimitra Zacharaki and Roza Lagoudaki contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table 1
Primer pair sequences designed for real-time PCR of stromal markers and histone-modifying genes. The table displays the corresponding primer sequences (forward and reverse). The size of each primer in base pairs (bp) is reported next to the corresponding gene name, within brackets. (PDF 121 kb)
Rights and permissions
About this article
Cite this article
Zacharaki, D., Lagoudaki, R., Touloumi, O. et al. Characterization of In Vitro Expanded Bone Marrow-Derived Mesenchymal Stem Cells Isolated from Experimental Autoimmune Encephalomyelitis Mice. J Mol Neurosci 51, 282–297 (2013). https://doi.org/10.1007/s12031-013-9992-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-013-9992-9