Abstract
Cyclin-dependent kinase (Cdk) 5 is critical for central nervous system development and neuron-specific functions including neurite outgrowth as well as synaptic function and plasticity. Cdk5 activity requires association with one of the two regulatory subunits, called p35 and p39. p35 redistribution as well as misregulation of Cdk5 activity is followed by cell death in several models of neurodegeneration. Posttranslational protein modification by small ubiquitin-related modifier (SUMO) proteins (sumoylation) has emerged as key regulator of protein targeting and protein/protein interaction. Under cell-free in vitro conditions, we found p35 covalently modified by SUMO1. Using both biochemical and FRET-/FLIM-based approaches, we demonstrated that SUMO2 is robustly conjugated to p35 in cells and identified the two major SUMO acceptor lysines in p35, K246 and K290. Furthermore, different degrees of oxidative stress resulted in differential p35 sumoylation, linking oxidative stress that is encountered in neurodegenerative diseases to the altered activity of Cdk5. Functionally, sumoylation of p35 increased the activity of the p35/Cdk5 complex. We thus identified a novel neuronal SUMO target and show that sumoylation is a likely candidate mechanism for the rapid modulation of p35/Cdk5 activity in physiological situations as well as in disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cdk5 as member of the Cdk family has a unique role not only due to its non-cyclin activators p35 and p39 but also because it fulfills functions distinct from the cell-cycle regulation by other Cdks (Dhavan and Tsai 2001; Su and Tsai 2011; Tang et al. 1997; van den Heuvel and Harlow 1993). Despite ubiquitous Cdk5 protein expression, Cdk5 activity is largely restricted to the central nervous system, because the homologous Cdk5 activators p35 and p39, which are the functional counterpart of the cyclins, are mostly expressed in neurons (Tang et al. 1995). Cdk5 is physiologically implicated in numerous neuron-specific functions including cytoskeletal dynamics, axonal outgrowth, synaptic plasticity, vesicle regulation, and neuronal migration (Su and Tsai 2011; Chae et al. 1997; Nikolic et al. 1996; Zukerberg et al. 2000; Hsiao et al. 2014; Verstegen et al. 2014).
Under pathological circumstances, Cdk5 turned out to be an inducer of neuronal cell death. Cdk5 neurotoxicity was first demonstrated in the context of amyloid-β toxicity, where proteolytic cleavage of p35 to the C-terminal fragment p25 led to redistribution and overactivation of Cdk5 after the formation of a neurotoxic p25/Cdk5 complex (Patrick et al. 1999; Patzke and Tsai 2002). Similarly, a central role of deregulated Cdk5 for neuronal demise could be demonstrated in many other in vitro and in vivo models for neurodegenerative diseases, e.g., amyotrophic lateral sclerosis, Parkinson’s disease, Huntington’s disease, or in stroke models (Su and Tsai 2011; Tan et al. 2003; Nguyen et al. 2001; Osuga et al. 2000; Smith et al. 2003; Weishaupt et al. 2003).
Few data exist on the molecular basis for posttranslational regulation of p35 or p25. It is known that Cdk5 phosphorylates p35, targeting its own activator for degradation (Patrick et al. 1998). p35 and its proteolytic fragment p25 are differentially distributed within the cell, due to a myristoylation site present only in the N-terminus of p35 that targets most of the p35 protein pool to cell membranes (Patrick et al. 1999; Asada et al. 2008). Beyond these studies, little is known about further p35 posttranslational modifications.
While SUMO can impact protein function through both covalent and non-covalent interactions, “sumoylation” generally refers to covalent modification. Four different SUMO proteins, SUMO1 to SUMO4, are encoded by the human genome. Like ubiquitination, sumoylation results in an isopeptide bond between the C-terminus of the SUMO protein and the ε-amino group of a lysine in the target protein. Sumoylation requires E1-activating enzymes, an E2-conjugating enzyme, and SUMO E3 ligases. Due to two classes of specific isopeptidases, called sentrin-specific proteases (SENP) and de-sumoylating isopeptidases (DeSI), sumoylation is a reversible and very dynamic process (Gong et al. 2000; Mukhopadhyay and Dasso 2007; Shin et al. 2012). SUMO conjugation was first described for the nuclear pore protein RanGAP1, (Mahajan et al. 1997; Matunis et al. 1996) and turned out to be necessary for nuclear translocation of numerous proteins. Meanwhile, we know that sumoylation provides regulatory mechanisms for controlling activity, stability, protein interactions, and subcellular localization of target proteins. Interestingly, several studies point to a role for SUMO during oxidative stress, a condition attributed to neurodegenerative diseases (Eckermann 2013; Feligioni and Nistico 2013; Krumova and Weishaupt 2013).
The dynamic regulation of the activity and subcellular distribution of the p35/Cdk5 complex as well as the presence of potential consensus SUMO acceptor sites (ψKxE, with ψ being an aliphatic branched amino acid; x being any amino acid) within the p35 sequence provoked us to test the hypothesis if p35/Cdk5 is regulated by sumoylation.
Materials and Methods
Antibodies and Chemicals
The following primary antibodies were used: anti-p35 (C-19), anti-Cdk5 (C-8), anti-myc (9E10), and normal mouse IgG (sc-2025) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-HA.11 (16B12) from Covance (Princeton, NJ), anti-SUMO2/3 (ab81371, clone 8A2), anti-Histone H1.0, and anti-Histone H1 (phospho) from abcam (Cambridge, UK). Secondary antibodies horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit IgGs were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Protease inhibitor mix was purchased from Roche (Basel, Switzerland).
In Vitro Sumoylation Assay with Recombinant p35
SUMO assays were performed as previously described (Krumova et al. 2011).
Cell Transfections and Oxidative Stress Experiments
HEK293 cells were transfected at 70–80 % confluency using the DNA–calcium phosphate precipitation method. In brief, DNA was diluted in 250 mM CaCl2, mixed with an equal volume of 2 × HBS solution (280 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM glucose monohydrate, 50 mM HEPES, pH 7.05), and incubated for 50 s at room temperature to allow the formation of precipitates. The reaction was stopped with pre-warmed Dulbecco’s Modified Eagle’s Medium supplemented with 2 % fetal bovine serum. 10–12 h after transfection, cells were cultured for additional 24 h in Dulbecco’s Modified Eagle’s Medium supplemented with 10 % fetal bovine serum and penicillin/streptomycin prior lysis.
CSM14.1 cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol.
For oxidative stress experiments, cells were treated with different hydrogen peroxide concentrations (0.005, 0.01, 0.1, 1, 10 mM) in serum-free Dulbecco’s Modified Eagle’s Medium for 30 min prior lysis or fixation.
Nickel–Nitrilotriacetic Acid (Ni2+–NTA) Affinity Chromatography
HEK293 cells expressing His6-tagged SUMO1 or SUMO2 and wild type (wt) or mutant p35 were lysed in guanidine buffer (6 M Guanidine-HCl, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl, pH 8, 10 mM imidazole, 10 mM N-Ethylmaleimide, 1 µg/ml Aprotinin, 1 µg/ml Pepstatin A, 1 µg/ml Leupeptin) 36 h after transfection. Ni2+–NTA affinity chromatography was performed as previously described (Jaffray and Hay 2006) with minor modifications. Lysates were incubated with Ni2+–NTA beads (Qiagen, Valencia, CA) for 2 h. Washing buffers were supplemented with 10 mM imidazole, 10 mM N-Ethylmaleimide, 1 µg/ml Aprotinin, 1 µg/ml Pepstatin A, and 1 µg/ml Leupeptin. Proteins were recovered in elution buffer (8 M Urea, 100 mM NaH2PO4/Na2HPO4, 10 mM Tris–HCl, pH 6.3, 10 mM N-Ethylmaleimide, 1 µg/ml Aprotinin, 1 µg/ml Pepstatin A, 1 µg/ml Leupeptin, 250 mM imidazole).
Co-immunoprecipitation
We used Cdk5 and p35wt or various p35 mutants for transfections. HEK293 cells were lysed in IP buffer (phosphate buffered saline (PBS) pH 7.4, 1 % Nonidet P-40, 0.5 % sodium deoxycholate, 1 mM EDTA, 1 mM EGTA, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol and protease inhibitors). Lysates were incubated for 10 min on ice and centrifuged for 5 min at 6,000×g and 4 °C. Protein concentrations were measured by bicinchoninic acid protein assay (Pierce, Rockford, USA). For preclearance, 400 µg of protein was incubated with 60 µl protein G-Sepharose 4B beads (Invitrogen, Carlsbad, CA) for 1 h at 4 °C. The immunoprecipitation was performed with 3 µg anti-p35 antibody for 4 h at 4 °C. Protein G-Sepharose prewashed in RIPA buffer (20 mM NaH2PO4/Na2HPO4 pH 7.4, 150 mM NaCl, 1 % Triton X-100, 0.5 % sodium deoxycholate, 0.1 % sodium dodecyl sulfate (SDS) and protease inhibitors) was incubated overnight at 4 °C with the antibody–protein mix. Beads were washed three times in RIPA buffer and boiled with 40 µl SDS-containing sample buffer for 5 min at 95 °C. Similarly, co-immunoprecipitation from untransfected HeLa cells was performed using 2 mg protein, SUMO2/3 antibody at 1:100 dilution, and normal mouse IgG at 1:50 dilution. Cell lysis, preclearance, and washing of protein G-Sepharose were performed in native buffer (300 mM NaCl, 50 mM NaH2PO4/Na2HPO4 pH 7.4, 10 mM N-Ethylmaleimide, 0.5 % Triton X-100, and protease inhibitors).
Cdk5 Activity Assay
His6-Cdk5 in complex with HA-p35wt, HA-p35-SUMO2, or HA-SUMO2-p35 fusion proteins was purified by Ni2+–NTA affinity chromatography under native and kinase activity-stabilizing conditions from HEK293 cell lysates. Unbound material was removed with washing buffer (40 mM 3-(N-morpholino)propanesulfonic acid, 1 mM EDTA, 1 mM Na3OV4, 1 mM phenylmethylsulfonyl fluoride, 10 mM imidazole, and protease inhibitors). p35/Cdk5 complexes were eluted with 20 mM Tris, 150 mM NaCl, and 250 mM imidazole. The activity assay was performed with 2.5 µg kinase complex in an assay volume of 25 µl. The assay buffer (10 mM Tris, 5 mM MgCl2, 25 mM HEPES, 0.025 % Nonidet P-40, 5 % Glycerol) was supplemented to final concentrations of 0.01 mg/ml bovine serum albumin, 0.5 mM dithiothreitol, 0.1 mg/ml Histone H1 (New England Biolabs, Ipswich, MA), protease, and phosphatase inhibitors. The reaction was initiated by addition of ATP (final concentration of 0.4 mM, New England Biolabs, Ipswich, MA) at 30 °C and stopped after 45 min with 2x SDS sample buffer.
SDS-PAGE and Western Blotting
Proteins were separated by sodium dodecylsulfate polyacrylamide gel electrophoresis on 12 % Tris–Glycine gels and detected by Western blotting on nitrocellulose membranes with antibodies of the following dilutions: Primary antibodies anti-p35 and anti-Cdk5 at 1:300, Histone H1 and phospho-Histone H1 at 1:500; secondary antibodies goat anti-mouse IgG-HRP at 1:4,000 and goat anti-rabbit IgG-HRP at 1:3,000 (Santa Cruz Biotechnology, Santa Cruz, CA).
Fluorescence Lifetime Imaging Microscopy (FLIM)/Förster Resonance Energy Transfer (FRET)
CSM14.1 cells were transfected with mTFP-p35 (monomeric teal fluorescent protein) constructs, Cdk5-mVenus, or mVenus-SUMO2 using Lipofectamine 2000 according to the manufacturer’s protocol. Two different SUMO constructs were used: the mature SUMO with C-terminal diglycine motif (mVenus-SUMO2) and the conjugation-deficient mutant (mVenus-SUMO2Δ7) as negative control. Cells were fixed 24 h posttransfection using PBS/4 % paraformaldehyde for 15 min, subsequently washed in PBS, and mounted in Mowiol. The fluorescence lifetime measurements were made using a Leica DMI 6000 inverted confocal microscope equipped with a Plan Fluor 63 X NA 1.32 Oil mm objective lens. The microscope was coupled to the Mai Tai HP with an integrated Spectra-Physics 14 W Millennia® pump laser provides more than 300 nm (690–1,040 nm) in useable tuning range with over 2.5 W of average power and a pulse width of less than 120 femtoseconds. Photons were acquired using TCSPC photon-counting module board (SPC-830, Becker and Hickl GmbH, Berlin, Germany). The photon collection was carried out using a high-sensitive HPM 100-50 hybrid PMT, with relatively less or no after pulse. mTFP donor was excited using 840 nm at a two-photon mode and the photons were collected using the filter set ET 480/40 M emission range. Photons were collected for approximately 120–180 s with the collection threshold of 1,000–1,500 Photons per pixel. The fluorescence transients were acquired using SPCIMAGE software (Becker and Hickl), exported, and analyzed using an in-house-developed ImageJ algorithm for single and multi-exponential decay. Histograms for the weighted mean average of changes in fluorescent lifetime were derived from ImageJ and statistically analyzed for the area under the curve and for frequency of distribution using Igor Pro (Wave matrics).
Results
In Vitro Sumoylation of p35
In order to investigate whether human p35 is a target for sumoylation, we first used a cell-free in vitro sumoylation assay. Recombinant glutathione S-transferase-tagged p35 in complex with Cdk5 (Millipore, Temecula, CA) was incubated with SUMO1 or SUMO2, E1-activating enzyme Aos1/Uba2 (Desterro et al. 1999), the E2-ubiquitin-conjugating enzyme 9 (Ubc9), and various E3 ligases (protein inhibitor of activator STAT (Pias)Xα, Pias1, Pias3, Ran binding protein 2 (BP2) or the catalytically active fragment of RanBP2 (IR1+M)) in the presence or absence of ATP. After incubation for 30 min at 30 °C, samples were analyzed by Western blotting. Incubation with BP2 or its catalytic fragment IR1 resulted in robust SUMO1 conjugation of p35 as indicated by the higher molecular weight species immunopositive for p35 (Fig. 1a, arrow). Weaker SUMO1 modification was also observed with the E3 ligase PiasXα. p35 can be phosphorylated by p35/Cdk5 activity at four phosphorylation sites (Patrick et al. 1998). Addition of ATP thus resulted in an expected minor band shift of p35 that most likely results from p35 phosphorylation (Fig. 1a, b). p35 was covalently modified by SUMO1, but not SUMO2 under the above in vitro conditions (Fig. 1b).
Sumoylation of the Cdk5 activator p35. Recombinant glutathione S-transferase-p35/Cdk5 was incubated with Aos1/Uba2 (E1-activating enzyme), Ubc9 (E2-conjugating enzyme), various E3 ligases and ATP in the presence of either a SUMO1 or b SUMO2, both with a molecular weight of 11 kDa. Samples were analyzed by SDS-PAGE and immunoblotting with anti-p35 antibody. Addition of ATP led to a minor band shift of p35 due to the expected phosphorylation of p35 by Cdk5. a Incubation with the E3 ligase BP2, its catalytic fragment IR1, and PiasXα resulted in an additional band shift due to conjugation of SUMO1 to p35 (arrow). b A covalent binding of SUMO2 to p35 could not be observed under in vitro conditions. c SUMO and SUMO conjugates were purified by denaturing Ni2+–NTA chromatography from lysates of HEK293 cells expressing His-SUMO1 or His-SUMO2 and p35wt. Samples were subjected to SDS-PAGE and analyzed with an anti-p35 antibody. The appearance of ~55 kDa species immunopositive for p35 (arrow) indicates covalent modification of p35 by SUMO2 in cells. d Co-immunoprecipitation from untransfected HeLa cells was performed in order to investigate endogenous sumoylation of human p35. 2 mg total protein from HeLa cell lysates were incubated with SUMO2 antibody, normal mouse IgG control or omitting antibody. Samples were subjected to SDS-PAGE and immunoblotting with p35 (C-19) antibody. To confirm the size of endogenous sumoylated p35, lysates from cells transfected with p35 and the fusion proteins SUMO2-p35 and p35-SUMO2 were separated in neighboring lanes
In the next step, we analyzed whether p35 is SUMO-conjugated in cells. We co-transfected HEK293 cells with plasmids encoding myc-tagged human p35 and His-tagged SUMO1 or SUMO2 and purified SUMO-conjugated species by Ni2+–NTA affinity chromatography under denaturing conditions. As shown in Fig. 1c, p35 was covalently modified by SUMO2 as indicated by the presence of higher molecular mass species (arrow) that are immunopositive for p35. Despite denaturing conditions and supplementation of imidazole, low non-specific binding of p35 to Ni2+–NTA beads was observed throughout all experiments.
Endogenous p35 Sumoylation
To further explore whether the interaction between human p35 and SUMO2 occurs endogenously, we performed co-immunoprecipitation (co-IP) from untransfected HeLa cells (Fig. 1d). We immunoprecipitated SUMO with a specific SUMO2/3 antibody and detected co-immunoprecipitated p35 with p35 (C-19) antibody (arrow). p35 is expressed only at very low levels in HeLa cells and was not detected in whole-cell lysates. To verify the presence and size of SUMO-modified p35 species compared to unsumoylated p35, we separated lysates from transfected cells expressing p35wt and fusion proteins SUMO2-p35 and p35-SUMO2 in neighboring lanes. Using p35 (C-19) antibody to immunoprecipitate p35, we failed to detect SUMO-positive p35 species with a SUMO2/3-specific antibody. This is most likely due to the very transient nature of SUMO modification, leading to only a small fraction of sumoylated p35 at a given timepoint and failure to detect p35-SUMO when using a pan-p35 antibody for immunoprecipitation, especially in cells with an overall low p35 expression.
Multiple Acceptor Sites in p35
p35 has 23 lysine residues some of which have a high probability of sumoylation according to the computational program SUMOplot™ (Abgent), but only one of them, K290, fully matches the consensus criteria for the SUMO acceptor site ψKxE (Tatham et al. 2001). A related motif with the second highest computed probability of sumoylation encloses K246. Interestingly, these two suspected sumoylation sites in p35 are largely conserved throughout evolution. We found that even in invertebrates for which p35 homologous proteins or transcripts are available (e.g., Drosophila melanogaster, Sphaerechinus granularis, or Caenorhabditis elegans), the two potential SUMO acceptor sites were either in exact keeping with (K246) or closely related to (K290) the canonical sumoylation motif.
Therefore, in order to experimentally find out whether a single of these two lysine residues serves as SUMO acceptor site in p35, we generated p35 lysine-to-arginine mutants, which were co-transfected with His-SUMO2 in HEK293 cells to determine their sumoylation status. In fact, single mutations of lysine 246 or 290 significantly reduced the conjugation of SUMO2 to the mutant variants of p35 compared with p35wt. Moreover, a double mutant with a combination of the two lysine-to-arginine mutations (named p35_2KR) abolished sumoylation when co-expressed with SUMO2 in HEK293 cells (Fig. 2a).
p35 has two main sumoylation sites. HEK293 cells co-expressing His-SUMO2 and p35wt or the indicated lysine-to-arginine mutants were used to identify the sumoylation sites of p35. Single mutation of lysine 246 or 290 resulted in a significant reduction of the sumoylation levels (arrow). A combination of both mutations in the double mutant 2KR fully abrogated the sumoylation. b Disrupting the sumoylation consensus motifs by exchange of the amino acid at the fourth position within both SUMO acceptor sites in the double mutant p35_2AA (F248A and E292A) also resulted in sumoylation-deficiency of p35 (arrow). c Plasmids encoding His-Ubiquitin and p35wt or p35_2KR were co-transfected in HEK293 cells. Ubiquitination levels (arrow: mono-ubiquitination, bracket: poly-ubiquitinated species) are comparable between p35wt and the SUMO-deficient mutant 2KR suggesting that ubiquitin is not targeted to the lysine residues 246 or 290. d Sumoylation of p35 was measured by FLIM in the neuronal CSM14.1 cell line. The distribution of the mTFP donor constructs (mTFP intensity pictures in gray) is shown in the upper panel. Lifetime images mapping spatial FRET in cells (lower panel) are depicted using a pseudocolor scale with warmer colors indicating lower lifetimes, that is, higher FRET efficiencies, and cooler colors indicating higher lifetimes, that is, lower FRET efficiencies. Typical mTFP fluorescence lifetimes (Day et al. 2008) were observed in cells expressing the mTFP-p35wt donor construct only. Co-expression of mTFP-p35wt with mVenus-SUMO2 resulted in strongly shortened lifetimes suggestive of interaction between p35 and SUMO2. Co-expression of the sumoylation-deficient mutants p35_2KR and p35_2AA or co-expression of p35wt with the conjugation-deficient mutant SUMO2Δ7 led only to a minor reduction in fluorescence lifetime. Statistical observations are represented in a distribution histogram. The FRET efficiencies are indicated as x-fold differences in the cumulative histogram for N = 10 images for each condition, integrated and normalized for Area under the curve. AU, arbitrary units. Scale bar 10 µm
Mutation of the acidic amino acid at the fourth position within the SUMO acceptor motif ψKxE was shown to reduce sumoylation (Sapetschnig et al. 2002). Accordingly, in addition to p35_2KR, we generated a mutant with alanine residues at positions 248 and 292 of p35 (named p35_2AA mutant). This mutant with preserved lysines which may be targeted by other posttranslational modifications but potential disruption of the SUMO acceptor site motif was tested for its sumoylation status. Indeed, this mutant showed a complete reduction in sumoylation as the lysine-to-arginine mutant p35_2KR (Fig. 2b). Importantly, we did not observe any difference in the ubiquitination levels between p35wt and the double mutant p35_2KR indicating that p35 lysine residues K246 or K290 are not major ubiquitination sites (Fig. 2c).
To further substantiate our data and to study p35 sumoylation in intact single cells with spatial resolution, we made use of the fluorescence lifetime imaging microscopy (FLIM) technique in the neuronal dopaminergic CSM14.1 cell line. For this FRET-based approach, mVenus, a monomeric variant of YFP, was fused N-terminally to SUMO2 as energy acceptor and cyan mTFP (monomeric teal fluorescent protein) was used as the energy donor fused N-terminally to p35wt or the SUMO-deficient p35 mutants (Fig. 2d; Day et al. 2008; Nagai et al. 2002). The changes in fluorescence lifetime of the donor mTFP in the presence or absence of acceptor mVenus reflect changes in FRET and allow spatial resolution of alterations in FRET. Cells were transfected with mTFP-p35 (wt, 2KR, or 2AA) in combination with mVenus-SUMO2, and protein interaction between these molecules was studied performing FRET/FLIM. Cells solely transfected with mTFP-p35wt were used as control for the basal non-interacting fluorescence lifetime of mTFP. Co-expression of mTFP-p35wt with mVenus-SUMO2 resulted in a strong reduction in fluorescence lifetime of mTFP through intensive FRET, indicating that p35wt robustly interacts with SUMO2 in intact neuronal cells. FRET was largely abolished when the sumoylation-deficient mutants mTFP-p35_2KR or mTFP-p35_2AA were co-expressed with mVenus-SUMO2. Similarly, when mTFP-p35wt was co-expressed with a SUMO2 mutant lacking the C-terminal diglycine motif necessary for covalent SUMO binding (mVenus-SUMO2Δ7), only a slight fluorescence lifetime reduction was observed compared with the very robust signal obtained with the conjugatable mVenus-SUMO2. Thus, the FRET/FLIM data are in agreement with our biochemical data and demonstrate covalent p35/SUMO2 binding in intact neuronal cells. The residual FRET signals obtained when using the sumoylation-deficient mutants mTFP-p35_2KR and mTFP-p35_2AA or mVenus-SUMO2Δ7 suggest also a minor component of non-covalent interaction that was not detectable in biochemical assays due to the denaturing conditions of our Ni2+–NTA affinity chromatography protocol.
Sumoylation of p35 is Affected by Oxidative Stress
Covalent SUMO modification is a highly dynamic process due to constant cycles of conjugation and deconjugation. It was shown to be regulated in a biphasic manner by reversible oxidation of SUMO-conjugating and -deconjugating enzymes (Bossis and Melchior 2006). To investigate the influence of oxidative stress, which is frequently encountered in neurodegenerative diseases, on our specific target, we treated HEK293 cells expressing p35wt and His-SUMO2 with different concentrations of hydrogen peroxide (H2O2) and determined the p35 sumoylation levels (Fig. 3a). p35 sumoylation levels decreased with increasing concentrations of hydrogen peroxide reaching a minimum at 1 mM H2O2. Further rise of oxidative stress obviously reversed the effect as illustrated by increased p35 sumoylation at 10 mM H2O2. FLIM analysis of cells corroborated the evidence for a regulation of p35 sumoylation by oxidative stress (Fig. 3b). CSM14.1 cells were co-transfected with mTFP-p35wt and mVenus-SUMO2 and treated with 1 mM H2O2. Compared to non-treated cells with decreased fluorescence lifetimes, the treated cells showed lifetimes as high as donor control levels indicating that SUMO modification of p35 is markedly reduced under oxidative stress.
Sumoylation of p35 is affected by oxidative stress. a HEK293 cells co-expressing His-SUMO2 and p35wt were treated with different concentrations of hydrogen peroxide 30 min prior lysis. Sumoylated species were purified via Ni2+–NTA chromatography and analyzed by Western blotting with anti-p35 antibody. p35 sumoylation levels decreased with increasing concentrations of H2O2 from 0.005 to 1 mM (arrow). Incubation with 10 mM H2O2 reversed the effect and resulted in increased p35 SUMO conjugation (rightmost lane). b Sumoylation of p35 under oxidative stress conditions was measured by FLIM in CSM14.1 cells. mTFP intensity pictures are shown in the upper panel and representative fluorescence lifetime images in the lower panel. For further details, see legend of Fig. 2d and “Materials and Methods” section. Fluorescence lifetimes strongly declined after co-expression of mTFP-p35wt and mVenus-SUMO2 indicating FRET and interaction between p35 and SUMO2. Treatment with 1 mM H2O2 resulted in lifetimes comparable to mTFP-p35wt donor controls indicating that oxidative stress substantially decreases sumoylation of p35. FRET efficiencies in the presence or absence of oxidative stress are indicated as x-fold differences in the cumulative histogram using N = 10 images for each condition. AU, arbitrary units. Scale bar 10 µm
Sumoylation of p35 Does Not Modulate Its Binding to Cdk5
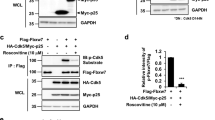
p35 is a regulatory protein that binds to and activates Cdk5 (Tsai et al. 1994). Since SUMO modification is a typical regulator of protein–protein interactions (Gill 2004), we investigated the influence of sumoylation on the binding of p35 to Cdk5. We performed co-immunoprecipitation experiments in order to determine the influence of sumoylation of p35 on its binding to Cdk5 (Fig. 4a). For that purpose, we used HEK293 cells overexpressing Cdk5 together with p35wt and SUMO-deficient p35_2KR. Additionally, to mimic constitutively sumoylated p35, p35 proteins fused N-terminally or C-terminally to conjugation-deficient SUMO2 via a two-alanine linker (p35-SUMO2 and SUMO2-p35, respectively) were generated. Finally, two previously described Cdk5 binding-deficient p35 mutants, p35(L151N,L152N) and p35(D288A,L289A), served as negative controls (Tang et al. 1997; Fu et al. 2006). Immunoprecipitation was performed with anti-p35 antibody, and the samples were subsequently analyzed by Western blotting with anti-Cdk5 and anti-p35 antibodies. We did not observe significant differences in Cdk5 binding between p35wt and the SUMO-deficient p35 mutant (Fig. 4a). Likewise, the fusion proteins p35-SUMO2 and SUMO2-p35, mimicking constitutive sumoylation of p35, showed similar binding properties to Cdk5 as p35wt. As expected, the mutants p35(L151N,L152N) and p35(D288A,L289A) did not co-immunoprecipitate with Cdk5 confirming their previously reported Cdk5 binding deficiency.
Sumoylation of p35 does not modulate the binding to its activator Cdk5. a Lysates from HEK293 cells co-expressing His-Cdk5 and various p35 constructs were subjected to immunoprecipitation (IP) with anti-p35 antibody (upper panels) followed by Western blotting (WB) with p35 and Cdk5 antibodies. Cell lysates (input) were immunoblotted as loading control (lower panels). No significant differences in Cdk5 binding were observed between p35wt, the SUMO-deficient mutant (2KR) or the sumoylation mimicking fusion proteins (p35-SUMO2 and SUMO2-p35). Note the higher molecular weight of the fusion proteins (arrow heads). In contrast, the Cdk5 binding-deficient mutants p35(L151N,L152N) and p35(D288A,L289A) failed to co-immunoprecipitate with Cdk5 as previously described (Fu et al. 2006; Tang et al. 1997). The p35 IgG heavy chain (approximately 55 kDa) is recognized by secondary antibodies in the p35 IP blotted for p35, and lower bands in p35-SUMO2 may represent cleavage products. b FLIM was performed in neuronal CSM14.1 cells in order to investigate the binding of p35 to Cdk5. Intensity pictures of the donor mTFP (gray scale) and representative fluorescence lifetime images (pseudocolor scale) are shown in the upper and lower panels, respectively. For further details, see legend of Fig. 2d and “Materials and Methods” section. Co-expression of mTFP-p35wt with Cdk5-mVenus resulted in decreased lifetimes indicative of interaction between p35 and Cdk5. Co-expression of the SUMO-deficient mutant p35_2KR resulted in similarly shortened fluorescence lifetimes compared to p35wt suggesting that Cdk5 binding was not dependent on the sumoylation status of p35. FRET efficiencies are indicated as x-fold difference in the cumulative histogram from N = 10 images. AU, arbitrary units. Scale bar 10 µm
FRET/FLIM studies confirmed that sumoylation does not alter the p35 binding to Cdk5 (Fig. 4b). To that end, the donor fluorophore mTFP was tagged to the N-terminus of p35 (wt and 2KR), and the FRET acceptor mVenus was tagged to the C-terminus of Cdk5. CSM14.1 cells were transfected as indicated and imaged 24 h later. We observed a significant decrease in fluorescence lifetime upon co-expression of p35wt and Cdk5 indicating an interaction between both proteins. Co-expression of the SUMO-deficient mutant p35_2KR and Cdk5 resulted in similar reductions in fluorescence lifetimes compared to p35wt. This further argues against a substantial impact of sumoylation on p35/Cdk5 interaction and confirms our results from the co-immunoprecipitation experiments.
Sumoylation of p35 Results in Increased Cdk5 Kinase Activity
Next, we aimed to find out whether p35 sumoylation had an influence on the enzymatic activity of the p35/Cdk5 complex (Fig. 5). We compared the kinase activity of Cdk5 in complex with its activator p35wt and fusion proteins mimicking constitutive sumoylation of p35 (p35-SUMO2 and SUMO2-p35). HEK293 cells were co-transfected with His-Cdk5 and p35 constructs. Subsequently, the p35/Cdk5 complexes were purified via Ni2+–NTA chromatography under native conditions to preserve enzymatic activity. Equal protein amounts between the Ni2+–NTA-purified samples were confirmed by Western blotting with anti-Cdk5 and anti-p35 antibodies (Fig. 5a). In the kinase activity assay, Histone H1 was used as substrate for the different p35/Cdk5 complexes in the presence or absence of ATP. Phosphorylation of Histone H1 was analyzed by Western blotting with a phosphorylation-dependent antibody (Fig. 5b). The fusion proteins p35-SUMO2 and SUMO2-p35 showed markedly increased levels of phosphorylated Histone H1 by approximately fivefold compared to p35wt indicative of significantly enhanced Cdk5 activity due to p35 sumoylation (Fig. 5c).
Sumoylation of p35 increases the kinase activity of the p35/Cdk5 complex. a Complexes of His-Cdk5 and p35wt or the indicated fusion proteins were purified by Ni2+–NTA affinity chromatography under native conditions to preserve enzymatic activity. Equal protein levels were confirmed by Western blotting with anti‐p35 and anti‐Cdk5 antibodies. b Purified complexes were applied to a kinase activity assay using Histone H1 as substrate and subjected to Western blotting with anti-Histone H1 and anti-phosphorylated Histone H1 antibodies. Levels of phosphorylated Histone H1 were significantly increased in conditions with sumoylated p35 (i.e., using p35‐SUMO2 or SUMO2‐p35 fusion protein mimicking constitutive sumoylation) compared with p35wt. Single transfections with p35wt or His-Cdk5 only (lane 1 + 2) as well as omitting ATP (lane 6) served as controls. c Quantification of phosphorylated Histone H1 reveals a fivefold higher Cdk5 activity of the sumoylated p35-mimicking fusion proteins p35-SUMO2 and SUMO2-p35 than p35wt. Data show the mean ± s.e.m. from five independent experiments; * P < 0.05
Discussion
In this study, we identify the Cdk5 activator p35 as a novel target for sumoylation and provide evidence that SUMO attachment to p35 modulates p35/Cdk5 activity.
Our initial in vitro sumoylation assays provided the first evidence that p35 is covalently conjugated to SUMO. While in cell-free assays, only modification of p35 by SUMO1 was detected, experiments in cells revealed that p35 is conjugated to SUMO2. We have so far no in vitro or in vivo evidence that Cdk5 is also a target for sumoylation, suggesting that the regulatory subunit of the p35/Cdk5 complex is subject to SUMO modification. Several studies showed that, although many substrates can be modified by both, SUMO1 and SUMO2/3, some substrates are preferentially modified by one isoform or the other. The functional importance of SUMO paralog-specific modifications is largely unknown. In cells, expression of SUMO2 is much better tolerated than SUMO1 raising the possibility that SUMO1 conjugation of p35 drops below detectable levels. SUMO2 was suggested to be involved in cellular responses to environmental stress that is encountered in neurodegenerative diseases (Saitoh and Hinchey 2000), which makes the functional investigation of SUMO2-conjugation to p35 highly interesting. Endogenous sumoylation of human p35 in untransfected HeLa cells further emphasizes the physiological relevance of this posttranslational modification.
Our mutagenesis analysis in cells revealed that lysine residues 246 and 290 are the main sumoylation sites of p35. The fact that both lysines are conserved in different species down to invertebrates suggested already the functional relevance of these sites. Single lysine-to-arginine mutations at position 246 or 290 led to similarly reduced SUMO conjugation which is indicative of remaining sumoylation when only one of these two sites is mutated. The mutation of either the lysines or the acidic amino acids at the fourth position of both sites together fully abrogates sumoylation. The complete absence of band shifts indicative of two SUMO proteins attached to p35wt argues in favor of an alternative mono-sumoylation at lysine 246 or 290 and against the possibility of simultaneous SUMO attachment to both sites. The existence of alternative sumoylation sites in p35 is highly reminiscent of a very similar recent finding in the Parkinson’s disease-related protein alpha-synuclein (Krumova et al. 2011). There, a similar non-simultaneous modification of K96 and K102 was shown to account for the majority of alpha-synuclein sumoylation. Thus, alternative and seemingly “redundant” sumoylation sites may be a general feature of sumoylation and could additionally point to the functional importance of such sites of posttranslational modification. Interestingly, lysines 246 and 290 are located in and at the end of alpha-helices, respectively, regarding the protein’s tertiary structure (Tarricone et al. 2001). Moreover, lysine residues buried within alpha-helical structure can usually not be subject to sumoylation due to structural hindrance (Bernier-Villamor et al. 2002; Kim et al. 2009; Macauley et al. 2006). Interestingly, in both p35 and alpha-synuclein, one of the two major sumoylation lysines is situated exactly at the end of an alpha-helix, suggesting that this might be a structural motif for several SUMO acceptor sites. Although SUMO2 has an internal sumoylation site at lysine residue 11 which enables it to form poly-SUMO chains, endogenous poly-sumoylated protein species are usually only detected after exposure of cells to stress stimuli such as heat shock, osmotic stress, and severe oxidative stress (Saitoh and Hinchey 2000; Golebiowski et al. 2009). The absence of p35-immunopositive high molecular mass species indicative of greater than two SUMO molecules let us conclude that p35 is not conjugated to poly-SUMO2 chains.
We applied FLIM measurements to assess the interaction of SUMO with a target protein. To our knowledge, this is the first FLIM-based demonstration of sumoylation in cells. The strong reduction in fluorescence lifetime that we observed is at least partly due to the spectrally optimized FRET pair mTFP/mVenus, but is also in accordance with the robust p35 sumoylation found in Ni2+–NTA-based purification experiments. The FLIM imaging technique thus confirmed p35 sumoylation at the level of single neuronal cells. In addition, the remaining FRET signal seen with the non-conjugatable SUMO2 mutant (SUMO2Δ7) or using the sumoylation-deficient p35_2KR and p35_2AA mutants, respectively, suggests that p35 and SUMO2 can interact non-covalently, e.g., through so-called SUMO interaction motifs (SIMs) or that p35 can be sumoylated at alternating sites with different FRET efficiencies (Hecker et al. 2006).
Levels of hydrogen peroxide exceed 100 µM in inflamed tissue, concentrations that also affect global sumoylation within cells (Bossis and Melchior 2006; Halliwell et al. 2000; Wittmann et al. 2012; Amor et al. 2014). Oxidative stress was shown to have a biphasic effect on sumoylation in that low concentrations of hydrogen peroxide reduce sumoylation by inhibiting SUMO-conjugating enzymes and high concentrations of hydrogen peroxide enhance sumoylation by blocking SUMO-deconjugating isopeptidases. We noticed decreasing sumoylation of the specific target p35 at increasing concentrations of hydrogen peroxide up to 1 mM. This negative correlation is reversed at concentrations higher than 10 mM analog to the biphasic effect described for global sumoylation levels (Bossis and Melchior 2006). Notably, the FRET/FLIM measurements clearly confirm that oxidative stress such as incubation with 1 mM hydrogen peroxide reduces the interaction between p35 and SUMO2. Taken together, we could confirm the biphasic effect of oxidative stress on sumoylation with regard to a specific neuronal target protein. In future studies, this finding may turn out to have important implications for a number of neuronal cell death paradigms, most of which have in common both increased oxidative stress and Cdk5 dysregulation (Shea et al. 2004; Sahlgren et al. 2006; Shukla et al. 2011; Sun et al. 2011).
Although the core function of p35 is its binding to and activation of Cdk5 and altered protein/protein interaction is a typical result of sumoylation, we found that Cdk5 binding to p35 is not altered upon SUMO modification of p35. In this respect, it is interesting that the p35 sumoylation sites are in fact located outside the region of amino acids 150–200, which was defined as the minimal Cdk5 binding domain of p35 using respective deletion mutants (Poon et al. 1997). However, one of the two main sumoylation sites (K290) is included in the residues between 279 and 291 that are needed for the full activation of Cdk5 by p35 (Poon et al. 1997). Therefore, it seems at least plausible that p35 SUMO-modified at K290 has an altered potential to activate Cdk5 despite unaltered Cdk5 binding affinity. Interestingly, FRET/FLIM analysis of p35wt and Cdk5 demonstrates a strong reduction in lifetime in cytoplasmic foci that looks close to the leading edge of the cells. It has been shown that p35/Cdk5 concentrates at the leading edges of axonal growth cones and regulates neurite outgrowth, axon guidance and cell migration in cortical neurons in culture (Nikolic et al. 1996). Sumoylation-deficient p35_2KR shows a similar localization in the complex with Cdk5 supplying further evidence that sumoylation of p35 does not change the binding of p35 to Cdk5.
According to the abundance of physiological and pathophysiological functions attributed to p35/Cdk5, the impact of SUMO modification on p35/Cdk5 activity may have a wide variety of cell biological implications. Depending on its localization, the p35/Cdk5 complex is able to execute different functions such as a pro-survival in the cytoplasm or cell-cycle arrest and transcriptional regulation in the nucleus (O’Hare et al. 2005; Li et al. 2004). The short half-life of SUMO conjugates is due to the constant interplay of sumoylating and desumoylating enzymes, which may explain the usually low steady-state level of target modification. To overcome the detection limit, we designed fusion proteins to mimic constitutive sumoylation and to allow the functional analysis of p35 sumoylation. Although not identical with physiologically sumoylated p35, the formation of Cdk5 complexes with the p35/SUMO2 fusion proteins clearly enhanced the activity of Cdk5 compared to p35/Cdk5, suggesting a modulatory impact of p35 sumoylation on Cdk5 activity. In contrast to p25/Cdk5, which also shows increased Cdk5 activity, sumoylated p35 can still be phosphorylated at S8, leading to subsequent ubiquitination and degradation of p35 via the proteasome (Patrick et al. 1998; Choe et al. 2007) and resulting in down-regulation of Cdk5 activity. Although speculative, it is conceivable that increased Cdk5 activity in complex with p35-SUMO2 leads to a transiently augmented substrate turnover with enhanced phosphorylation of p35 at S8, thereby providing a negative feedback mechanism and protecting the cell from aberrant Cdk5 activity.
In summary, we show that p35 regulatory subunit of Cdk5 is a novel neuronal SUMO target and identify its two major SUMO acceptor lysines, K246 and K290. Sumoylation of p35 is sensitive to oxidative stress in a manner similar to global sumoylation under oxidative stress that was previously shown. SUMO fusion on either N-terminus or C-terminus of p35 to mimic constitutive sumoylation does not interfere with the binding to Cdk5 but increases Cdk5 kinase activity in vitro. Our study suggests that sumoylation of p35 provides a regulatory mechanism in cells to dynamically modulate Cdk5 activity in a spatial and temporal manner and to react toward varying extrinsic or intrinsic conditions in development, differentiation or under stress.
References
Amor, S., Peferoen, L. A., Vogel, D. Y., Breur, M., van der Valk, P., Baker, D., et al. (2014). Inflammation in neurodegenerative diseases–an update. Immunology, 142(2), 151–166.
Asada, A., Yamamoto, N., Gohda, M., Saito, T., Hayashi, N., & Hisanaga, S. (2008). Myristoylation of p39 and p35 is a determinant of cytoplasmic or nuclear localization of active cyclin-dependent kinase 5 complexes. Journal of Neurochemistry, 106(3), 1325–1336.
Bernier-Villamor, V., Sampson, D. A., Matunis, M. J., & Lima, C. D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell, 108(3), 345–356.
Bossis, G., & Melchior, F. (2006). Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Molecular Cell, 21(3), 349–357.
Chae, T., Kwon, Y. T., Bronson, R., Dikkes, P., Li, E., & Tsai, L. H. (1997). Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron, 18(1), 29–42.
Choe, E. A., Liao, L., Zhou, J. Y., Cheng, D., Duong, D. M., Jin, P., et al. (2007). Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. Journal of Neuroscience, 27(35), 9503–9512.
Day, R. N., Booker, C. F., & Periasamy, A. (2008). Characterization of an improved donor fluorescent protein for Forster resonance energy transfer microscopy. Journal of Biomedial Optics, 13(3), 031203.
Desterro, J. M., Rodriguez, M. S., Kemp, G. D., & Hay, R. T. (1999). Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. Journal of Biological Chemistry, 274(15), 10618–10624.
Dhavan, R., & Tsai, L. H. (2001). A decade of CDK5. Nature Reviews Molecular Cell Biology, 2(10), 749–759.
Eckermann, K. (2013). SUMO and parkinson’s disease. Neuromolecular Med, 15(4), 737–759.
Feligioni, M., & Nistico, R. (2013). SUMO: a (oxidative) stressed protein. Neuromolecular Medicine, 15(4), 707–719.
Fu, X., Choi, Y. K., Qu, D., Yu, Y., Cheung, N. S., & Qi, R. Z. (2006). Identification of nuclear import mechanisms for the neuronal Cdk5 activator. Journal of Biological Chemistry, 281(51), 39014–39021.
Gill, G. (2004). SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes & Development, 18(17), 2046–2059.
Golebiowski, F., Matic, I., Tatham, M. H., Cole, C., Yin, Y., Nakamura, A., et al. (2009). System-wide changes to SUMO modifications in response to heat shock. Science Signaling, 2(72), ra24.
Gong, L., Millas, S., Maul, G. G., & Yeh, E. T. (2000). Differential regulation of sentrinized proteins by a novel sentrin-specific protease. Journal of Biological Chemistry, 275(5), 3355–3359.
Halliwell, B., Clement, M. V., & Long, L. H. (2000). Hydrogen peroxide in the human body. FEBS Letters, 486(1), 10–13.
Hecker, C. M., Rabiller, M., Haglund, K., Bayer, P., & Dikic, I. (2006). Specification of SUMO1- and SUMO2-interacting motifs. Journal of Biological Chemistry, 281(23), 16117–16127.
Hsiao, K., Bozdagi, O., & Benson, D. L. (2014). Axonal cap-dependent translation regulates presynaptic p35. Developmental Neurobiology, 74(3), 351–364.
Jaffray, E. G., & Hay, R. T. (2006). Detection of modification by ubiquitin-like proteins. Methods, 38(1), 35–38.
Kim, E. T., Kim, K. K., Matunis, M. J., & Ahn, J. H. (2009). Enhanced SUMOylation of proteins containing a SUMO-interacting motif by SUMO-Ubc9 fusion. Biochemical and Biophysical Research Communications, 388(1), 41–45.
Krumova, P., Meulmeester, E., Garrido, M., Tirard, M., Hsiao, H. H., Bossis, G., et al. (2011). Sumoylation inhibits alpha-synuclein aggregation and toxicity. Journal of Cell Biology, 194(1), 49–60.
Krumova, P., & Weishaupt, J. H. (2013). Sumoylation in neurodegenerative diseases. Cellular and Molecular Life Sciences, 70(12), 2123–2138.
Li, Z., David, G., Hung, K. W., DePinho, R. A., Fu, A. K., & Ip, N. Y. (2004). Cdk5/p35 phosphorylates mSds3 and regulates mSds3-mediated repression of transcription. Journal of Biological Chemistry, 279(52), 54438–54444.
Macauley, M. S., Errington, W. J., Scharpf, M., Mackereth, C. D., Blaszczak, A. G., Graves, B. J., et al. (2006). Beads-on-a-string, characterization of ETS-1 sumoylated within its flexible N-terminal sequence. Journal of Biological Chemistry, 281(7), 4164–4172.
Mahajan, R., Delphin, C., Guan, T., Gerace, L., & Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88(1), 97–107.
Matunis, M. J., Coutavas, E., & Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. Journal of Cell Biology, 135(6 Pt 1), 1457–1470.
Mukhopadhyay, D., & Dasso, M. (2007). Modification in reverse: the SUMO proteases. Trends in Biochemical Sciences, 32(6), 286–295.
Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K., & Miyawaki, A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnology, 20(1), 87–90.
Nguyen, M. D., Lariviere, R. C., & Julien, J. P. (2001). Deregulation of Cdk5 in a mouse model of ALS: toxicity alleviated by perikaryal neurofilament inclusions. Neuron, 30(1), 135–147.
Nikolic, M., Dudek, H., Kwon, Y. T., Ramos, Y. F., & Tsai, L. H. (1996). The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes & Development, 10(7), 816–825.
O’Hare, M. J., Kushwaha, N., Zhang, Y., Aleyasin, H., Callaghan, S. M., Slack, R. S., et al. (2005). Differential roles of nuclear and cytoplasmic cyclin-dependent kinase 5 in apoptotic and excitotoxic neuronal death. Journal of Neuroscience, 25(39), 8954–8966.
Osuga, H., Osuga, S., Wang, F., Fetni, R., Hogan, M. J., Slack, R. S., et al. (2000). Cyclin-dependent kinases as a therapeutic target for stroke. Proceedings of the National Academy of Sciences USA, 97(18), 10254–10259.
Patrick, G. N., Zhou, P., Kwon, Y. T., Howley, P. M., & Tsai, L. H. (1998). p35, the neuronal-specific activator of cyclin-dependent kinase 5 (Cdk5) is degraded by the ubiquitin-proteasome pathway. Journal of Biological Chemistry, 273(37), 24057–24064.
Patrick, G. N., Zukerberg, L., Nikolic, M., de la Monte, S., Dikkes, P., & Tsai, L. H. (1999). Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature, 402(6762), 615–622.
Patzke, H., & Tsai, L. H. (2002). Calpain-mediated cleavage of the cyclin-dependent kinase-5 activator p39 to p29. Journal of Biological Chemistry, 277(10), 8054–8060.
Poon, R. Y., Lew, J., & Hunter, T. (1997). Identification of functional domains in the neuronal Cdk5 activator protein. Journal of Biological Chemistry, 272(9), 5703–5708.
Sahlgren, C. M., Pallari, H. M., He, T., Chou, Y. H., Goldman, R. D., & Eriksson, J. E. (2006). A nestin scaffold links Cdk5/p35 signaling to oxidant-induced cell death. EMBO Journal, 25(20), 4808–4819.
Saitoh, H., & Hinchey, J. (2000). Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. Journal of Biological Chemistry, 275(9), 6252–6258.
Sapetschnig, A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F., et al. (2002). Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO Journal, 21(19), 5206–5215.
Shea, T. B., Zheng, Y. L., Ortiz, D., & Pant, H. C. (2004). Cyclin-dependent kinase 5 increases perikaryal neurofilament phosphorylation and inhibits neurofilament axonal transport in response to oxidative stress. Journal of Neuroscience Research, 76(6), 795–800.
Shin, E. J., Shin, H. M., Nam, E., Kim, W. S., Kim, J. H., Oh, B. H., et al. (2012). DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Reports, 13(4), 339–346.
Shukla, V., Mishra, S. K., & Pant, H. C. (2011). Oxidative stress in neurodegeneration. Advance Pharmacology Science, 2011, 572634.
Smith, P. D., Crocker, S. J., Jackson-Lewis, V., Jordan-Sciutto, K. L., Hayley, S., Mount, M. P., et al. (2003). Cyclin-dependent kinase 5 is a mediator of dopaminergic neuron loss in a mouse model of Parkinson’s disease. Proceedings of the National Academy of Sciences USA, 100(23), 13650–13655.
Su, S. C., & Tsai, L. H. (2011). Cyclin-dependent kinases in brain development and disease. Annual Review of Cell and Developmental Biology, 27, 465–491.
Sun, K. H., Chang, K. H., Clawson, S., Ghosh, S., Mirzaei, H., Regnier, F., et al. (2011). Glutathione-S-transferase P1 is a critical regulator of Cdk5 kinase activity. Journal of Neurochemistry, 118(5), 902–914.
Tan, T. C., Valova, V. A., Malladi, C. S., Graham, M. E., Berven, L. A., Jupp, O. J., et al. (2003). Cdk5 is essential for synaptic vesicle endocytosis. Nature Cell Biology, 5(8), 701–710.
Tang, D., Chun, A. C., Zhang, M., & Wang, J. H. (1997). Cyclin-dependent kinase 5 (Cdk5) activation domain of neuronal Cdk5 activator. Evidence of the existence of cyclin fold in neuronal Cdk5a activator. Journal of Biological Chemistry, 272(19), 12318–12327.
Tang, D., Yeung, J., Lee, K. Y., Matsushita, M., Matsui, H., Tomizawa, K., et al. (1995). An isoform of the neuronal cyclin-dependent kinase 5 (Cdk5) activator. Journal of Biological Chemistry, 270(45), 26897–26903.
Tarricone, C., Dhavan, R., Peng, J., Areces, L. B., Tsai, L. H., & Musacchio, A. (2001). Structure and regulation of the CDK5-p25(nck5a) complex. Molecular Cell, 8(3), 657–669.
Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H., et al. (2001). Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. Journal of Biological Chemistry, 276(38), 35368–35374.
Tsai, L. H., Delalle, I., Caviness, V. S, Jr, Chae, T., & Harlow, E. (1994). p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature, 371(6496), 419–423.
van den Heuvel, S., & Harlow, E. (1993). Distinct roles for cyclin-dependent kinases in cell cycle control. Science, 262(5142), 2050–2054.
Verstegen, A. M., Tagliatti, E., Lignani, G., Marte, A., Stolero, T., Atias, M., et al. (2014). Phosphorylation of synapsin I by cyclin-dependent kinase-5 sets the ratio between the resting and recycling pools of synaptic vesicles at hippocampal synapses. Journal of Neuroscience, 34(21), 7266–7280.
Weishaupt, J. H., Kussmaul, L., Grotsch, P., Heckel, A., Rohde, G., Romig, H., et al. (2003). Inhibition of CDK5 is protective in necrotic and apoptotic paradigms of neuronal cell death and prevents mitochondrial dysfunction. Molecular and Cellular Neuroscience, 24(2), 489–502.
Wittmann, C., Chockley, P., Singh, S. K., Pase, L., Lieschke, G. J., & Grabher, C. (2012). Hydrogen peroxide in inflammation: messenger, guide, and assassin. Advances in Hematology, 2012, 541471.
Zukerberg, L. R., Patrick, G. N., Nikolic, M., Humbert, S., Wu, C. L., Lanier, L. M., et al. (2000). Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron, 26(3), 633–646.
Acknowledgments
We thank Christine Poser and Claudia Fokken for excellent technical support. We thank Ron Hay (University of Dundee, Dundee, UK) for providing us with the His6-SUMO1 and His6-SUMO2 plasmids, Frauke Melchior (ZMBH, Heidelberg, Germany) for the components used for the in vitro sumoylation assay and Gertrude Bunt (University Medical Center Göttingen, Göttingen, Germany) for mVenus and mTFP plasmids. This work was supported by the Cluster of Excellence and DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain (CNMPB).
Conflict of Interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Anja Büchner, Petranka Krumova, Katrin Eckermann, and Jochen H. Weishaupt have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Büchner, A., Krumova, P., Ganesan, S. et al. Sumoylation of p35 Modulates p35/Cyclin-Dependent Kinase (Cdk) 5 Complex Activity. Neuromol Med 17, 12–23 (2015). https://doi.org/10.1007/s12017-014-8336-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-014-8336-4