Abstract
Glutamate over-activation and the consequent neuronal excitotoxicity have been identified as crucial players in brain dysfunctions such as status epilepticus (SE). Owing to the central function of 2-amino-3-(hydroxyl-5-methylisoxazole-4-yl) propionic acid receptors (AMPARs) in fast excitatory neurotransmission, these receptors have been recognized to play a prominent role in the development and generation of epileptic seizure. This study was undertaken to investigate both the early changes that affect glutamatergic neurons in the rat cerebral cortex and hippocampus and the level and channel properties of AMPARs in response to SE. The results obtained after 3 h of pilocarpine (PILO)-induced SE showed a disorganization of glutamatergic neurons in the CA3 and a thinner neuronal cell layer in the dentate gyrus (DG) region as compared with controls. A significant increase in AMPAR GluA2 protein expression, a decrease in GluA1, GluA3, and GluA4 expression, and a reduction in the phosphorylation of Ser831-GluA1 and Ser880-GluA2 were also observed. In addition, we report a downregulation of R/G editing levels and of Flip splicing isoforms, with a prominent effect on the hippocampus of PILO-treated rats. Our results suggest the presence of an attenuation of AMPARs’ post-synaptic excitatory response to glutamate after PILO treatment, thus conferring neuronal protection from the excitotoxic conditions observed in the SE. This study suggests a role for AMPARs in alterations of the glutamatergic pathway during the onset and early progression of epilepsy, thus indicating additional targets for potential therapeutic interventions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic administration of pilocarpine (PILO) in rodents is a known chemical-inductive model of epilepsy since it reproduces the main features of the human disease and has been widely used in several experimental studies to reproduce the epileptic disorder (Bracey et al. 2009; Cavalheiro et al. 1991; Curia et al. 2008; Leite et al. 1990). PILO is a muscarinic cholinergic agonist that, by activating M1 and M2 receptors, induces seizures, leading to a severe and widespread cell loss in several brain regions, with a predominant effect on the hippocampus (Costa et al. 2004; Lehmann et al. 2001; Scorza et al. 2009). The initial neuronal damage takes place within a few minutes and persists up to a few hours after PILO injection, leading to an extended process of neurodegeneration that may continue for weeks and months (Cavalheiro 1995; Leite et al. 1990). Hippocampal neuropathological changes caused by PILO-induced SE include an increase in glutamate release (Cavalheiro 1995; Costa et al. 2004; de Oliveira et al. 2011), neuronal loss (Carvalho et al. 2011; Tang et al. 2005), reactive astrocytosis (Cavalheiro 1995; Ding et al. 2007), and mossy fibers sprouting (McAuliffe et al. 2011).
Over-activation of glutamatergic neurotransmission has been implicated in many acute and chronic brain dysfunctions, including SE (Carvalho et al. 2011; Meldrum 1993; Pellegrini-Giampietro et al. 1997), and it is considered to be a crucial player in the development and maintenance of epileptic seizures (Costa et al. 2004). Several studies have reported an increase in glutamate release and a disruption of glutamate homeostasis following PILO-induced SE (Cavalheiro 1995; Smolders et al. 1997). Additionally, modifications of glutamate receptor expression and function have been associated with glutamate excitotoxicity and neuronal death during SE (Bracey et al. 2009; Ding et al. 2007; Millan et al. 1993; Zhang et al. 2004). In particular, animal models of SE have presented alterations in synaptic functions, modifications in the membrane properties of neurons, and changes in the composition of glutamate receptors (Kortenbruck et al. 2001; Maas et al. 2006; Sanchez et al. 2001; Su et al. 2002; Zhang et al. 2004).
Among the three classes of ionotropic glutamate receptors, AMPARs mediate the majority of fast excitatory synaptic transmissions (Dingledine et al. 1999). AMPARs are ligand-gated ion channels, characterized by fast-gating kinetics and rapid desensitization, and they respond rapidly to synaptic inputs and facilitate prompt plastic changes in synaptic strength (Ashby et al. 2006). AMPARs are heteromeric complexes of four subunits, GluA1 to GluA4, which combine in different stoichiometry to form distinct receptor subtypes (Shepherd and Huganir 2007). GluA1, GluA3, and GluA4 all display a strong, inwardly rectifying current–voltage relation and calcium permeability, whereas the GluA2 subunit eliminates calcium permeability (Geiger et al. 1995; Jonas and Burnashev 1995).
The functional properties of the four AMPA subunits can be modulated by post-transcriptional modifications, such as alternative splicing and RNA editing (Barbon and Barlati 2011), which represent sophisticated mechanisms that allow for the modification of receptor subunits in positions that are key in regulating the permeability of the channel, the kinetics of pore opening, the time of desensitization of the receptor, and the time of recovery from desensitization (Seeburg 2002; Seeburg and Hartner 2003). Alternative splicing generates the Flip and Flop isoforms of AMPAR subunits, which possess different desensitization characteristics and expression patterns (Monyer et al. 1991; Seeburg et al. 1998). Conversely, RNA editing is a peculiar enzymatic reaction that varies single nucleotides on the coding RNA molecules, modifying specific amino acids and leading to functionally distinct proteins (Barbon and Barlati 2011; Tariq and Jantsch 2012). The predominant editing change is an adenosine-to-inosine modification, catalyzed by an adenosine deaminase acting on RNA (ADAR) enzymes (Bass 2002; Orlandi et al. 2012; Reenan 2001). GluA2, GluA3, and GluA4 may be edited at different sites, which are distinguished by the type of amino acid substitution that occurs: the Q/R site in AMPA receptors’ GluA2, and the R/G site in GluA2, GluA3, and GluA4 (Barbon and Barlati 2011; Seeburg et al. 1998). The GluA2 Q/R-edited subunits confer the capacity for linear current–voltage relationships (Boulter et al. 1990) and impermeability to Ca2+ (Isaac et al. 2007). Furthermore, the R/G editing site modulates the kinetic properties of the AMPAR channel (Lomeli et al. 1994), thus determining the time course of desensitization and resensitization (Krampfl et al. 2002; Lomeli et al. 1994).
Due to the central function of AMPARs in fast excitatory neurotransmission, these receptors might play a prominent role in the development and generation of seizures (Bracey et al. 2009; Kortenbruck et al. 2001; Maas et al. 2006; Porter et al. 2006). Accordingly, the aim of our study was to investigate the early changes in AMPARs that may affect the channel properties in response to SE. Three hours after PILO-induced SE, we observed modifications in RNA editing, splicing processes, and subunit protein levels. These results indicate that AMPARs are involved in the early alterations in glutamatergic neurotransmission in response to SE, leading to a significant reduction in AMPARs’ responsiveness.
Materials and Methods
PILO-induced Status Epilepticus
All animal procedures were performed following the guidelines issued by the European Community Council Directive 86/609/EEC and approved by the Italian Ministry of Health (Project ID: 320/2010). Adult male Sprague–Dawley rats were injected with a single intraperitoneal dose of the muscarinic agonist pilocarpine dissolved in 0.9 % NaCl (250 mg/Kg, i.p.; Sigma-Aldrich, St. Louis, MO, USA); peripheral muscarinic effects were reduced by previous administration of methyl-scopolamine dissolved in PBS (1 mg/kg, i.p.; 30 min before injection of pilocarpine; Sigma-Aldrich) (Turski et al. 1989). The SE onset was characterized by the presence of repeated head cloning movements, salivation, intensive masticatory movements, rearing and falling (Costa et al. 2004). Only rats that developed generalized seizure activity within an hour of PILO injection were used (Wu et al. 2011). At 3 h following PILO administration, at least four treated and control rats were deeply anesthetized with Zoletil (60 mg/kg, ip, Virbac, France) and euthanized. Experimental procedures were then performed.
Perfusion and Tissue Processing
Pilocarpine-treated and control rats were deeply anesthetized with intraperitoneal injection of Zoletil (60 mg/Kg, Virbac, France) and transcardially perfused with 4 % paraformaldehyde 7.4 pH (PFA), freshly prepared as follows: H2O treated with 0.1 % diethylpyrocarbonate (DEPC, Sigma-Aldrich), 4 % PFA (Sigma-Aldrich), 0.4 % NaOH (Sigma-Aldrich), 2.5 % sodium dihydrogen phosphate–monohydrate (Sigma-Aldrich), and 1 mM MgCl2. Brains were post-fixed in 4 % PFA for 24 h and placed in 30 % sucrose before sectioning. Coronal sections (40 μm) were stored in cryoprotectant solution (30 % ethylene glycol, 30 % glycerol, 0.05 M phosphate buffer) at −20 °C until processing.
Immunohistochemistry and Immunofluorescence
Immunohistochemistry and immunofluorescence staining were performed on free-floating 40-μm coronal sections. The slices were permeabilized and saturated with blocking solution (5 % normal goat serum, 0.1 % Triton-X in phosphate-buffered solution (PBS)) for 1 h at room temperature (RT). Subsequently, the slices were incubated with primary antibodies, diluted in the blocking solution, at 4 °C overnight (ON). The primary antibodies used were mouse anti-NeuN (1:1000, Chemicon) and rabbit anti-GFAP (1:800, Millipore), rabbit anti-GluA1 (1:200, Millipore), and rabbit anti-GluA2 (1:200, Chemicon). For the immunofluorescence staining, following several washes, the slices were incubated with secondary antibody Alexa-fluor 488 (1:200, Invitrogen). The slices were then mounted using SlowFade Gold antifade (Invitrogen) reagent.
For the immunohistochemistry experiments, the peroxidase method (ABC system) was used. Specifically, the slices were incubated for 1 h with biotinylated secondary antibody (Vector Laboratories, Vectastain Elite, Burlingame, CA, USA) at RT. With intermittent rinses in PBST, the avidin–biotin–peroxidase complex was applied for 30 min, followed by peroxidase detection for 1 min (DAB, 3,3′-diaminobenzidine tetrahydrochloride; enhanced liquid substrates system, 1:30; Sigma-Aldrich). Sections were mounted on slides and completely dried. Then, sections were rinsed for 1 min in distilled water, subsequently followed by dehydration with graded concentrations of alcohol (50, 70, 90, and 100 %; 1 min each). The sections were immersed in xylene and cover-slipped with mounting media. Images of immunohistochemistry staining were obtained with an Olympus BX50 microscope, while the images of immunofluorescence staining were captured using a Zeiss Axioplan2 microscope.
Riboprobes Preparation and In Situ Hybridization
Digoxigenin (DIG)-labeled riboprobes detecting GluA1 and pan-GluA2 were generated from PCR templates adapted with SP6 and T7 RNA polymerase sites, using the following primer sequences (GluA1: T7-for GAAATTAATACGACTCACTATAGGGAGACCTGCAGCAGGTGCGCTTCGA and Sp6-rev ACGATTTAGGTGACACTATAGAAGCACGTCTGCTCTTCCATAGACCAGT; panGluA2: T7-for GAAATTAATACGACTCACTATAGGGAGAGTTTCGCAGTCACCAATGCTTTCTGCTCC and Sp6-rev ACGATTTAGGTGACACTATAGAAGCATGTTTTCCAATGGTAATAACCTGGTCC). Riboprobes were transcribed using a DIG RNA Labeling Kit (Life technologies) according to the manufacturer’s instructions. Specifically, the 20-μl transcription mixture included 200 ng of template purified cDNA, 1 × Transcription Buffer, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.17 mM UTP, 0.33 mM DIG-UTP (Life Technologies), and 40 U of either T7 or SP6 RNA polymerase. The reaction was incubated for 2 h at 37 °C and stopped by adding 0.5 M EDTA. The riboprobes were then purified in NucAway™ spin columns (Ambion) and quantified with Nanodrop 1000 (Thermo Scientific).
In situ hybridization experiments on brain slices were carried out under RNAse-free conditions, and in this regard, all the solutions were prepared with DEPC-treated H2O. In situ hybridization was developed using the peroxidase method (Vectastain Elite, Vector Laboratories). Free-floating sections were post-fixated for 3 h in 4 % PFA at RT, washed twice in 0.1 Tween 20 in 1 × PBS (PBST) at RT for 5 min, and then were treated with 0.3 % H2O2 (Sigma-Aldrich) in H2O for 20 min to block endogenous peroxidase. Following 3 washes for 5 min in PBST, the slices were quickly washed in H2O, and then permeabilized with 2.3 % sodium meta-periodate (Sigma-Aldrich) in H2O at RT for 5 min and quickly washed in H2O. After these steps, the sections were incubated in 1 % sodium borohydride (Sigma-Aldrich) in 0.1 M Tris–HCl buffer pH 7.5 at RT for 10 min, and then washed twice in PBST at RT for 3 min. The slices were digested with 8 μg/ml proteinase K (Sigma-Aldrich) in PBST at RT for 20 min and washed twice in PBST at RT for 5 min. After digestion, the tissue slices were fixed in 4 % PFA at RT for 5 min and washed three times in PBST at RT for 10 min. Slices were incubated at 55° C for 90 min in prehybridization solution containing 20 mM Tris–HCl (pH7.5) (Sigma-Aldrich), 1 mM EDTA (Gibco), 1 × Denhardt’s solution (Invitrogen), 300 mM NaCl, 100 mM DTT (Fluka), 0.5 mg/ml salmon sperm DNA (Gibco), 0.5 mg/ml polyadenylic acid (Sigma-Aldrich), and 50 % formamide (Sigma-Aldrich). Slices were then incubated on at 55° C in the hybridization solution, composed of the prehybridization solution supplemented with 10 % dextransulfate and 100 ng/ml DIG-labeled riboprobes. The next day, the slices were washed twice in 2 × saline sodium citrate (Molecular Biology), 0.1 % Tween 20 (SSCT), and 50 % deionized formamide at 55° C for 30 min; for 20 min in 2 × SSCT at 55 °C and twice in 0.2 × SSCT at 60 °C for 30 min. Subsequently, the slices were detected using the peroxidase method (ABC system) with biotinylated donkey anti-mouse IgG antibodies and diaminobenzidine as the chromogen (Vector Laboratories, Vectastain Elite). Sections were mounted as described for immunohistochemistry. Images of the in situ hybridization were detected using an Olympus BX50 microscope, and the intensity of the hybridization signal was quantified as optical density per pixel using the specific function in Adobe Photoshop software. The intensity was quantified in CA1, CA3 and DG regions, analyzing areas with defined dimensions, of different slices from control and PILO-treated rats.
RNA Extraction and Retro-transcription
RNA from the hippocampal and cortical tissue of PILO-treated and control rats were extracted using a Qiagen Micro Kit according to the manufacturer’s instructions (Qiagen). RNA quantification and quality controls were performed using spectrophotometric analysis and the AGILENT Bioanalyzer 2100 lab-on-a-chip technology (Agilent Technologies).
Retro-transcription (RT) was carried out using Moloney murine leukemia virus-reverse transcriptase (MMLV-RT), provided by Invitrogen. Briefly, RNA were mixed with 2.2 μl of 0.2-ng/μl random hexamer (Invitrogen), 10 μl of 5X buffer (Invitrogen), 10 μl of 2-mM dNTPs, 1 μl of 1-mM DTT (Invitrogen), 0.4 μl of 33-U/μl RNasin (Promega), and 2 μl MMLV-RT (200 u/μl) in a final volume of 50 μl. The reaction mixture was incubated at 37° C for 2 h, and then the enzyme was inactivated at 95° C for 10 min.
Quantitative Real-time PCR (TaqMan)
To analyze the RNA expression pattern of the target genes, we used the Applied Biosystems 7500 Real-time PCR system (Foster City, CA, USA) with TaqMan probes, following the manufacturer’s instructions. We used the following probes: GluA1 (ID: Rn00709588_m1), GluA2 (ID: Rn00568514_m1), GluA3 (ID: Rn00583547_m1), and GluA4 (ID: Rn00568544_m1). The expression ratio of the target genes was calculated as previously described (Pfaffl et al. 2002), using the geometric mean of the β-actin (ID: Rn00667869_m1), GAPDH (ID: Rn99999916_s1) and H2afz (ID: Rn00821133_g1) as reference.
Flip/Flop Alternative Splicing and Editing Quantification
The levels of the Flip/Flop splicing variants were evaluated by sequence analysis as previously described (Barbon et al. 2010). Briefly, for each AMPA receptor subunits, both isoforms were amplified and then sequenced, in the electropherogram the flip and flop exons appeared as peaks superimposed. Flip and Flop exons exhibit mismatches of A/G in specific locations. The relative expression level of the Flip exon was thus calculated as the ratio peak areas of G (Flip) and the sum of the peak areas A + G (Flip + Flop), using Discovery Studio (DS) Gene 1.5 (Accelrys Inc.).
The editing levels of AMPA receptor GluA2, GluA3, and GluA4 editing sites were quantified by sequence analysis, as previously reported in detail (Barbon et al. 2010) (Barbon et al. 2003).
Protein Extraction and Western Blot
Proteins from hippocampal and cortical tissue of PILO-treated and control rats were extracted with RIPA (50 mM Tris–HCl, 150 mM NaCl, 1 mM EDTA, 0.25 % NaDOC, 1 % IGEPAL, 0.1 % SDS, 2 % CHAPS, and 1X protease inhibitor tablets (Roche Applied Science)), sonicated, and then quantified using a BCA protein concentration assay (Sigma-Aldrich). Proteins were separated by electrophoresis on polyacrylamide gels (4–12 % NuPAGE Bis–Tris gels, Invitrogen) and then transferred to a Hybond-P PVDF membrane (GE Healthcare). Membranes were then incubated ON at 4° C with the following antibodies: rabbit anti-GluA1 (1:200, Millipore), rabbit anti-GluA2 (1:3000, Chemicon), rabbit anti-GluA3 (1:500, Alomone), rabbit anti-GluA4 (1:200, Chemicon), rabbit anti-GluA1 phospho Ser 831 (1:10000, Abcam), rabbit anti-GluA2 phospho Ser 880 (1:1000, Abcam), and mouse anti-GAPDH (1:30000, Millipore) as a loading control. Subsequently, the membranes were incubated for 1 h at RT, with AP-conjugated secondary antibodies (Santa Cruz Biotechnology), and then incubated with CDP star blotting detection reagents (Roche Applied Science). For each investigated protein, the intensity of the immunoreactive bands was analyzed with Image Pro Plus. Data are presented as the ratio of the optical density of the investigated protein band to the GAPDH band and are expressed as a percentage of controls. For the GluA phosphorylated subunits, the data are presented as the ratio of the optical density of the phosphorylated band to the total subunit band, previously normalized on the GAPDH band, and are expressed as a percentage of total GluA levels.
Statistical Analysis
All data are expressed as the mean ± SEM. Statistical significance was assessed with t-tests using GraphPad Prism, and the significance was set at values of p < 0.05.
Results
Hippocampal Histological Alterations Caused by PILO-induced SE
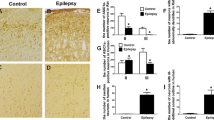
To investigate the possible neuronal damages induced by PILO treatment, we performed NeuN immunohistochemistry on coronal sections of control and treated rats 3 h after PILO injection (Zhang et al. 2009b). In particular, we focused our attention on the hippocampus, which is the main brain region affected by PILO injection-induced SE (Scorza et al. 2009). Comparing NeuN staining between PILO-injected and control rats, we observed important histological modifications in the hippocampus of PILO-treated rats (Fig. 1a, b); in particular, the CA3 region reported a disorganization in the glutamatergic neurons pattern (Fig. 1c, d) (Szabadics et al. 2010); the dentate gyrus (DG) showed a thinner neuronal cell layer as compared with control rats (Fig. 1e, f) (Amaral et al. 2007), while DG hilus seems do not reported alterations. We did not observe histological alterations in the cortical organization of PILO-treated rats (data not shown).
Histological alterations caused by PILO-induced SE. Representative photomicrographs of NeuN immunoreactivity in the hippocampus subfield of control (a, c, e) and PILO-injected rats (b, d, f). High magnification of representative photomicrographs of NeuN immunoreactivity in the CA3 (c and d) and DG regions (e and f). Scale bars = 50 μm and high magnification scale bars = 50 μm
Subsequently, we performed GFAP staining to investigate the changes in astrocytes in response to PILO-induced SE. PILO caused a robust activation of glial cells within the hippocampus, including CA3, CA1, and the hilus of the DG (Fig. 2). Intense GFAP-immunoreactive glial cells, with enhanced staining intensity, retracted processes, and amoeboid appearance, were observed in treated rats (Fig. 2b, d, f). While in the hippocampus of control rats, GFAP-immunoreactive cells maintained their morphology with small cell bodies as well as thin and ramified processes (Fig. 2a, c, e). No particular morphological changes were observed in astrocytes in the cortex of PILO-treated rats (data not shown).
Morphological astrocytic changes caused by PILO-induced SE. Representative photomicrographs of GFAP immunoreactivity in the hippocampus subfield of control (a, c, e) and PILO-injected rats (b, d, f). Images represent glial cells in the CA1 region (a, b), the CA3 region (c, d), and in the hilus of the DG (e, f) of control and PILO-treated rats, respectively. Scale bars = 50 μm
Modulation of AMPA Subunit Protein Expression Following PILO-induced SE
To evaluate the effects of PILO-induced SE on the modulation of AMPA subunit protein expression, we performed western blot experiments for each subunit on cortical and hippocampal regions of treated and control rats (Fig. 3a). As compared with control rats, PILO administration induced important reductions in GluA1 subunit in the hippocampus (100 ± 7.93 vs. 74 ± 6.09, p < 0.05), GluA3 both in the cortex (100 ± 13.15 vs. 59.63 ± 7.60, p < 0.05) and in the hippocampus (100 ± 5.35 vs. 48.65 ± 2.34, p < 0.01), and GluA4 both in the cortex (100 ± 9.51 vs. 69.45 ± 3.66, p < 0.05) and in the hippocampus (100 ± 4.86 vs. 52 ± 6.41, p < 0.01). As well, we observed a significant increase in GluA2 subunit expression in the hippocampus (100 ± 5.25 vs. 153.84 ± 6.44, p < 0.01) relative to control rats (Fig. 3a). In addition, we performed GluA1 and GluA2 immunohistochemistry on coronal slices of control and treated rats (Fig. S1, S2, respectively) to evaluate whether AMPA subunits modulation occurred in all the hippocampal regions. We focused our attention on GluA1 and GluA2 subunits as they are expressed at higher levels in the hippocampus as compared to the GluA3 and GluA4 subunits. We observed that after PILO-induced SE, GluA1 (Fig. S1 A, B), and GluA2 (Fig. S2 A, B) immunostaining reported a similar staining pattern among the different hippocampal regions analyzed. This indicates that, after pilocarpine injection, AMPA receptor subunits undergo the same modulation in all the hippocampal subfields.
Modulation of AMPA subunit proteins following PILO-induced SE. a. The left panel reports the relative expression of GluA subunits in the cortex and in the hippocampus of control and PILO-injected rats. The right panel conveys the representative western blots showing GluA1, GluA2, GluA3, and GluA4; GAPDH was used as loading control. b. Relative expression of GluA1 phopsho Ser 831 and GluA2 phospho Ser 880 relative to the total expression of subunits in the hippocampus of control and PILO-injected rats, as evidenced by the representative western blots. The results are expressed as the mean ± SE (n ≥ 4 per group). Data were analyzed using t-tests, and significance was set at p < 0.05
As the phosphorylation of GluA1 Ser 831 is involved in channel conductance and synaptic activity (Lee et al. 2000; Yuen et al. 2007), and GluA2 Ser 880 promotes endocytosis and degradation of the GluA2 subunit from synaptic surface (Carvalho et al. 2000; Wyszynski et al. 1999), we analyzed the phosphorylation levels of these residues in the hippocampal region. We found that PILO treatment induced relevant reductions in GluA1 phospho Ser 831 (100 ± 6.67 vs. 69.57 ± 13.42, p < 0.05) and GluA2 phospho Ser 880 relative to control rats (100 ± 9.12 vs. 45.4 ± 7.47, p < 0.01) (Fig. 3b).
AMPA Receptors’ RNA Splicing Modifications Following PILO-induced SE
To evaluate the post-transcriptional modifications of AMPA receptors caused by PILO-induced SE, we also quantified the relative expression of Flip/Flop isoform of GluA1-4. PILO administration mildly but significantly affected the splicing process of specific GluA1 and 2 subunits in the hippocampus and in the cortex (Fig. 4). Specifically, 3 h after PILO injection, a reduction in Flip expression for GluA1 (44 ± 0.70 vs. 37 ± 1.56, p < 0.01) and GluA2 (50.62 ± 1.84 vs. 40.04 ± 0.29, p < 0.001) in the hippocampus and in the cortex (44.7 ± 0.02 vs. 37.8 ± 4.35, p < 0.05; 48.95 ± 1.32 vs. 41.59 ± 0.82, p < 0.01; GluA1 and GluA2, respectively) was observed in relation to the control rats. On the contrary, GluA3 and GluA4 splicing variants were unaffected by the PILO treatment in both analyzed regions (Fig. 4).
AMPA receptor RNA splicing modifications following PILO-induced SE. Quantification of relative expression of the Flip isoform of GluR subunits in the cortex and in the hippocampus of control and PILO-treated rats. The results are expressed as the mean ± SE (n ≥ 4 per group). Data were analyzed using t-tests, and significance was set at p < 0.05
AMPA Receptors’ RNA Editing Modifications Following PILO-induced SE
Subsequently, we analyzed the editing levels of GluR subunits in combination with the splicing variants Flip and Flop. We found that, in both cortical and hippocampal regions of PILO-treated rats, the level of editing at the Q/R site of GluA2 was always 100 %, as observed in the control brains (Fig. 5). Interestingly, PILO-injected rats showed a reduction in the R/G editing site only for the Flip isoforms of GluA2 (55.32 ± 2.13 vs. 48.93 ± 0.34, p < 0.05) and GluA3 (80.89 ± 0.62 vs. 75.1 ± 0.41, p < 0.001) in the hippocampus, and of GluA4 both in the hippocampus and in the cortex (42.76 ± 1.52 vs. 32.08 ± 0.98, p < 0.001 and 47.42 ± 1 vs. 36.48 ± 1.18, p < 0.001, respectively) (Fig. 5). PILO treatment did not have any effects on the Flop isoform of all GluR subunits in both regions analyzed (data not shown).
Modulation of AMPA Receptor Subunit mRNAs Following PILO-induced SE
To evaluate the effects of PILO-induced SE on the expression of AMPA receptor mRNAs, we performed quantitative RT-PCR for each AMPA subunit on cortical and hippocampal regions of control and treated rats. We observed that PILO administration affected hippocampal mRNA expression with no effect on the cortex. Specifically, GluA1, GluA3, and GluA4 subunit mRNA expression were increased relative to the levels observed in the hippocampus of control rats (1 ± 0.06 vs. 1.61 ± 0.11, p < 0.01; 1 ± 0.02 vs. 1.36 ± 0.09, p < 0.05; 1 ± 0.03 vs. 1.22 ± 0.04, p < 0.05, respectively) (Fig. 6). No variations were detected for the GluA2 mRNA subunit following PILO treatment, both in the cortex and in the hippocampus (Fig. 6).
Quantitative real-time PCR analysis of AMPA receptor subunit mRNA. mRNA expression of GluA1, GluA2, GluA3, and GluA4 subunits in the cortex and hippocampus of control and PILO-injected rats. The results are expressed as the mean ± SE (n ≥ 4 per group). Data were analyzed using t-tests, and the significance was set at p < 0.05
GluA1 and GluA2 RNA Hybridization Densities Following PILO-induced SE
AMPA receptor mRNA changes were analyzed also by in situ hybridization on coronal sections of PILO-treated and control rats. As for the immunohistochemistry, we focused our attention on GluA1 and GluA2 mRNAs. Compared to control rats, PILO-injected rats showed a higher hybridization signal for GluA1 mRNA in all the hippocampal subfields (Fig. 7). Conversely, no significant differences were observed for GluA2 mRNA in all the hippocampal subfields analyzed (Fig. 8).
In situ hybridization of GluA1 mRNA. Images reporting mRNA hybridization of GluA1 in the hippocampus of control a and PILO-injected rats b Scale bar = 200 μm. Magnification images of CA1, CA3, and DG are reported Scale bar = 50 μm. c Hilus of control and PILO-treated rats. Scale bar = 200 μm. d Quantification of the hybridization signal of GluA1 mRNA in CA1, CA3, and DG of control and PILO-treated rats. The results are expressed as the mean ± SE. Data were analyzed using t-tests, and the significance was set at p < 0.05
In situ hybridization of GluA2 mRNA. Images reporting mRNA hybridization of GluA2 in the hippocampus of control (a) and PILO-injected rats (b). Scale bar = 200 μm. Magnification images of CA1, CA3, and DG are reported. Scale bar = 50 μm. (c) Hilus of control and Pilo-treated rats. Scale bar = 50 μm. (d) Quantification of the hybridization signal of GluA1 mRNA in CA1, CA3, and DG of control and PILO-treated rats. The results are expressed as the mean ± SE. Data were analyzed using t-tests, and the significance was set at p < 0.05
Discussion
In this study, we investigated the changes that affect the number and the channel properties of AMPARs in early phases following PILO-induced SE. We found that PILO injection significantly alters AMPAR protein expression. Additionally, we report the involvement of AMPARs’ RNA editing and splicing processes in response to PILO treatment; in particular, we found a decrease in the R/G editing levels and in the Flip splicing isoform for specific subunits. All these changes appear to be specific regulatory modifications of AMPARs in response to SE because treatment with PILO specifically affects the hippocampus and particular GluR subunits. Taken together, these data suggest that AMPARs are involved in fast alterations in glutamatergic neurotransmission in response to SE, leading to the attenuation of the post-synaptic excitatory response to glutamate, thus conferring neuronal protection to the excitotoxic conditions.
Systemic PILO administration in rodents induces SE that can last for up to 12 h (Cavalheiro 1995; Turski et al. 1983) and may continue for weeks to months, thus causing a possible chronic epileptic condition (Cavalheiro 1995; Leite et al. 1990). PILO-induced SE triggers a series of morphological and biochemical changes associated with spontaneous seizures, such as neuronal cell loss, astrogliosis, axonal sprouting, and a hyper-excitable state, predominantly in the hippocampal region (Cavalheiro 1995, 1991; Tang et al. 2005). In particular, it has been reported that SE induced an extensive loss of vulnerable interneurons within the hilus as well as pyramidal neurons within the DG, CA1, and CA3 regions (Cavalheiro 1995; Grooms et al. 2000; Mello et al. 1993; Mello and Covolan 1996; Rangel et al. 2005; Szabadics et al. 2010). However, the loss of hilar neurons, which has been described in many epilepsy models (Buckmaster and Dudek 1997; Mello et al. 1993), appears to be correlated with the development of spontaneous and recurrent seizures (Borges et al. 2003).
In accordance with these results, we report that, 3 h following SE, important histological modifications were present in the hippocampus of PILO-treated rats, with no effects on the cortex. In particular, as compared with control rats, we observed a robust activation of glial cells within CA3, CA1, and the hilus of the DG; in addition, PILO treatment induces a disorganization of the glutamatergic neurons pattern in the CA3 region and a thinner neuronal cell layers in the DG, while DG hilus did not report any change. These data may indicate an alteration in the vulnerable neurons, accompanied with an inflammatory reaction, in response to the excitotoxic conditions following the onset of SE (Cavalheiro 1995; de Oliveira et al. 2011).
SE-induced pathological alterations are usually associated with over-stimulation of the glutamatergic pathway due to increased levels of glutamate in the synaptic cleft and excessive activation of ionotropic receptors (Clifford et al. 1990; Costa et al. 2004; Fujikawa 1995). Several studies report that the acute phase of PILO-induced SE is characterized by an increased glutamate release in the hippocampus, which becomes extremely toxic to neuronal survival (Cavalheiro 1995; Costa et al. 2004; Scorza et al. 2009). Excessive glutamate levels induce cell death by increasing the [Ca2+] in neurons, thereby leading to the generation of free radicals and the activation of enzymes that degrade proteins, membranes, and nucleic acids (Coyle and Puttfarcken 1993; Dong et al. 2009; Schreiber and Baudry 1995; Siesjo and Bengtsson 1989). The high increase in cytosolic Ca2+ evoked by the activation of glutamate receptors, including AMPARs, seems to be involved in the process of excitotoxicity and neuronal degeneration during SE (Carvalho et al. 2011). This hypothesis is corroborated by studies that show that the administration of AMPAR antagonists in animal models gives rise to an improvement in seizures, attenuating long-term increases in seizure susceptibility and seizure-induced neuronal injury in the hippocampus, thus supporting a critical role for AMPARs in epileptogenesis (Chang et al. 2012; Koh et al. 2004; Rakhade et al. 2008).
Modulation of glutamate receptors, including AMPARs, has been reported in various animal models of SE, although these studies reported apparently conflicting results with decreased, increased, or unchanged specific AMPA subunits at the mRNA and/or protein level (Condorelli et al. 1994; Grooms et al. 2000; Hu et al. 2012; Mathern et al. 1998; Sanchez et al. 2001; Maas et al. 2006). This variability could be ascribed to the different methodologies used to induce the SE, including chemical-inductive SE using PILO, kainate, and LiCl-PILO drugs (Bracey et al. 2009; Condorelli et al. 1994; de Oliveira et al. 2011), or prolonged electrical stimulation or hypoxia-inductive SE (Brandt et al. 2003; Rakhade et al. 2008). In addition, the variable drug concentrations utilized or the different experimental time points used following the induction of SE should be taken into account.
In this regard, Condorelli et al. (1994) showed a reduction in GluA1 and GluA3 subunits in the hippocampus in two different animal models of (Condorelli et al. 1994). More recently, decreases in the GluA1, GluA3, and GluA4 subunits have been reported following “status convulsion,” induced by lithium and PILO systemic injection, in adult and young animals (Hu et al. 2012). At the same time, an increase in GluA2 subunit protein expression has been evidenced in adult and young hippocampus of different animal models of SE induction (Condorelli et al. 1994; Hu et al. 2012; Porter et al. 2006), as well as in the hippocampus of post-mortem epileptic brain patients (Blumcke et al. 1996; Suzuki et al. 2000). In addition, concerning PILO injection, it has been reported that alterations in AMPARs might last for several weeks after treatment (Hu et al. 2012; Porter et al. 2006). In agreement with these data, our protein analyses revealed a reduction in the GluA1, GluA3, and GluA4 subunits, with a parallel increase in the GluA2 subunit in the hippocampus of PILO-treated rats; in particular, as shown by immunohistochemistry experiments, AMPA subunits reported similar expression levels among the various hippocampal subfields.
In addition, we also report that phosphorylation at GluA1 Ser 831 was reduced following PILO treatment, thus indicating an inhibition of channel conductance and of GluA1 subunit delivery to the synaptic surface (Lee et al. 2000; Yuen et al. 2007). This was also combined with reduction in phosphorylation of GluA2 Ser 880, which has been linked to the internalization and disruption of the GluR2 subunit from the synaptic surface (Carvalho et al. 2000; Wyszynski et al. 1999). Taken together, these findings indicate that, following SE, hippocampal neurons give rise to the formation of AMPARs that contain fewer GluA1 and GluA3 subunits, leading to a lower synaptic activity of the channels. At the same time, an increase in the GluA2 subunit, which determines the Ca2+ impermeability of the channels, should give rise to a reduction of Ca2+ influx, thus conferring protection from the excitotoxic effects of the increased glutamate levels induced by PILO-induced SE. This hypothesis is strengthened by GluA1 and GluA2 phosphorylation patterns that point to a decreased delivery of GluA1 and an increased stabilization of the GluA2 subunit.
However, when we analyzed AMPA subunit mRNA expression in response to SE, an unexpected increase in GluA1-, GluA3-, and GluA4-coding mRNAs was observed. Concerning GluA1 and GluA2 mRNAs, the results were confirmed also by in situ hybridization in all the hippocampal subfields analyzed. These differences between mRNAs and proteins might be attributable to several different biological processes, since there is growing evidence of a difference in transcriptional and translational regulation, thus indicating a possible widespread uncoupling between the two functions (Tebaldi et al. 2012). Translational control has been increasingly recognized as a very general feature of eukaryotic cells; this process is orchestrated by a large amount of translational factors that might affect translational initiation, ribosomal scanning, and translational elongation (Groppo and Richter 2009). Moreover, a growing number of mRNA-binding proteins (RBP) and non-coding RNAs such us microRNA might modulate translation levels (Anderson and Kedersha 2009; Fabian et al. 2010). In this contest, the amount of macromolecules potentially able to operate at the interface between mRNA and proteins might be extremely high and could explain the uncoupling between mRNA and protein levels (Groppo and Richter 2009; Jackson et al. 2010). In addition, the mechanism of proteasome-mediated degradation of the receptor subunits, in response to the pathological conditions, should be taken into account (Patrick et al. 2003; Zhang et al. 2009a). Specifically, in response to PILO-induced SE, GluA2-coding mRNA might have undergone an important translational induction, thus leading to an increase in the translated protein, dampening glutamate excitotoxicity. Furthermore, GluA1-, GluA3-, and GluA4-coding mRNA may have been subjected to an increase in transcription, possibly to compensate for the internalization and degradation of GluR subunit proteins, thus recovering the cellular homeostasis.
In response to pathological conditions, the functional properties of AMPARs can also be modulated by RNA alternative splicing and editing processes (Barbon and Barlati 2011). In this regard, several studies have shown that the R/G editing site, in association with the Flip/Flop variant, is important for the modulation of the kinetic properties of AMPAR channels and for the time required for desensitization and resensitization (Krampfl et al. 2002; Lomeli et al. 1994). In particular, GluA2 subunits with the Flip variant take longer to desensitize than those with the Flop variant; moreover, GluA2 subunits with an R/G-edited site report a faster recovery rate from desensitization compared to unedited subunits (Krampfl et al. 2002; Lomeli et al. 1994). In this study, we observed a significant reduction in the GluA2 and GluA4 R/G editing levels for the Flip isoform, together with a decrease in GluA1 and GluA2 Flip relative expression, indicating the formation of channels with faster kinetics of desensitization and slower kinetics of resensitization. This should contribute to the attenuation of AMPARs’ post-synaptic excitatory response to increased glutamate levels, thus protecting neurons against the SE excitotoxic conditions. In support of this, the GluA2 Q/R editing site is still fully edited following PILO-induced SE, thus promoting a reduced conductance of the channels containing the GluA2 Q/R-edited subunit (Hollmann et al. 1991; Swanson et al. 1997).
Overall, the data reported in the present study support the hypothesis that the early lasting changes that occur following PILO-induced SE involve specific modifications of AMPARs’ properties at transcriptional and translational levels, with a prominent effect on the hippocampus. Our findings indicate a possible relevant contribution of AMPARs in the alterations in the glutamatergic pathway in the onset and progression of epilepsy. Understanding the role of AMPARs in the molecular mechanisms that contribute to the onset of SE can be critical in the development of potential therapeutic interventions.
References
Amaral, D. G., Scharfman, H. E., & Lavenex, P. (2007). The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies). Progress in Brain Research, 163, 3–22.
Anderson, P., & Kedersha, N. (2009). RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nature Reviews Molecular Cell Biology, 10, 430–436.
Ashby, M. C., Maier, S. R., Nishimune, A., & Henley, J. M. (2006). Lateral diffusion drives constitutive exchange of AMPA receptors at dendritic spines and is regulated by spine morphology. Journal of Neuroscience, 26, 7046–7055.
Barbon, A., & Barlati, S. (2011). Glutamate receptor RNA editing in health and disease. Biochemistry (Mosc), 76, 882–889.
Barbon, A., Fumagalli, F., Caracciolo, L., Madaschi, L., Lesma, E., Mora, C., et al. (2010). Acute spinal cord injury persistently reduces R/G RNA editing of AMPA receptors. Journal of Neurochemistry, 114, 397–407.
Barbon, A., Vallini, I., La Via, L., Marchina, E., & Barlati, S. (2003). Glutamate receptor RNA editing: a molecular analysis of GluR2, GluR5 and GluR6 in human brain tissues and in NT2 cells following in vitro neural differentiation. Brain Research. Molecular Brain Research, 117, 168–178.
Bass, B. L. (2002). RNA editing by adenosine deaminases that act on RNA. Annual Review of Biochemistry, 71, 817–846.
Blumcke, I., Beck, H., Scheffler, B., Hof, P. R., Morrison, J. H., Wolf, H. K., et al. (1996). Altered distribution of the alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor subunit GluR2(4) and the N-methyl-D-aspartate receptor subunit NMDAR1 in the hippocampus of patients with temporal lobe epilepsy. Acta Neuropathologica, 92, 576–587.
Borges, K., Gearing, M., McDermott, D. L., Smith, A. B., Almonte, A. G., Wainer, B. H., et al. (2003). Neuronal and glial pathological changes during epileptogenesis in the mouse pilocarpine model. Experimental Neurology, 182, 21–34.
Boulter, J., Hollmann, M., O’Shea-Greenfield, A., Hartley, M., Deneris, E., Maron, C., et al. (1990). Molecular cloning and functional expression of glutamate receptor subunit genes. Science, 249, 1033–1037.
Bracey, J. M., Kurz, J. E., Low, B., & Churn, S. B. (2009). Prolonged seizure activity leads to increased Protein Kinase A activation in the rat pilocarpine model of status epilepticus. Brain Research, 1283, 167–176.
Brandt, C., Glien, M., Potschka, H., Volk, H., & Loscher, W. (2003). Epileptogenesis and neuropathology after different types of status epilepticus induced by prolonged electrical stimulation of the basolateral amygdala in rats. Epilepsy Research, 55, 83–103.
Buckmaster, P. S., & Dudek, F. E. (1997). Neuron loss, granule cell axon reorganization, and functional changes in the dentate gyrus of epileptic kainate-treated rats. The Journal of Comparative Neurology, 385, 385–404.
Carvalho, A. L., Duarte, C. B., & Carvalho, A. P. (2000). Regulation of AMPA receptors by phosphorylation. Neurochemical Research, 25, 1245–1255.
Carvalho, A. S., Torres, L. B., Persike, D. S., Fernandes, M. J., Amado, D., Naffah-Mazzacoratti, M. D. G., et al. (2011). Neuroprotective effect of pyruvate and oxaloacetate during pilocarpine induced status epilepticus in rats. Neurochemistry International, 58, 385–390.
Cavalheiro, E. A. (1995). The pilocarpine model of epilepsy. Italian Journal of Neurological Sciences, 16, 33–37.
Cavalheiro, E. A., Leite, J. P., Bortolotto, Z. A., Turski, W. A., Ikonomidou, C., & Turski, L. (1991). Long-term effects of pilocarpine in rats: structural damage of the brain triggers kindling and spontaneous recurrent seizures. Epilepsia, 32, 778–782.
Chang, P. K., Verbich, D., & McKinney, R. A. (2012). AMPA receptors as drug targets in neurological disease—advantages, caveats, and future outlook. European Journal of Neuroscience, 35, 1908–1916.
Clifford, D. B., Olney, J. W., Benz, A. M., Fuller, T. A., & Zorumski, C. F. (1990). Ketamine, phencyclidine, and MK-801 protect against kainic acid-induced seizure-related brain damage. Epilepsia, 31, 382–390.
Condorelli, D. F., Belluardo, N., Mudo, G., Dell’Albani, P., Jiang, X., & Giuffrida-Stella, A. M. (1994). Changes in gene expression of AMPA-selective glutamate receptor subunits induced by status epilepticus in rat brain. Neurochemistry International, 25, 367–376.
Costa, M. S., Rocha, J. B., Perosa, S. R., Cavalheiro, E. A., & Naffah-Mazzacoratti, M. D. G. (2004). Pilocarpine-induced status epilepticus increases glutamate release in rat hippocampal synaptosomes. Neuroscience Letters, 356, 41–44.
Coyle, J. T., & Puttfarcken, P. (1993). Oxidative stress, glutamate, and neurodegenerative disorders. Science, 262, 689–695.
Curia, G., Longo, D., Biagini, G., Jones, R. S., & Avoli, M. (2008). The pilocarpine model of temporal lobe epilepsy. Journal of Neuroscience Methods, 172, 143–157.
de Oliveira, D. L., Bavaresco, C., Mussulini, B. H., Fischer, A., Souza, D. O., Wyse, A. T., et al. (2011). Early life LiCl-pilocarpine-induced status epilepticus reduces acutely hippocampal glutamate uptake and Na +/K + ATPase activity. Brain Research, 1369, 167–172.
Ding, S., Fellin, T., Zhu, Y., Lee, S. Y., Auberson, Y. P., Meaney, D. F., et al. (2007). Enhanced astrocytic Ca2 + signals contribute to neuronal excitotoxicity after status epilepticus. Journal of Neuroscience, 27, 10674–10684.
Dingledine, R., Borges, K., Bowie, D., & Traynelis, S. F. (1999). The glutamate receptor ion channels. Pharmacological Reviews, 51, 7–61.
Dong, X. X., Wang, Y., & Qin, Z. H. (2009). Molecular mechanisms of excitotoxicity and their relevance to pathogenesis of neurodegenerative diseases. Acta Pharmacologica Sinica, 30, 379–387.
Fabian, M. R., Sundermeier, T. R., & Sonenberg, N. (2010). Understanding how miRNAs post-transcriptionally regulate gene expression. Progress in Molecular and Subcellular Biology, 50, 1–20.
Fujikawa, D. G. (1995). Neuroprotective effect of ketamine administered after status epilepticus onset. Epilepsia, 36, 186–195.
Geiger, J. R., Melcher, T., Koh, D. S., Sakmann, B., Seeburg, P. H., Jonas, P., et al. (1995). Relative abundance of subunit mRNAs determines gating and Ca2 + permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron, 15, 193–204.
Grooms, S. Y., Opitz, T., Bennett, M. V., & Zukin, R. S. (2000). Status epilepticus decreases glutamate receptor 2 mRNA and protein expression in hippocampal pyramidal cells before neuronal death. Proc Natl Acad Sci USA, 97, 3631–3636.
Groppo, R., & Richter, J. D. (2009). Translational control from head to tail. Current Opinion in Cell Biology, 21, 444–451.
Hollmann, M., Hartley, M., & Heinemann, S. (1991). Ca2 + permeability of KA-AMPA–gated glutamate receptor channels depends on subunit composition. Science, 252, 851–853.
Hu, Y., Jiang, L., Chen, H., & Zhang, X. (2012). Expression of AMPA receptor subunits in hippocampus after status convulsion. Childs Nervous System, 28, 911–918.
Isaac, J. T., Ashby, M. C., & McBain, C. J. (2007). The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron, 54, 859–871.
Jackson, R. J., Hellen, C. U., & Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology, 11, 113–127.
Jonas, P., & Burnashev, N. (1995). Molecular mechanisms controlling calcium entry through AMPA-type glutamate receptor channels. Neuron, 15, 987–990.
Koh, S., Tibayan, F. D., Simpson, J. N., & Jensen, F. E. (2004). NBQX or topiramate treatment after perinatal hypoxia-induced seizures prevents later increases in seizure-induced neuronal injury. Epilepsia, 45, 569–575.
Kortenbruck, G., Berger, E., Speckmann, E. J., & Musshoff, U. (2001). RNA editing at the Q/R site for the glutamate receptor subunits GLUR2, GLUR5, and GLUR6 in hippocampus and temporal cortex from epileptic patients. Neurobiology of Diseases, 8, 459–468.
Krampfl, K., Schlesinger, F., Zorner, A., Kappler, M., Dengler, R., & Bufler, J. (2002). Control of kinetic properties of GluR2 flop AMPA-type channels: impact of R/G nuclear editing. European Journal of Neuroscience, 15, 51–62.
Lee, H. K., Barbarosie, M., Kameyama, K., Bear, M. F., & Huganir, R. L. (2000). Regulation of distinct AMPA receptor phosphorylation sites during bidirectional synaptic plasticity. Nature, 405, 955–959.
Lehmann, T. N., Gabriel, S., Eilers, A., Njunting, M., Kovacs, R., Schulze, K., et al. (2001). Fluorescent tracer in pilocarpine-treated rats shows widespread aberrant hippocampal neuronal connectivity. European Journal of Neuroscience, 14, 83–95.
Leite, J. P., Bortolotto, Z. A., & Cavalheiro, E. A. (1990). Spontaneous recurrent seizures in rats: an experimental model of partial epilepsy. Neuroscience and Biobehavioral Reviews, 14, 511–517.
Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J. R., Kuner, T., et al. (1994). Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science, 266, 1709–1713.
Maas, S., Kawahara, Y., Tamburro, K. M., & Nishikura, K. (2006). A-to-I RNA editing and human disease. RNA Biology, 3, 1–9.
Mathern, G. W., Pretorius, J. K., Mendoza, D., Lozada, A., & Kornblum, H. I. (1998). Hippocampal AMPA and NMDA mRNA levels correlate with aberrant fascia dentata mossy fiber sprouting in the pilocarpine model of spontaneous limbic epilepsy. Journal of Neuroscience Research, 54, 734–753.
McAuliffe, J. J., Bronson, S. L., Hester, M. S., Murphy, B. L., Dahlquist-Topala, R., Richards, D. A., et al. (2011). Altered patterning of dentate granule cell mossy fiber inputs onto CA3 pyramidal cells in limbic epilepsy. Hippocampus, 21, 93–107.
Meldrum, B. S. (1993). Excitotoxicity and selective neuronal loss in epilepsy. Brain Pathology, 3, 405–412.
Mello, L. E., Cavalheiro, E. A., Tan, A. M., Kupfer, W. R., Pretorius, J. K., Babb, T. L., et al. (1993). Circuit mechanisms of seizures in the pilocarpine model of chronic epilepsy: cell loss and mossy fiber sprouting. Epilepsia, 34, 985–995.
Mello, L. E., & Covolan, L. (1996). Spontaneous seizures preferentially injure interneurons in the pilocarpine model of chronic spontaneous seizures. Epilepsy Research, 26, 123–129.
Millan, M. H., Chapman, A. G., & Meldrum, B. S. (1993). Extracellular amino acid levels in hippocampus during pilocarpine-induced seizures. Epilepsy Research, 14, 139–148.
Monyer, H., Seeburg, P. H., & Wisden, W. (1991). Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron, 6, 799–810.
Orlandi, C., Barbon, A., & Barlati, S. (2012). Activity regulation of adenosine deaminases acting on RNA (ADARs). Molecular Neurobiology, 45, 61–75.
Patrick, G. N., Bingol, B., Weld, H. A., & Schuman, E. M. (2003). Ubiquitin-mediated proteasome activity is required for agonist-induced endocytosis of GluRs. Current Biology, 13, 2073–2081.
Pellegrini-Giampietro, D. E., Gorter, J. A., Bennett, M. V., & Zukin, R. S. (1997). The GluR2 (GluR-B) hypothesis: Ca(2 +)-permeable AMPA receptors in neurological disorders. Trends in Neurosciences, 20, 464–470.
Pfaffl, M. W., Horgan, G. W., & Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research, 30, e36.
Porter, B. E., Cui, X. N., & Brooks-Kayal, A. R. (2006). Status epilepticus differentially alters AMPA and kainate receptor subunit expression in mature and immature dentate granule neurons. European Journal of Neuroscience, 23, 2857–2863.
Rakhade, S. N., Zhou, C., Aujla, P. K., Fishman, R., Sucher, N. J., & Jensen, F. E. (2008). Early alterations of AMPA receptors mediate synaptic potentiation induced by neonatal seizures. Journal of Neuroscience, 28, 7979–7990.
Rangel, P., Cysneiros, R. M., Arida, R. M., de Albuquerque, M., Colugnati, D. B., Scorza, C. A., et al. (2005). Lovastatin reduces neuronal cell death in hippocampal CA1 subfield after pilocarpine-induced status epilepticus: preliminary results. Arquivos de Neuro-Psiquiatria, 63, 972–976.
Reenan, R. A. (2001). The RNA world meets behavior: A– > I pre-mRNA editing in animals. Trends in Genetics, 17, 53–56.
Sanchez, R. M., Koh, S., Rio, C., Wang, C., Lamperti, E. D., Sharma, D., et al. (2001). Decreased glutamate receptor 2 expression and enhanced epileptogenesis in immature rat hippocampus after perinatal hypoxia-induced seizures. Journal of Neuroscience, 21, 8154–8163.
Schreiber, S. S., & Baudry, M. (1995). Selective neuronal vulnerability in the hippocampus–a role for gene expression? Trends in Neurosciences, 18, 446–451.
Scorza, F. A., Arida, R. M., Naffah-Mazzacoratti, M. D. G., Scerni, D. A., Calderazzo, L., & Cavalheiro, E. A. (2009). The pilocarpine model of epilepsy: what have we learned? Anais da Academia Brasileira de Ciências, 81, 345–365.
Seeburg, P. H. (2002). A-to-I editing: new and old sites, functions and speculations. Neuron, 35, 17–20.
Seeburg, P. H., & Hartner, J. (2003). Regulation of ion channel/neurotransmitter receptor function by RNA editing. Current Opinion in Neurobiology, 13, 279–283.
Seeburg, P. H., Higuchi, M., & Sprengel, R. (1998). RNA editing of brain glutamate receptor channels: mechanism and physiology. Brain Research. Brain Research Reviews, 26, 217–229.
Shepherd, J. D., & Huganir, R. L. (2007). The cell biology of synaptic plasticity: AMPA receptor trafficking. Annual Review of Cell and Developmental Biology, 23, 613–643.
Siesjo, B. K., & Bengtsson, F. (1989). Calcium fluxes, calcium antagonists, and calcium-related pathology in brain ischemia, hypoglycemia, and spreading depression: a unifying hypothesis. Journal of Cerebral Blood Flow and Metabolism, 9, 127–140.
Smolders, I., Khan, G. M., Lindekens, H., Prikken, S., Marvin, C. A., Manil, J., et al. (1997). Effectiveness of vigabatrin against focally evoked pilocarpine-induced seizures and concomitant changes in extracellular hippocampal and cerebellar glutamate, gamma-aminobutyric acid and dopamine levels, a microdialysis-electrocorticography study in freely moving rats. Journal of Pharmacology and Experimental Therapeutics, 283, 1239–1248.
Su, H., Sochivko, D., Becker, A., Chen, J., Jiang, Y., Yaari, Y., et al. (2002). Upregulation of a T-type Ca2 + channel causes a long-lasting modification of neuronal firing mode after status epilepticus. Journal of Neuroscience, 22, 3645–3655.
Suzuki, F., Hirai, H., Onteniente, B., Riban, V., Matsuda, M., & Kurokawa, K. (2000). Long-term increase of GluR2 alpha-amino-3-hydroxy-5-methylisoxazole-4-propionate receptor subunit in the dispersed dentate gyrus after intrahippocampal kainate injection in the mouse. Neuroscience, 101, 41–50.
Swanson, G. T., Kamboj, S. K., & Cull-Candy, S. G. (1997). Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. Journal of Neuroscience, 17, 58–69.
Szabadics, J., Varga, C., Brunner, J., Chen, K., & Soltesz, I. (2010). Granule cells in the CA3 area. Journal of Neuroscience, 30, 8296–8307.
Tang, F. R., Chia, S. C., Zhang, S., Chen, P. M., Gao, H., Liu, C. P., et al. (2005). Glutamate receptor 1-immunopositive neurons in the gliotic CA1 area of the mouse hippocampus after pilocarpine-induced status epilepticus. European Journal of Neuroscience, 21, 2361–2374.
Tariq, A., & Jantsch, M. F. (2012). Transcript Diversification in the Nervous System: A to I RNA Editing in CNS Function and Disease Development. Frontiers in Neuroscience, 6, 99.
Tebaldi, T., Re, A., Viero, G., Pegoretti, I., Passerini, A., Blanzieri, E., et al. (2012). Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics, 13, 220.
Turski, W. A., Cavalheiro, E. A., Schwarz, M., Czuczwar, S. J., Kleinrok, Z., & Turski, L. (1983). Limbic seizures produced by pilocarpine in rats: behavioural, electroencephalographic and neuropathological study. Behavioural Brain Research, 9, 315–335.
Turski, L., Ikonomidou, C., Turski, W. A., Bortolotto, Z. A., & Cavalheiro, E. A. (1989). Review: cholinergic mechanisms and epileptogenesis. The seizures induced by pilocarpine: a novel experimental model of intractable epilepsy. Synapse (New York, N. Y.), 3, 154–171.
Wu, Y. C., Williamson, R., Li, Z., Vicario, A., Xu, J., Kasai, M., et al. (2011). Dendritic trafficking of brain-derived neurotrophic factor mRNA: regulation by translin-dependent and -independent mechanisms. Journal of Neurochemistry, 116, 1112–1121.
Wyszynski, M., Valtschanoff, J. G., Naisbitt, S., Dunah, A. W., Kim, E., Standaert, D. G., et al. (1999). Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo. Journal of Neuroscience, 19, 6528–6537.
Yuen, E. Y., Liu, W., & Yan, Z. (2007). The phosphorylation state of GluR1 subunits determines the susceptibility of AMPA receptors to calpain cleavage. Journal of Biological Chemistry, 282, 16434–16440.
Zhang, D., Hou, Q., Wang, M., Lin, A., Jarzylo, L., Navis, A., et al. (2009a). Na, K-ATPase activity regulates AMPA receptor turnover through proteasome-mediated proteolysis. Journal of Neuroscience, 29, 4498–4511.
Zhang, S., Khanna, S., & Tang, F. R. (2009b). Patterns of hippocampal neuronal loss and axon reorganization of the dentate gyrus in the mouse pilocarpine model of temporal lobe epilepsy. Journal of Neuroscience Research, 87, 1135–1149.
Zhang, G., Raol, Y. S., Hsu, F. C., & Brooks-Kayal, A. R. (2004). Long-term alterations in glutamate receptor and transporter expression following early-life seizures are associated with increased seizure susceptibility. Journal of Neurochemistry, 88, 91–101.
Acknowledgments
This work was supported through grants from MIUR (PRIN 2009BRMW4 W) and the NEDD project Regione Lombardia (ID 14546-A SAL7). We thank Nature Publishing Group Language Editing for assistance with the manuscript. The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Russo, I., Bonini, D., Via, L.L. et al. AMPA Receptor Properties are Modulated in the Early Stages Following Pilocarpine-induced Status Epilepticus. Neuromol Med 15, 324–338 (2013). https://doi.org/10.1007/s12017-013-8221-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-013-8221-6