Abstract
Schizophrenia (SCZ) is a complex psychiatric disease with a lifetime morbidity rate of 0.5–1.0 %. To date, aberrant DNA methylation in SCZ has been reported in several studies. However, no comprehensive studies using medication-free subjects with SCZ have been conducted. In addition, most of these studies have been limited to the analysis of the CpG sites in CpG islands (CGIs) in the gene promoter regions, so little is known about the DNA methylation signatures across the whole genome in SCZ. Genome-wide DNA methylation profiling (485,764 CpG sites) of peripheral leukocytes was conducted in the first set of samples (24 medication-free patients with SCZ and 23 non-psychiatric controls) using Infinium HumanMethylation450 Beadchips. Second, a monozygotic twin study was performed using three pairs of monozygotic twins that were discordant for SCZ. Finally, the data from these two independent cohorts were compared. A total of 234 differentially methylated CpG sites that were common between these two cohorts were identified. Of the 234 CpG sites, 153 sites (65.4 %) were located in the CGIs and in the regions flanking CGIs (CGI: 40.6 %; CGI shore: 13.3 %; CGI shelf: 11.5 %). Of the 95 differently methylated CpG sites in the CGIs, most of them were located in the promoter regions (promoter: 75.8 %; gene body: 14.7 %; 3′-UTR: 2.1 %). Aberrant DNA methylation in SCZ was identified at numerous loci across the whole genome in peripheral leukocytes using two independent sets of samples. These findings support the notion that altered DNA methylation could be involved in the pathophysiology of SCZ.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schizophrenia (SCZ) is a mental disease characterized by auditory hallucinations, delusional ideas, and cognitive impairments. Its reported lifetime morbidity risk is 7.2 per 1,000 (Bhugra et al. 2005). SCZ is a complex disorder that results from genetic and environmental etiological influences, and its heritability is estimated to exceed 80 % (Sullivan et al. 2003). Although candidate gene approaches, genome-wide association studies, and copy number variant studies have been carried out for SCZ (Harrison and Weinberger 2005; International Schizophrenia Consortium 2008; Purcell et al. 2009; Rees et al. 2011; Shi et al. 2009; Stefansson et al. 2009), the effects of each individual genetic factor are not large.
Epigenetics is defined as the study of mitotically or meiotically heritable variations in gene function that cannot be explained by changes in DNA sequence (Petronis et al. 2000). The 41–65 % concordance rate of SCZ in monozygotic twins, non-Mendelian inheritance, the presence of sporadic cases, sexual dimorphism, and parental origin effects suggest that epigenetic components are involved in the etiology of SCZ (Cardno and Gottesman 2000). DNA methylation, which is the addition of a methyl group to the cytosine in a CpG dinucleotide, is a major epigenetic mechanism, and attention to its role in SCZ has recently increased. To date, aberrant DNA methylation in SCZ has been reported in several studies (Abdolmaleky et al. 2006; Carrard et al. 2011; Chen et al. 2011; Dempster et al. 2011; Grayson et al. 2005; Iwamoto et al. 2005; Melas et al. 2012; Mill et al. 2008; Nohesara et al. 2011). Although antipsychotic drugs are known to influence DNA methylation (Dong et al. 2008; Melas et al. 2012; Mill et al. 2008; Shimabukuro et al. 2006; Tremolizzo et al. 2005), no comprehensive studies using medication-free subjects with SCZ have been conducted. In addition, most of previous studies have been limited to the analysis of the CpG sites in CpG islands (CGIs) in the gene promoter regions, so little is known about the DNA methylation signatures across the whole genome in SCZ.
In this study, first, genome-wide DNA methylation profiling (485,764 CpG dinucleotides) of peripheral leukocytes both in the first set of samples (24 medication-free SCZ patients and 23 non-psychiatric controls) and in the second set of samples (3 pairs of monozygotic twins discordant for SCZ) was conducted. Then, the data from these two independent cohorts were compared, and common changes in DNA methylation between the cohorts were detected.
Materials and Methods
Subjects
For the first set of samples, twenty-four medication-free patients with SCZ (11 males and 13 females, mean age: 30.9 ± 10.5 y) were recruited from Tokushima and Kochi University Hospitals in Japan. The diagnosis of SCZ was made by at least two experienced psychiatrists according to DSM-IV criteria on the basis of extensive clinical interviews and a review of medical records. None of the patients with SCZ had any psychiatric comorbidity. Among the twenty-four patients, sixteen of patients had no history of taking antipsychotics, and of the other eight patients, seven had not taken any antipsychotics for at least 2 months. Twenty-three control subjects (10 males and 13 females, mean age: 31.9 ± 9.7 year) were selected from volunteers who were recruited from hospital staff, students, and company employees documented to be free from psychiatric problems, a past history of mental illness, and medications. For the second set of samples, three pairs of monozygotic twins discordant for SCZ were recruited from Nagasaki University Hospital. All of the twins were males, and their mean age was 52.7 ± 10.4 year. These three pairs of twins were also reported in a previous study (Ono et al. 2010). All affected individuals among the twins were treated with various psychotic drugs. Demographic data of all samples analyzed in this study are presented in Supplementary Table S1. All subjects who participated in this study were of unrelated Japanese origin and signed written informed consent approved by the institutional ethics committees of the University of Tokushima Graduate School, Kochi Medical School, and Nagasaki University Graduate School of Biomedical Science to participate in this study.
DNA Methylation Methods
Genomic DNA was extracted from peripheral blood using the phenol–chloroform method. Bisulfite conversion of 500 ng genomic DNA was performed using the EZ DNA methylation kit (Zymo Research). DNA methylation level was assessed according to the manufacturer’s instructions using Infinium® HumanMethylation450 Beadchips (Illumina Inc.). The technical schemes, the accuracy, and the high reproducibility of this array have been described in previous papers (Bibikova et al. 2011; Dedeurwaerder et al. 2011; Sandoval et al. 2011). Quantitative measurements of DNA methylation were determined for 485,764 CpG dinucleotides, which covered 99 % of the RefSeq genes and were distributed across the whole gene regions, including promoter, gene body, and 3′-untranslated regions (UTRs). They also covered 96 % of CGIs from the UCSC database with additional coverage in CGI shores (0–2 kb from CGI) and CGI shelves (2–4 kb from CGI). Detailed information on the contents of the array is available in the Infinium HumanMethylation450 User Guide and HumanMethylation450 manifest (www.illumina.com) and in recent papers (Bibikova et al. 2011; Sandoval et al. 2011). DNA methylation data were analyzed with the methylation analysis module within the BeadStudio software (Illumina Inc.). DNA methylation status of the CpG sites was calculated as the ratio of the signal from a methylated probe relative to the sum of both methylated and unmethylated probes. This value, known as β, ranges from 0 (completely unmethylated) to 1 (fully methylated). For intra-chip normalization of probe intensities, colored balance and back ground corrections in every set of twelve samples from the same chip were performed using internal control probes. X chromosome CpG sites in the CGIs in the AR gene in this array as well as the internal control probes were checked to validate the DNA methylation measurements, as done in a previous study (Siegmund et al. 2007), and large sex differences were observed at all of these CpG sites (Supplementary Figure S1).
Statistical Methods
In the first set of samples, surrogate variable analysis (Leek and Storey 2007) was used to identify CpG loci showing significant differences in DNA methylation between medication-free patients with SCZ and the controls. This analysis is useful in clinical studies, where a large number of clinical variables, including known and unknown factors, have a complicated joint impact on microarray data, as applied in previous studies (Colantuoni et al. 2011; Numata et al. 2012). A false discovery rate (FDR) correction was applied at the 0.05 level for multiple testing. In the second set of samples, a paired t test was used to assess the significance of DNA methylation differences between the affected and unaffected twin subjects. P values < 0.05 and average DNA methylation differences between two groups >0.01 were considered significant differential methylation.
Results
Diagnostic Differences in DNA Methylation Between Medication-Free Patients With SCZ and Controls
DNA methylation levels were compared between 24 medication-free patients with SCZ and 23 control subjects using Infinium® HumanMethylation450 BeadChips. Of 485,764 CpG sites, significant diagnostic differences in DNA methylation were observed at 10,747 CpG sites at FDR 5 % correction. The top 100-ranking differentially methylated CpG sites are shown in Supplementary Table S2.
DNA Methylation Differences in Monozygotic Twins Discordant for SCZ
Genome-wide DNA methylation profiling of three pairs of monozygotic twins that were discordant for SCZ using the same Illumina methylation arrays was also conducted. Of 485,764 CpG sites, significant diagnostic differences in DNA methylation were observed at 15,872 CpG sites (P < 0.05 and average Δβ > 0.01). The top 100 ranking differentially methylated CpG sites are shown in Supplementary Table S3. In addition, a list of the CpG sites that showed a Δβ > 0.3 within each individual twin pair is shown in Supplementary Table S4.
Common Changes in DNA Methylation in SCZ Between the Two Independent Cohorts
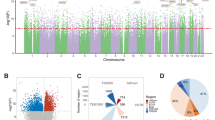
The data from these two independent cohorts were compared, and a total of 234 differentially methylated CpG sites that were common between the cohorts were identified. Of these 234 CpG sites, 215 sites (92.4 %) demonstrated higher DNA methylation in SCZ compared to controls. When these 234 differentially methylated CpG sites were classified into four categories (CGI, CGI shore, CGI shelf, and others) according to the CpG content in the genes, 153 sites (65.4 %) were located in the CGIs and in the regions flanking CGIs (CGI shore and CGI shelf) (Supplementary Table S5). Although the proportions of CpG sites in the CGIs and in the regions flanking CGIs in this array were respectively 31 and 33 %, fewer changes in DNA methylation in SCZ in the regions flanking CGIs were observed than in the CGIs. Ninety five sites (40.6 %) were located in the CGIs, 31 sites (13.3 %) in CGI shores, and 27 sites (11.5 %) in CGI shelves (Fig. 1a). When these 234 differentially methylated CpG sites were classified into four different categories (promoter region, gene body, 3′-UTR, and intergenic region) according to their location in the genes, 109 sites (46.6 %) were located in the promoter regions, 64 sites (27.4 %) in gene bodies, and 6 sites (2.6 %) in 3′-UTRs (Fig. 1b). Of the 95 differentially methylated CpG sites in the CGIs, most of them were located in the promoter regions. Seventy two sites (75.8 %) were located in the promoter regions, 14 sites (14.7 %) in gene bodies, and 2 sites (2.1 %) in 3′-UTRs (Fig. 1c). Examples include two differentially methylated CpG sites in the CGIs in the promoter regions in the B3GAT2 and HDAC4 genes, which have been implicated in SCZ (Kähler et al. 2011; Kim et al. 2010) (Fig. 2).
The percentage of the CpG sites associated with schizophrenia (SCZ) between the two cohorts. a The percentage of the CpG sites associated with SCZ according to their CpG contents in the genes. Of the 234 CpG sites significantly associated with SCZ, 95 sites (40.6 %) were located in the CGIs, 31 sites (13.3 %) in CGI shores, and 27 sites (11.5 %) in CGI shelves. b The percentage of the CpG sites associated with SCZ according to their location in the genes. Of the 234 CpG sites significantly associated with SCZ, 109 sites (46.6 %) were located in the promoter regions, 64 sites (27.4 %) in gene bodies, and 6 sites (2.6 %) in 3′-UTRs. c The percentage of the CpG sites in the CGIs associated with SCZ according to their location in the genes. Of the 95 CpG sites significantly associated with SCZ, 72 sites (75.8 %) were located in the promoter regions, 14 sites (14.7 %) in gene bodies, and 2 sites (2.1 %) in 3′-UTRs
DNA methylation signatures of two genes (B3GAT2 and HDAC4). DNA methylation levels are shown on the y-axis. Patients with SCZ are shown in blue, and the controls are shown in red. The CpG sites of B3GAT2 (cg19273746) and HDAC4 (cg15142485) demonstrated significant differences in DNA methylation between SCZ and controls both in the first set of samples (24 medication-free SCZ patients and 23 controls) and in the second set of samples (3 pairs of monozygotic twins discordant for SCZ)
Discussion
In this study, first, genome-wide DNA methylation profiling of peripheral leukocytes was conducted in the first set of samples (24 medication-free patients with SCZ and 23 non-psychiatric controls) using Infinium HumanMethylation450 Beadchips. To our knowledge, this study is the first to use medication-free samples with SCZ for DNA methylation profiling. Second, a monozygotic twin study was performed using three pairs of monozygotic twins that were discordant for SCZ. Although DNA methylation is associated with genotypic variants (Numata et al. 2012), a twin study is a useful method for investigating DNA methylation differences between disease phenotypes without the influence of genetic discordance. In fact, this approach has been applied successfully to identify epigenetic differences in complex diseases, such as autoimmune disease, type-1 diabetes, psoriasis, and bipolar disorder (Gervin et al. 2012; Javierre et al. 2010; Kuratomi et al. 2008; Rakyan et al. 2011). Finally, a total of 234 differentially methylated CpG sites that were common between the cohorts were identified.
The present study demonstrated that altered DNA methylation in SCZ occurred at CpG sites not only in the CGIs but also in CGI shores and CGI shelves. As shown in Fig. 1, aberrant DNA methylation in SCZ was mostly observed at CpG sites in the CGIs (40.6 %). Interestingly, of the 95 differently methylated CpG sites in the CGIs, most of them were located in the promoter regions (75.8 %). Among these 72 differentially methylated CpG sites in the CGIs in the promoter regions, several genes, such as B3GAT2, HDAC4, and DGKI, have been implicated in SCZ (Kähler et al. 2011; Kim et al. 2010; Moskvina et al. 2009). When we compared to previous methylation studies using peripheral blood samples (Carrard et al. 2011; Chen et al. 2011; Melas et al. 2012), we could not replicate altered DNA methylation changes in SCZ in the COMT, HTR1A, and MAOA genes. The lack of replications between studies may be due to differences in sample size, CpG sites examined, and the demographical features of samples (age, sex, race, medications, clinical subtypes, or illness severity). In particular, antipsychotic drugs are well known to influence DNA methylation (Dong et al. 2008; Melas et al. 2012; Mill et al. 2008; Shimabukuro et al. 2006; Tremolizzo et al. 2005). Irizarry et al. demonstrated that altered DNA methylation in cancer occurred in CGI shores rather than in the CGIs, and DNA methylation changes in CGI shores were strongly related to gene expression (Irizarry et al. 2009). In the present study, 15 differentially methylated CpG sites in the regions flanking CGIs in the promoter regions were identified, and several genes, such as PCM1 and INSIG2, have been implicated in SCZ (Datta et al. 2010; Gurling et al. 2006; Lett et al. 2011). However, we did not observe more variable DNA methylation changes in SCZ in the regions flanking CGIs than in the CGIs. This observation is consistent with the findings of Deaton et al.’s report in the immune system (Deaton et al. 2011).
The present study also demonstrated that altered DNA methylation in SCZ occurred at CpG sites not only in the promoter regions but also in gene bodies. The role of DNA methylation in gene bodies is still unclear. Shann et al. demonstrated the correlation between intragenic hypomethylation and gene silencing in cancer cell lines (Shann et al. 2008), and Ball et al. demonstrated that gene body DNA methylation in highly expressed genes is a consistent phenomenon in human cells (Ball et al. 2009). Recently, it became apparent that CGIs in gene bodies act as alternative promoters (Illingworth et al. 2010; Maunakea et al. 2010) and that tissue-specific or cell type-specific CGI methylation is prevalent in gene bodies (Deaton et al. 2011; Maunakea et al. 2010). In the present study, 14 differentially methylated CpG sites in the CGIs in the gene bodies were identified. GFRA2 is one such gene of interest. The GFRA2 protein is a cell-surface receptor for GDNF and neurturin, and GDNF is a neurotrophic factor of dopaminergic neurons. The variants in this gene have been associated with tardive dyskinesia in patients with SCZ and antipsychotic responses in SCZ (Lavedan et al. 2009; Souza et al. 2010a, b).
There are several limitations to the present study. First, the sample size was not large. Replication studies will be needed in larger samples, including chronic patients with SCZ who are taking psychotic drugs. Second, the analyzed CpG sites were limited in number, although the 450 K microarray is one of the most powerful and cost-effective tools currently available for assessing methylation changes. Third, we demonstrated DNA methylation signatures of only peripheral leukocytes, not brain tissues. However, DNA methylation changes in major psychosis in the brain were also found in peripheral samples in particular genes (Kaminsky et al. 2012; Kuratomi et al. 2008). Hypermethylation of the RAI1 gene in SCZ in our study was also observed in a previous comprehensive DNA methylation study using post-mortem brain tissues (Mill et al. 2008). Finally, it is not possible to differentiate methylation from 5-hydroxymethylation of cytosine, which also plays a critical role in gene regulation (Bhutani et al. 2011).
In summary, aberrant DNA methylation in SCZ was identified at numerous CpG sites across the whole genome in peripheral leukocytes using two independent sets of samples. Of the differently methylated CpG sites in the CGIs, most of them were located in the promoter regions. These findings support the hypothesis that altered DNA methylation could be involved in the pathophysiology of SCZ.
References
Abdolmaleky, H. M., Cheng, K. H., Faraone, S. V., Wilcox, M., Glatt, S. J., Gao, F., et al. (2006). Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Human Molecular Genetics, 15, 3132–3145.
Ball, M. P., Li, J. B., Gao, Y., Lee, J., LeProust, E. M., Park, I., et al. (2009). Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature Biotechnology, 27, 361–368.
Bhugra, D. (2005). The global prevalence of schizophrenia. PLoS Medicine, 2, e151–e175.
Bhutani, N., Burns, D. M., & Blau, H. M. (2011). DNA demethylation dynamics. Cell, 146, 866–872.
Bibikova, M., Barnes, B., Tsan, C., Ho, V., Klotzle, B., Le, J. M., et al. (2011). High density DNA methylation array with single CpG site resolution. Genomics, 98, 288–295.
Cardno, A. G., & Gottesman, I. I. (2000). Twin studies of schizophrenia: From bow-and-arrowconcordances to star wars Mx and functional genomics. American Journal of Medical Genetics, 97, 12–17.
Carrard, A., Salzmann, A., Malafosse, A., & Karege, F. (2011). Increased DNA methylation status of the serotonin receptor 5HTR1A gene promoter in schizophrenia and bipolar disorder. Journal of Affective Disorders, 132, 450–453.
Chen, Y., Zhang, J., Zhang, L., Shen, Y., Xu, Q. (2011). Effects of MAOA promoter methylation on susceptibility to paranoid schizophrenia. Human Genetics [Epub ahead of print].
Colantuoni, C., Lipska, B. K., Ye, T., Hyde, T. M., Tao, R., Leek, J. T., et al. (2011). Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature, 478, 519–523.
Datta, S. R., McQuillin, A., Rizig, M., Blaveri, E., Thirumalai, S., Kalsi, G., et al. (2010). A threonine to isoleucine missense mutation in the pericentriolar material 1 gene is strongly associated with schizophrenia. Molecular Psychiatry, 15, 615–628.
Deaton, A. M., Webb, S., Kerr, A. R. W., Illingworth, R. S., Guy, J., Andrews, R., et al. (2011). Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Research, 21, 1074–1086.
Dedeurwaerder, S., Defrance, M., Calonne, E., & Sotiriou, C. (2011). Evaluation of the Infinium 450 K technology. Epigenomics, 3, 771–784.
Dempster, E. L., Pidsley, R., Schalkwyk, L. C., Owens, S., Georgiades, A., Kane, F., et al. (2011). Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human Molecular Genetics, 20, 4786–4796.
Dong, E., Nelson, M., Grayson, D. R., Costa, E., & Guidotti, A. (2008). Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proceedings of National Academy of Sciences of the United States of America, 105, 13614–13619.
Gervin, K., Vigeland, M. D., Mattingsdal, M., Hammerø, M., Nygård, H., Olsen, A. O., et al. (2012). DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: Identification of epigenetically dysregulated genes. PLoS Genetics, 8, e1002454.
Grayson, D. R., Jia, X., Chen, Y., Sharma, R. P., Mitchell, C. P., Guidotti, A., et al. (2005). Reelin promoter hypermethylation in schizophrenia. Proceedings of National Academy of Sciences of the United States of America, 10, 9341–9346.
Gurling, H. M., Critchley, H., Datta, S. R., McQuillin, A., Blaveri, E., Thirumalai, S., et al. (2006). Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Archives of General Psychiatry, 63, 844–854.
Harrison, P. J., & Weinberger, D. R. (2005). Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Molecular Psychiatry, 10, 40–68.
Illingworth, R. S., Gruenewald-Schneider, U., Webb, S., Kerr, A. R., James, K. D., Turner, D. J., et al. (2010). Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genetics, 6, e1001134.
International Schizophrenia Consortium. (2008). Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature, 455, 237–241.
Irizarry, R. A., Ladd-Acosta, C., Wen, B., Wu, Z., Montano, C., Onyango, P., et al. (2009). The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genetics, 41, 178–186.
Iwamoto, K., Bundo, M., Yamada, K., Takao, H., Iwayama-Shigeno, Y., Yoshikawa, T., et al. (2005). DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. The Journal of Neuroscience, 25, 5376–5381.
Javierre, B. M., Fernandez, A. F., Richter, J., Al-Shahrour, F., Martin-Subero, J. I., Rodriguez-Ubreva, J., et al. (2010). Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Research, 20, 170–179.
Kähler, A. K., Djurovic, S., Rimol, L. M., Brown, A. A., Athanasiu, L., Jönsson, E. G., et al. (2011). Candidate gene analysis of the human natural killer-1 carbohydrate pathway and perineuronal nets in schizophrenia: B3GAT2 is associated with disease risk and cortical surface area. Biological Psychiatry, 69, 90–96.
Kaminsky, Z., Tochigi, M., Jia, P., Pal, M., Mill, J., Kwan, A., et al. (2012). A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Molecular Psychiatry, 17(7), 728–740.
Kim, T., Park, J. K., Kim, H. J., Chung, J. H., & Kim, J. W. (2010). Association of histone deacetylase genes with schizophrenia in Korean population. Psychiatry Research, 178, 266–269.
Kuratomi, G., Iwamoto, K., Bundo, M., Kusumi, I., Kato, N., Iwata, N., et al. (2008). Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Molecular Psychiatry, 13, 429–441.
Lavedan, C., Licamele, L., Volpi, S., Hamilton, J., Heaton, C., Mack, K., et al. (2009). Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Molecular Psychiatry, 14, 804–819.
Leek, J. T., & Storey, J. D. (2007). Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genetics, 3, 1724–1735.
Lett, T. A., Wallance, T. J. M., Chowdhur, N. I., Tiwari, A. K., Kennedy, J. L., & Muller, D. J. (2011). Pharmacogenetics of antipsychotic-induced weight gain: Review and clinical implications. Molecular Psychiatry, 17, 242–266.
Maunakea, A. K., Nagarajan, R. P., Bilenky, M., Ballinger, T. J., D’souza, C., Fouse, S. D., et al. (2010). Conserved role of intragenic DNA mehtylaion in regulating alternative promoters. Nature, 466, 253–257.
Melas, P. A., Rogdaki, M., Osby, U., Challing, M., Lavebratt, C., & Ekstrom, T. J. (2012). Epigenetic aberrations in leukocytes of patients with schizophrenia: Association of global DNA methylation with antipsychotic drug treatment and disease onset. The FASEB Journal, 26, 2712–2718.
Mill, J., Tang, T., Kaminsky, Z., Khare, T., Yazdanpanah, S., Bouchard, L., et al. (2008). Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American Journal of Human Genetics, 82, 696–711.
Moskvina, V., Craddock, P., Nikolov, I., Pahwa, J. S., Green, E., Wellcome Trust Case Control Consortium, et al. (2009). Gene-wide analysis of genome-wide association data sets: Evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Molecular Psychiatry, 14, 252–260.
Nohesara, S., Ghadirivasfi, M., Mostafavi, S., Eskandari, M. R., Ahmadkhaniha, H., Thiagalingam, S., et al. (2011). DNA hypomethylation of MB-COMT promoter in the DNA derived from saliva in schizophrenia and bipolar disorder. Journal of Psychiatic Research, 45, 1432–1438.
Numata, S., Ye, T., Hyde, T. M., Guitart-Navarro, X., Tao, R., Wininger, M., et al. (2012). DNA methylation signatures in development and aging of the human prefrontal cortex. American Journal of Human Genetics, 90, 260–272.
Ono, S., Imamura, A., Tasaki, S., Kurotaki, N., Ozawa, H., Yoshiura, K., et al. (2010). Failure to confirm CNVs as of aetiological significance in twin pairs discordant for schizophrenia. Twin Research and Human Genetics, 13, 455–460.
Petronis, A., Gottesman, I. I., Crow, T. J., DeLisi, L. E., Klar, A. J., Macciardi, F., et al. (2000). Psychiatric epigenetics: A new focus for the new century. Molecular Psychiatry, 5, 34234–34236.
Purcell, S. M., Wray, N. R., Stone, J. L., Visscher, P. M., O’Donovan, M. C., Sullivan, P. F., et al. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460, 748–752.
Rakyan, V. K., Beyan, H., Down, T., Hawa, M. I., Maslau, S., Aden, D., et al. (2011). Identification of type 1 diabetes–associated DNA methylation variable positions that precede disease diagnosis. PLoS Genetics, 7, e1002300.
Rees, E., Moskvina, V., Owen, M. J., O’Donovan, M. C., & Kirov, G. (2011). De novo rates and selection of schizophrenia-associated copy number variations. Biological Psychiatry, 70, 1109–1114.
Sandoval, J., Heyn, H., Moran, S., Serra-Musach, J., Pujana, M. A., Bibikova, M., et al. (2011). Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics, 6, 692–702.
Shann, Y. J., Cheng, C., Chiao, C. H., Chen, D. T., Li, P. H., & Hsu, M. T. (2008). Genome-wide mapping and characterization of hypomethylated sites in human tissues and breast cancer cell lines. Genome Research, 18, 791–801.
Shi, J., Levinson, D. F., Duan, J., Sanders, A. R., Zheng, Y., Pe’er, I., et al. (2009). Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature, 460, 753–757.
Shimabukuro, M., Jinno, Y., Fuke, C., & Okazaki, Y. (2006). Haloperidol treatment induces tissue- and sex-specific changes in DHA methylation: A control study using rats. Behavioral and Brain Functions, 2, 37.
Siegmund, K. D., Connor, C. M., Campan, M., Long, T. I., Weisenberger, D. J., Biniszkiewicz, D., et al. (2007). DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE, 2(9), e895.
Souza, R. P., de Luca, V., Remington, G., Lieberman, J. A., Meltzer, H. Y., Kennedy, J. L., et al. (2010a). Glial cell line-derived neurotrophic factor alpha 2 (GFRA2) gene is associated with tardive dyskinesia. Psychopharmacology (Berl), 210, 347–354.
Souza, R. P., Romano-Silva, M. A., Lieberman, J. A., Meltzer, H. Y., MacNeil, L. T., Culotti, J. G., et al. (2010b). Genetic association of the GDNF alpha-receptor genes with schizophrenia and clozapine response. Journal of Psychiatry Research, 44, 700–706.
Stefansson, H., Ophoff, R. A., Steinberg, S., Andreassen, O. A., Cichon, S., Rujescu, D., et al. (2009). Common variants conferring risk of schizophrenia. Nature, 460, 744–747.
Sullivan, P. F., Kendler, K. S., & Neale, M. C. (2003). Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Archives of General Psychiatry, 60, 1187–1192.
Tremolizzo, L., Doueiri, M. S., Dong, E., Grayson, D. R., Davis, J., Pinna, G., et al. (2005). Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biological Psychiatry, 57, 500–550.
Acknowledgments
The authors would like to thank all the volunteers who understood our study purpose and participated in this study, and the physicians who helped us to collect clinical data and blood samples at the mental hospitals. The authors would also like to thank Mrs. Akemi Okada for her technical assistance. This work was supported by Japan Science and Technology Agency, CREST and by a Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science and Technology. The all authors report no biomedical financial interests or potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Makoto Kinoshita and Shusuke Numata contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kinoshita, M., Numata, S., Tajima, A. et al. DNA Methylation Signatures of Peripheral Leukocytes in Schizophrenia. Neuromol Med 15, 95–101 (2013). https://doi.org/10.1007/s12017-012-8198-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-012-8198-6