Abstract
We sought to determine the expression levels of hypoxia-inducible factor-1α (HIF-1α) in the bone marrow chronic myelogenous leukemia (CML) patients. We also tried to determine the roles HIF-1α in the proliferation of CML cells by small interfering RNA (siRNA) knockdown. Real-time PCR was performed to determine the expression levels of HIF-1α in the bone marrows of CML patients and healthy volunteers. HIF-1α knockdown by siRNA in K562 cells was confirmed by RT-PCR. Proliferation and colony formation of the treated cells were determined by CCK8 after HIF-1α knockdown. RT-PCR and western blotting were performed to detect mRNA and protein levels of p21 and p53 in K562 cells. HIF-1α mRNA expression in the bone marrow of CML patients was significantly higher than that in the control, which was statistically significant (P < 0.05). HIF-1α knockdown dramatically reduced the proliferation of K562 cells, which was also statistically significant (P < 0.05). HIF-1α knockdown markedly reduced the colony formation ability of K562 cells, which was also statistically significant (P < 0.05). The mRNA and protein expression of p21 were significantly reduced in K562 cell after HIF-1α knockdown with affecting the mRNA and protein levels of p53. HIF-α promotes chronic CML cell proliferation by up-regulating p21 expression
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chronic myelogenous leukemia (CML) is a disease derived from hematopoietic stem cells. The main feature of the disease at the molecular level is the generation of Philadelphia chromosome and BCR/ABL onco-protein by t(9;22) chromosomal translocation. The fusion protein BCR/ABL exhibits very high tyrosine kinase activity, which can induce malignant transformation of cells. Hypoxia-inducible factor-1α (HIF-1α) is a nuclear protein with transcriptional activity, which performs a variety of biological functions. It exhibits especially high expression during tumor growth in a low oxygen environment [1]. However, the expression level of HIF-1α in patients with chronic myeloid leukemia and how it is correlated with the pathogenesis of chronic myeloid leukemia remain to be understood. Here, we detected and analyzed the expression levels of HIF-1α in patients with CML. We also knocked down the expression of HIF-1α by small interfering in CML cell lines and analyzed how HIF-1α impacts on the proliferation of these cell lines to explore the possible molecular mechanisms of HIF-1α during the pathogenesis of CML.
Materials and Methods
Patient Samples
Five bone marrow samples were collected from CML patients and three from healthy volunteers. All the samples were obtained by bone marrow biopsy. Diagnosis and staging were according to “The diagnosis and staging of blood disorders” by Dr. Zhinan Zhang. Healthy controls were relative, that is to say, slight change in the number of blood cells by blood test, but they were confirmed as normal by biopsies.
Experimental Materials and Methods
K562 CML cell line was used in all the experiments. Incomplete low glucose DMEM medium and fetal bovine serum (FBS) were purchased from Gibco (USA). Trizol kit was from Invitrogen (Shanghai) and cDNA reverse transcription kit was purchased from Femantes (USA). PCR kit, DNA Marker, DEPC, penicillin, and streptomycin were purchased from Shengxing Bio limited (Nanjing).
Transfection
Lipofectamine™ 2000 was used for the transfection of K562 cells by siRNA-HIF-1α and HIF-1α plasmid (Shanghai Genepharma). HIF-1α siRNA sequence was 5′-TACGTTGTGAGTGGTATTATT and control sequence was 5′-AATTCTCCGAACGTGTCACGT. Transfection efficiency was determined by RT-PCR.
Total RNA Extraction and Reverse Transcription
0.2 ml Trizol was added to the collected K562 cell 48 h after transfection. Total RNA was extracted according to the manufacturer’s instructions and dissolved in 30 μl DEPC-treated water. The concentration and purity was determined by UV photo spectrometer (NanoDrop ND-1000). RNA was further reverse transcribed into cDNA using Femantes reverse transcription kit and stored at −20 °C. Real-Time PCR was performed with Real-Time PCR 7300 machine (ABI). PCR condition was as follows: denature at 95 °C for 20 s, 60 °C for 20 s, and 70 °C for 10 s, the cycle was repeated 40 times. Data were quantitated with \(2_{{}}^{{ - \Delta \Delta C_{{\text{t}}} }}\) method.
Primer sequences
Gene | Primer sequence (F: forward R: reverse) |
|---|---|
p21 | F: 5-AGCAGAGGAAGACCATGTGG |
R:5-GGGTATGTACATGAGGAGGT | |
p53 | F: 5-CAGCCAAGTCTGTGACTTGCACGTAC |
R: 5-CTATGTCAAAAAGTGTTTCTGTCATC | |
GAPDH | F: 5-TGACGGGGTCACCCACACTGTGCCCATCTA |
R: 5-CTAGAAGCATTTGCGGTGGACGATGGAGGG |
MCF-7 Cell Proliferation Analysis by CCK-8
K562 cells were transfected with control and HIF-1α siRNA at 50–60 % confluency. DMEM and Cell Counting Kit-8 (CCK-8, cell signaling, USA) were mixed in the ratio of 1:10 and added into each well 0, 24, 48, and 72 h after transfection. OD was measured at 450 nm by photo spectrometer, and cell growth curve was graphed with the data.
Western Blotting
K562 cells were collected 48 h after HIF-1α knockdown and lysed with 1× SDS cell lysis buffer. The samples were subjected to SDS-PAGE and transferred to membrane at 110 V for 90 min. The membrane was blocked at 37 °C for 2 h and incubated with p21 antibody (Biolegend) and p53 antibody (Biolegend) at 4 °C for 12 h. HRP-conjugated mouse-anti-rabbit secondary antibody (Nanjin Shengxing Bio, 1:1000 dilution) was incubated at 37 °C for 40 min and ECL was used for detected. β-Actin (Sigma Aldrich) was used as internal control.
Statistics and Graphing
All data were analyzed using SPSS17.0 statistical software, measurement data were expressed as mean ± standard deviation, and the two groups were compared student’s t test. P < 0.05 was considered as statistically significant.
Results
Increased HIF-1α Expression in Bone Marrows of CML Patients
By quantitative real-time PCR, we compared the mRNA expression of HIF-1α in the bone marrow samples of five CML patients and three relatively normal patients. Compared with the normal controls, HIF-1α expression increased significantly in bone marrows from CML patients (P < 0.05, Fig. 1). Our results showed that abnormally high HIF-1α expression occurs in the diseased bone marrow microenvironment and HIF-1α could be an important regulator of bone marrow hematopoietic microenvironment in CML.
HIF-1α Knockdown Reduced the Proliferation of K562 CML Cells
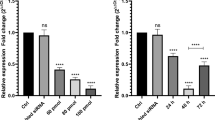
Compared with controls, HIF-1α knockdown in K562 cells by siRNA led to marked reduction of HIF-1α mRNA (Fig. 2a). Significantly reduced proliferation of K562 cells was observed after HIF-1α knockdown (Fig. 2b), which was statistically significant (P < 0.05). Our results indicated that HIF-1α positively promoted CML cell proliferation.
HIF-1α Knockdown Decreased the Colony Formation of K562 CML Cells
Compared with controls, HIF-1α siRNA transfected K562 cells displayed dramatically reduced colonies (Fig. 3), which was statistically significant (P < 0.05). These results suggested that HIF-1α is required for the colony formation of CML cells.
HIF-1α Knockdown Resulted in Decreased p21 Expression
We further examined apoptosis-related gene expression after HIF-1α knockdown. Our results showed that HIF-1α knockdown significantly decreased p21 mRNA level in K562 cell (Fig. 4a). However, p53 mRNA level was not noticeably affected (Fig. 4c). Moreover, the reduction in p21 protein level was also confirmed after HIF-1α knockdown (Fig. 4b). Besides, p53 protein level was not affected (Fig. 4d). These results showed that transcriptional activation of p21 by HIF-1α was required for the proliferation of CML cells.
Discussion
Hypoxia represents a basic feature of tumor microenvironment in solid tumors, which can trigger the malignant transformation or even the metastasis of tumors. Accumulating clinical data have shown that hypoxia inside tumors is an important indicator for poor prognosis. The effects of hypoxia are executed by a class of proteins named hypoxia-inducible factor-1. Among them, HIF-1α is main subclass with important activity. Its expression can be induced and kept stable under hypoxia condition [2–4]. Recent studies have identified key roles of HIF-1α during the occurrence, development, and lymph node metastasis, and it is also an important indicator for cancer treatment and prognosis. HIF-1α has been shown to regulate multiple cancer-related gene expression upon activation, such as factors related to tumor invasion and metastasis, angiogenesis, proliferation, and apoptosis [5–8]. HIF-1α helps cells to adapt to hypoxia environment.
Myelogenous leukemia is a malignant proliferative disease derived from hematopoietic stem cells, which takes up more than 20 % of all the adult leukemia. Currently, translocation of proto-oncogene c-ab1 on chromosome 9 to the end of chromosome 22 to form the fused BCR/ABL gene is one of the major causes of the disease [9], but other relevant cause and regulatory mechanisms remain to be discovered. Under normoxia, HIF-1α expression is kept low. However, HIF-1α expression is induced under hypoxia to complex with other cytoplasmic protein for signal transduction [10]. During tumor growth, rapid proliferation of tumor cells leads to local hypoxia and induce high expression of HIF-1α [11, 12]. Thus, we compared HIF-1α expression in the bone marrows of CML patients and healthy volunteers. We also performed loss-of-function assays by siRNA knockout to further determine the roles of HIF-1α in CML.
Our results showed significantly increased HIF-1α in the bone marrows of CML patients compared with controls, which suggested important roles HIF-1α in the pathogenesis of CML. Using siRNA interference, we were able to knock down HIF-1α expression in CML cell line and found reduced proliferation and colony formation ability, which further confirmed the value of HIF-1α expression levels in the diagnosis and prognosis of myelogenous leukemias. Our observation is in line with the proposed roles of HIF-1α in other malignancies [13–15]. Previous studies have shown that p53 is an important gene to maintain the normal cell growth, inhibit malignant proliferation and monitor genomic DNA integrity. When genomic DNA is damaged, p53 is activated to transcribe p21 and arrest cell cycle at G1 phase for the repair of the DNA damage. If the repair fails, p53 immediately initiates apoptosis program to prevent the formation of mutant cells and subsequent malignant transformation [16, 17]. Our results showed that HIF-1α knockdown could significantly inhibit the mRNA and protein expression of p21 in CML cells without affecting the mRNA and protein expression of p53. These observations suggested that HIF-1α could promote CML cell proliferation by modulating p21 expression [18].
In summary, the expression level of HIF-1α plays an important role in the pathogenesis of CML, which could promote the proliferation of CML cell by modulating p21 expression. Further understanding of HIF-1α functions in CML can be of great clinical value for the prognosis of CML.
References
Esteban, M. A., & Maxwell, P. H. (2005). HIF, a missing link between metabolism and cancer. Nature Medicine, 11, 1047–1048.
Brogi, E., Schatteman, G., Wu, T., Kim, E. A., Varticovski, L., Keyt, B., & Isner, J. M. (1996). Hypoxia-induced paracrine regulation of vascular endothelial growth factor receptor expression. Journal of Clinical Investigation, 97, 469–476.
Janssen, H. L., Haustermans, K. M., Sprong, D., Blommestijn, G., Hofland, I., Hoebers, F. J., et al. (2002). HIF-1A, pimonidazole, and iododeoxyuridine to estimate hypoxia and perfusion in human head-and-neck tumors. International Journal of Radiation Oncology Biology Physics, 54, 1537–1549.
Kitajima, Y., & Miyazaki, K. (2013). The critical impact of HIF-1a on gastric cancer biology. Cancers (Basel), 5, 15–26.
Kelly, B. D., Hackett, S. F., Hirota, K., Oshima, Y., Cai, Z., Berg-Dixon, S., et al. (2003). Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circulation Research, 93, 1074–1081.
Huang, J. H., Lee, F. S., Pasha, T. L., Sammel, M. D., Karakousis, G., Xu, G., et al. (2010). Analysis of HIF-1a and its regulator, PHD2, in retroperitoneal sarcomas: Clinico-pathologic implications. Cancer Biology & Therapy, 9, 303–311.
Oladipupo, S. S., Hu, S., Santeford, A. C., Yao, J., Kovalski, J. R., Shohet, R. V., et al. (2011). Conditional HIF-1 induction produces multistage neovascularization with stage-specific sensitivity to VEGFR inhibitors and myeloid cell independence. Blood, 117, 4142–4153.
Kaidi, A., Williams, A. C., & Paraskeva, C. (2007). Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nature Cell Biology, 9, 210–217.
Chomel, J. C., Sorel, N., Guilhot, J., Guilhot, F., & Turhan, A. G. (2012). BCR-ABL expression in leukemic progenitors and primitive stem cells of patients with chronic myeloid leukemia. Blood, 119, 2964–2965. author reply 2965–2966.
Makino, Y., Uenishi, R., Okamoto, K., Isoe, T., Hosono, O., Tanaka, H., et al. (2007). Transcriptional up-regulation of inhibitory PAS domain protein gene expression by hypoxia-inducible factor 1 (HIF-1): A negative feedback regulatory circuit in HIF-1-mediated signaling in hypoxic cells. Journal of Biological Chemistry, 282, 14073–14082.
O’Donnell, J. L., Joyce, M. R., Shannon, A. M., Harmey, J., Geraghty, J., & Bouchier-Hayes, D. (2006). Oncological implications of hypoxia inducible factor-1alpha (HIF-1alpha) expression. Cancer Treatment Reviews, 32, 407–416.
Gort, E. H., Groot, A. J., Derks van de Ven, T. L., van der Groep, P., Verlaan, I., van Laar, T., et al. (2006). Hypoxia-inducible factor-1alpha expression requires PI 3-kinase activity and correlates with Akt1 phosphorylation in invasive breast carcinomas. Oncogene, 25, 6123–6127.
Lee, J. Y., Choi, J. Y., Lee, K. M., Park, S. K., Han, S. H., Noh, D. Y., et al. (2008). Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clinica Chimica Acta, 389, 167–170.
Bardos, J. I., & Ashcroft, M. (2004). Hypoxia-inducible factor-1 and oncogenic signalling. BioEssays, 26, 262–269.
Jiang, Y. A., Fan, L. F., Jiang, C. Q., Zhang, Y. Y., Luo, H. S., Tang, Z. J., et al. (2003). Expression and significance of PTEN, hypoxia-inducible factor-1 alpha in colorectal adenoma and adenocarcinoma. World Journal of Gastroenterology, 9, 491–494.
Nakatsuka, A., Wada, J., Hida, K., Hida, A., Eguchi, J., Teshigawara, S., et al. (2012). RXR antagonism induces G0/G1 cell cycle arrest and ameliorates obesity by up-regulating the p53-p21(Cip1) pathway in adipocytes. Journal of Pathology, 226, 784–795.
Yang, X., Wang, W., Qin, J. J., Wang, M. H., Sharma, H., Buolamwini, J. K., et al. (2012). JKA97, a novel benzylidene analog of harmine, exerts anti-cancer effects by inducing G1 arrest, apoptosis, and p53-independent up-regulation of p21. PLoS One, 7, e34303.
Wu, G., Lin, N., Xu, L., Liu, B., & Feitelson, M. A. (2013). UCN-01 induces S and G2/M cell cycle arrest through the p53/p21(waf1) or CHK2/CDC25C pathways and can suppress invasion in human hepatoma cell lines. BMC Cancer, 13, 167.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, H., Shen, Y., Gong, F. et al. HIF-α Promotes Chronic Myelogenous Leukemia Cell Proliferation by Upregulating p21 Expression. Cell Biochem Biophys 72, 179–183 (2015). https://doi.org/10.1007/s12013-014-0434-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-0434-2