Abstract
Although fluoride is known to reduce the incidence of caries, chronic excessive fluoride exposure can impair human health, even resulting in fluorosis. Now the underlying mechanisms of fluoride-induced toxicity are not fully understood. So, we conducted this study with the purpose of investigating the effect of sodium fluoride (NaF) in human RPMI8226 cells. In this experiment, human RPMI8226 cells were cultured with varied doses of fluoride (10, 20, 40, 80, 160, 320 μM). After 48 h exposure, the change of cell viability was examined by CCK-8 assay, and also the messenger RNA (mRNA) expression of relevant genes was assessed by QRT-PCR. Compared to the control group, fluoride exposure increased the human RPMI8226 cells viability at relatively lower levels (10–160 μM); however, when the concentration reached to 320 μM, the cell proliferation was significantly inhibited (p < 0.05). In addition, the genes mRNA expression, including ANKRD1, CRSP6, KLF2, SBNO2, ZNF649, FANCM, PDGFA, RNF152, CDK10, and CETN2 changed in a concentration-dependent manner and increased with fluoride exposure concentration. The results suggest that overexposure to fluoride (160–320 μM) can induce cytotoxicity and regulate relevant genes expression. Our findings provide novel insights into the mechanisms of action of fluoride-induced toxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fluorides are naturally present in soil, water, and food, which exist only in combination with other elements as compounds; hence, pathways of fluoride exposure are related with dental products, food, pesticides, and water. According to previous study [1], although fluorides are normally used in low concentrations to reduce the incidence of caries, chronic excessive fluoride exposure can impair human health, even resulting in fluorosis, a disease caused by deposition of excessive fluoride in the body, which has been reported in endemic areas including 29 provinces, municipalities, and autonomous regions of China [2–4]. Fluorosis can cause damage not only to skeletal tissue and teeth, but also to soft tissues, such as brain, liver, kidney, pancreas, and spinal cord [5]. Early research [6] reported that, F− concentrations from 16 to 64 mg/kg are lethal to human adults while lower concentrations (3–16 mg/kg) are deleterious for infants. It has been subsequently established that the minimal risk level for daily oral F- uptake is 0.05 mg/kg/d. In fact, it has been confirmed that the exposure level of fluoride higher than 0.15 mg/kg/d can result in fluorosis.
In order to investigate the mechanism of fluoride-induced toxicity, several researches have been done which confirm that the sensitivity to these effects can vary, depending on cells type, exposure concentration, and so on [7–9].
First of all, the growth of cells in culture was inhibited by sodium fluoride exposure. Je-Seon Song et al. demonstrated that at higher concentrations (100–250 ppm, equivalent to 2.4–6.0 μM)), NaF induced HL-60 cell viability and inhibited its growth, also anti-proliferative effects of NaF appeared to be attributable to its induction of apoptotic cell death [10]. Another investigation revealed that excessive fluoride (1000–5000 μM) inhibited cell proliferation of ameloblasts-like LS8 cells. After 72 h treatment, cell proliferation was significantly reduced compared with 24 and 48-h treatment at both low (<2000 μM mM) and high dose (>2000 μM) exposure [11].
Furthermore, a lot of studies have been focused on apoptosis induced by fluorosis [12–16]. Yan-Jie Liu et al. reported that exposure to excessive fluoride (50 ppm, equivalent to 1.2 μM) for 6 months resulted in the increase of apoptosis in rat brains and SH-SY5Y cells, in which one of the mechanism might be activating JNK phosphorylation [17]. As for the osteoblasts, Shiyong Yang et al. found that NaF reduced the viability of MC3T3-E1 osteoblastic cells in a temporal and concentration-dependent manner and promoted apoptosis even at low concentrations (0.01 μM). This increased apoptosis was due to alternation in the expression of both pro-apoptosis and anti-apoptosis proteins which suggested that its mechanism was the direct effects of fluoride on expression of Bcl-2 family members [18].
Fluoride cytotoxicity has also been associated with general changes in DNA, RNA, and protein biosynthesis [19]. Je-Seon Song et al. found that at higher concentration (100–250 ppm, equivalent to 2.4–6.0 μM), NaF reduced cell viability and decreased DNA and protein biosynthesis capability in cultured HL-60 cells. Then, the NaF treatment also decreased the expression of the anti-apoptosis protein Bcl-2 and increased activation of caspase-3 and cleavage of poly (ADP-ribose) polymerase which might be the important mechanism by which NaF mediates cytotoxicity and apoptosis [10]. Another research showed that exposure to 4000 μM NaF for 24 h induced apoptosis in odontoblast-lineage cell, which is mediated via a mitochondria-dependent mechanism characterized by cytochrome c release as well as changes in the expression of Bcl-2 family. In this process, the JNK-MARK pathway also played an important role in the apoptosis [20]. In addition to this, many other experiments also came to the same conclusion that high amount of fluoride can alter the expression of Bcl-2 family members [11, 21]. Bcl-2 family members include both pro-(bax/bad proteins) and anti-(bcl-2, bcl-XL, bcl-W) apoptotic members and the ratio between apoptotic promotors and inhibitors determines whether, or not, a cell undergoes apoptosis [22]. Hence, it is considered that Bcl-2 family members might be involved in this process of cell apoptosis [11].
On the other side, after the incubation with various concentrations (20, 40, 80 mg/L, equivalent to 0.48, 0.95, 1.9 μM) of sodium fluoride, gene expression of NF-kB in rat hippocampal neurons was found enhanced in a dose-dependent manner. Also, fluoride could result in S-phase cell-cycle arrest and DNA damage in primary rat hippocampal neurons. Though fluoride may exhibit neurotoxic effects on cells, the detailed mechanisms need further investigation [5]. In another experiment, compared to the relevant controls, a significant decrease of the concentration of Dikkopf-1 (Dkk-1) and Sclerostin (SOST) in rat skin fibroblasts, belonging to the inhibitors of Wnt/β-catenin pathway, was observed as the fluoride concentration increased (0–20 mg/L, equivalent to 0–0.48 μM). So, they were treated as important indicators for diagnosing bone metabolism changes caused by fluoride exposure [23]. Recently, the expression of core-binding factorα (Cbfa1) and osteocalcin (OCN) were found increased in fibroblasts and osteoblasts after exposure to fluoride (0–20 mg/L, equivalent to 0–0.48 μM) and results suggested that the osteogenic function of fibroblasts induced by fluoride could play an important role in the development of extraperiosteal ossification during skeletal fluorosis [24].
Though different levels of fluoride-induced toxicity have been studied, the data is not enough to explain the molecular mechanism. Now, the effective therapy of fluoride-induced diseases is in an urgent need. So, based on the previous researches, we carried out further study to explore the clear mechanism of fluoride toxicity. In the present study, the human RPMI8226 cells were exposed to various concentration (10–320 uM) NaF and its proliferation and expressions of relevant genes in cultured cells were investigated through CCK-8 and QRT-PCR, respectively.
Materials and Methods
Materials and Chemicals
Human RPMI8226 cells were purchased from Peking Union Medical College (Beijing, China). RPMI8226 cell line is one of the most widely used human cell lines for studying the cellular and molecular effect induced by exogenous substances. Sodium fluoride was purchased from Sinopharm Chemical Regent Co., Ltd. Culture media RPMI-1640 and Phosphate Buffered Saline (PBS) were purchased from Hyclone (Thermo Scientific, Logan, UT, USA). Fetal bovine serum (FBS) was purchased from Gibco® life technologies. The Cell Counting Kit-8 (CCK-8 kit) was purchased from Dojindo Molecular Technologies, Inc. The E.Z.N.A.TM Total RNA KitIIkits were purchased from Omega Bio-Tek Inc. The complementary DNA (cDNA) reverse transcription kits were purchased from Applied Biosystems®. SYBR® Premix Ex Taq TM II was obtained from the Takara Biotechnology Company (Dalian, China). All quantitative real-time polymerase chain reaction (QRT-PCR) primers were synthesized by Sangon Biotech (Shanghai, China) Co.,Ltd. β-actin was also purchased from Sangon Biotech (Shanghai, China). All of other chemicals used in the experiment were of analytical grade.
Cell Culture and Treatment
Human RPMI8226 cells were cultured in RPMI 1640 media supplemented with 10 % fetal bovine serum (FBS) in humidified incubators with 5 % CO2/95 % air at 37 °C. The cells were maintained in culture for 7 days before treatment and the initial culture period allowed for a moderate increase in cell numbers. After that, exponentially growing human RPMI8226 cells were exposed to NaF dissolved in phosphate-buffered saline (PBS). Cells in the control group were incubated with PBS only. The concentrations of treated groups in contemporary study were set as follows: 10, 20, 40, 80, 160, and 320 μM. For individual experiments, the cells were subcultured with various concentrations of NaF for up to 48 h.
Cell Proliferation Assay
Effect of fluoride on cell proliferation was measured by Cell Counting Kit-8 (CCK-8) assay. Exponentially growing human RPMI8226 cells were collected through centrifuging at a speed of 800 r/min for 8 min. The number of cells was counted; then cells were seeded at 1 × 105 cells/ml in 96-well plates with 100 μL per well. Cells were exposed to NaF with the final concentrations of 10, 20, 40, 80, 160, and 320 μM. After 48 h, 10 μL CCK-8 reagent per well was added and then incubated for 2 h. Fluorescence signals were recorded using an enzyme labeled instrument Model 680 (Bio-Rad, USA) at 450 nm wavelength. The cell viability was calculated according to the following formula:
ODdose is the optical density of treated group, while ODPBS represents the optical density of the control group. Experiments were performed in triplicate.
RNA Extraction and Quantitative Real-time Polymerase Chain Reaction (QRT-PCR)
After 48 h exposure, total RNA was extracted from RPMI8226 cells using E.Z.N.A.TM Total RNA Kit, following the manufacturer’s instructions. Then, the RNA purity was tested by spectrophotometry, and the OD260/OD280 ratio between 1.8 and 2.0 was accepted. Five nanograms of total RNA from each sample was reverse-transcribed into single-stranded cDNA using the cDNA reverse transcription kits. The synthesized cDNA with primer and SYBR Green PCR master mix was then amplified by RT-PCR. β-actin was used as an internal control. The thermal cycling conditions were 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, Tm for 30 s, 72 °C for 1 min, and, finally, ending with 72 °C for 5 min, 94 °C for 1 min, and 61 °C for 10 s. The instrument used for RT-PCR is Bio-Rad, IQ5 Multicolor Real-Time PCR Detection System (575BR, USA). All the assays were performed in triplicate. Melting curve analysis was included to assure that only one PCR product was formed. The relative amount of RNA was calculated by the 2−△△Ct method.
Statistical Analysis
The data were expressed as means ± S.D. The difference between control and treated groups were assessed by one-way ANOVA followed by the Tukey–Kramer multiple comparison test. A probability of p < 0.05 was considered as significant difference.
Results
Effect of Fluoride on the Cell Proliferation
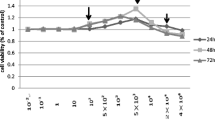
In this experiment, RPMI8226 cells were treated with various concentration of NaF. After 48-h treatment, the effect of fluoride on the cell viability was examined by CCK-8 assay. As shown in Fig. 1, the cell viability presented obvious changes with the exposure concentration. As the exposure concentration increased, the cell viability in treatment groups presented the situation that increased firstly and then decreased. In the treatment groups (cells were treated with 10, 20, 40, 80, 160 μM NaF, respectively), the cell viability were all shown to be higher than that of the control group; especially the groups with 10, 20, 40, 160 μM NaF displayed a significant increase compared to the control group (p < 0.01, p < 0.01, p < 0.05, p < 0.01, respectively). However, when the exposure concentration was increased to 320 μM, the cell viability exhibited significant decrease in comparison to the control group (p < 0.05).
Effect of Fluoride on the mRNA Extraction
In our previous experiment, the genes which presented obvious up-regulation or down-regulation after exposure to various NaF concentration have been already filtered by the microarray analysis technology. In order to test and verify the microarray analysis results, the changes of messenger RNA (mRNA) extraction after NaF exposure were examined by QRT-PCR. Figures 2, 3, and 4 showed the results.
The Changes of mRNA Extraction Relating to Transcription
According to the previous microarray analysis results, we selected seven genes relating to transcription including ankyrin repeat domain 1 (ANKRD1), nuclear receptor binding factor 2 (NRBF2), cofactor required for Sp1 transcriptional activation, subunit 6 (CRSP6), Kruppel-like factor 2 (KLF2), myeloid/lymphoid or mixed-lineage leukemia 3 (MLL3), strawberry notch homolog 2 (SBNO2), and zinc finger protein 649 (ZNF649). Figure 2 displayed the changes of these genes mRNA extraction after exposure to different concentration. As indicated in Fig. 2, the relative mRNA expression of ANKRD1 in high concentration treatment groups (cells were treated with 160, 320 μM NaF, respectively) was found increased significantly compared to the control group (p < 0.01). As for the CRSP6, the relative mRNA expression showed increasing trend with exposure concentration. The amount of mRNA in treatment groups (cells were treated with 10, 40, 160, 320 μM NaF, respectively) were shown to be higher than the control group (p < 0.01), as well as the same trend was found in the group of 20 μM exposure concentration (p < 0.05). Also, in comparison to the control group, the relative mRNA expression of KLF2 in groups of 80 and 320 μM exposure concentration significantly increased (p < 0.01). Meanwhile, the relative mRNA expression of SBNO2 and ZNF649 in the highest group of 320 μM NaF was found significantly increased compared to the control group (p < 0.01). However, the comparison between the treatment groups and control group of another two genes, NRBF2 and MLL3, were not found significant different.
The Changes of mRNA Extraction Relating to Protein Biosynthesis
The changes of mRNA extraction relating to protein biosynthesis were also examined by the QRT-PCR, including SH3-domain binding protein 2 (SH3BP2), ring finger protein 8, E3 ubiquitin protein ligase (RNF8), Fanconi anemia, complementation group M (FANCM), and melanoma antigen family F, 1 (MAGEF1). The relative mRNA expressions of the four genes were shown in Fig. 3. Among the four genes, only the relative mRNA expression of FANCM in high treatment groups (cells were exposed to 80 and 320 μM NaF, respectively) was found elevated significantly compared to the control group. As for another three genes, the relative mRNA expression presented no obvious different as the exposure concentration increased. The results revealed that only FANCM expression was affected by high fluoride exposure, while the expression of SH3BP2, RNF8, and MAGEF1 were not influenced significantly.
The Changes of mRNA Extraction Relating to Cell Proliferation, Apoptosis, and Cycle
In order to explore the clear cytotoxicity mechanism, the relative mRNA expressions of platelet-derived growth factor alpha polypeptide (PDGFA), ring finger protein 152 (RNF152), cyclin-dependent kinase 10 (CDK10), and centrin, EF-hand protein, 2 (CETN2) were analyzed by QRT-PCR. As depicted in Fig. 4, the relative mRNA expression of PDGFA in higher exposure concentration groups (cells were treated with 160 and 320 μM NaF, respectively) were significantly increased compared to the control group (p < 0.01). Also, the relative mRNA expression of RNF152 in three treatment groups (cells were 20, 160, and 320 μM NaF, respectively) were significantly higher than the control group (p < 0.01). Moreover, the CDK10 relative expression levels in treatment groups (cells were 10, 160, and 320 μM NaF, respectively) exhibited increase significantly compared to the control group (p < 0.01, p < 0.05, p < 0.01, respectively). In addition, the CETN2 relative expression levels were found significantly elevated in treatment groups (cells were 10, 40, 160, and 320 μM NaF, respectively) in comparison to the control group.
Discussion
As we all know, fluoride is an essential trace element for bone and dental health in human beings and animals; however, chronic excessive intake is known to impair health. Previous research result [10] revealed that the increased use of fluorides in industry, medicine, and dentistry makes human beings and animals exposed to even greater levels of fluoride compounds. In the early researches, the mechanism by which fluoride-induced toxicity still remains unclear. Therefore, we attempted to analyze the cytotoxicity of human RPMI8226 cells induced by NaF at various concentrations ranging from 0 to 320 μM.
In the present study, the effect of NaF treatment on proliferation of human RPMI8226 cells was observed; also, the changes of relevant genes mRNA expression induced by fluoride were investigated. Results demonstrated that both the cell proliferation and relevant genes mRNA expression were affected dramatically by relative high concentration of NaF.
According to the analysis result of Fig. 1, little cytotoxicity of NaF was observed at lower concentrations, in terms of cell viability. In fact, after 48-h treatment, we observed a slight increase in cell viability at the concentration of NaF < 160 μM (compared to the control group). However, when the exposure concentration increased to 320 μM, the cell proliferation was significantly inhibited. Actually, in the previous study, it has been reported that when exposed to 100–200 ppm NaF (equivalent to 2.4–4.8 μM), cell proliferation can be inhibited and apoptosis can be induced in human promyelocytic leukemia HL-60 cells [10]. Also, another research pointed that concentrations ranging from 0.05–1.0 μM can reduce MC3T3-E1 osteoblast-like cells viability after 48-h exposure [18]. Compared with these early studies, our study showed that just when the concentration increased to a relative high level (>320 μM), cell proliferation was significantly inhibited. So, it can be confirmed that with the same culture time, different cell lines are sensitive to different NaF concentration. What’s more, an early study revealed that culture of osteoblasts in the presence of low concentrations of NaF (0.5 and 5 mg/L, equivalent to 0.01–0.11 μM) for 72 h resulted in a subtle, but non-significant increase in osteoblasts proliferation, while culture in the presence of higher concentrations (20 and 30 mg/l, equivalent to 0.48–0.71 μM) dramatically reduced osteoblasts proliferation [25].
At the same time, alteration of cell proliferation is a tightly regulated process, which might involve changes in the expression of relevant genes. In this study, the changes of relevant genes mRNA expression were observed, including three groups: (1) genes relating to transcription (ANKRD1, NRBF2, CRSP6, KLF2, MLL3, SBNO2, and ZNF649); (2) genes relating to protein biosynthesis (SH3BP2, RNF8, FANCM, and MAGEF1); (3) genes relating to the cell proliferation, apoptosis, and cycle (PDGFA, RNF152, CDK10, CETN2). It seems that few researches were conducted to explore the effect of fluoride exposure on these genes. So, our study was made up for this gap.
The genes in group 1 function as the transcription factors. The activation of gene transcription is a multistep process that is triggered by factors that recognize transcriptional enhancer sites in DNA. Our results indicated that mRNA expressions of these genes were up-regulated dramatically by high level exposure (320 μM) except the NRBF2 and MLL3 (see Fig. 2). According to the results, we inferred that high fluoride exposure affected the cell function to regulate the gene transcription.
Also, the changes of mRNA extraction relating to protein biosynthesis were measured. Among these genes, only FANCM mRNA expression was found increased in relative high exposure level (80, 320 μM) (see Fig. 3). FANCM belongs to the Fanconi anemia complementation group (FANC). Fanconi anemia complementation group M protein (FANCM), a component of the Fanconi anemia pathway, acts as DNA translocase that is essential during the repair of DNA interstrand cross-links [26]. The members of the Fanconi anemia complementation group do not share sequence similarity; they are related by their assembly into a common nuclear protein complex. This gene encodes the protein for complementation group M. The results revealed that the cell function to regulate the protein biosynthesis also be influenced.
In addition, the genes relating to cell proliferation, apoptosis, and cycle were studied in our experiment. From Fig. 4, all the four genes, including PDGFA, RNF152, CDK10, and CETN2, were regulated by high fluoride exposure (160, 320 μM). The mRNA expressions of these four genes were exhibited increase significantly compared to the control group. The changes of the genes were consistent with the cell viability (see Figs. 1 and 4) except 320 μM. So, it was confirmed that at relative lower concentrations, the cell viability and relevant genes mRNA expression all changed in a concentration-dependent manner and increased with the exposure concentration. However, when exposure concentration reached to 320 μM, the cell viability was found decreased significantly while the relevant genes still increased at the exposure level.
Conclusion
In summary, at relative lower levels (10–160 μM), fluoride exposure increased the human RPMI8226 cells viability; however, when the concentration reached to 320 μM, the cell proliferation was significantly inhibited. Besides, the genes mRNA expression, including ANKRD1, CRSP6, KLF2, SBNO2, ZNF649, FANCM, PDGFA, RNF152, CDK10, and CETN2 changed in a concentration-dependent manner and increased with fluoride exposure concentration. The results suggest that overexposure to fluoride (160–320 μM) can induce cytotoxicity and regulate relevant genes expression.
References
Gabuda SP, Gaidash AA, Kozlova SG, Allan NL (2006) Structural forms of fluorides in bone tissue of animals under chronic fluoride intoxication. J Struct Chem 47(2):258–266
Lianfang W, Jianzhong H (1995) Outline of control practice of endemic fluorosis in China. Soc Sci Med 41(8):1191–1195
Watanabe T, Kondo T, Asanuma S, Sakurai S, Tamura K, Ando M (1997) Endemic fluorosis in southern China: radiological findings. Nihon Igaku Hoshasen Gakkai Zasshi. Nippon Acta Radiol 57(7):425–426
Wei ZD, Wei Y (2002) Fluoridation in China: a clouded future. Fluoride 35(1):1–4
Ming Z, Aiguo W, Tao X, Ping H (2008) Effects of fluoride on DNA damage, S-phase cell-cycle arrest and the expression of NF-kB in primary cultured rat hippocampal neurons. Toxicol Lett 179:1–5
National Research Council (2006) Fluoride in drinking water: a scientific review of EPA’s standards. The National Academic Press, Washington D.C
Helgeland K, Leirsker J (1976) pH and the cytotoxity of fluoride in an animal cell culture system. Scand J Dent Res 84:37–45
Holland RI (1980) Cytotoxicity of fluoride. Acta Odontol Scand 38:69–79
Barbier O, Arreola ML, Del Razo LM (2010) Molecular mechanisms of fluoride toxicity. Chem-Biol Interact 188(2):319–333
Song JS, Lee HY, Lee E, Hwang HJ, Kim JH (2002) Cytotoxicity and apoptosis induction of sodium fluoride in human promyelocytic leukemia (HL-60) cells. Environ Toxicol Pharmacol 11(2):85–91
Yang T, Zhang Y, Li Y, Hao Y, Zhou M, Dong N, Duan X (2013) High amounts of fluoride induce apoptosis/cell death in matured ameloblast-like LS8 cells by downregulating Bcl-2. Arch Oral Biol 58(9):1165–1173
Zhan XA, Wang M, Xu ZR, Li WF, Li JX (2006) Evaluation of caspase-dependent apoptosis during fluoride-induced liver lesion in pigs. Arch Toxicol 80:74–80
Anarudha CD, Kanno S, Hirano S (2000) Fluoride induces apoptosis by caspase-3 activation in human leukemia HL-60 cells. Arch Toxicol 74:226–230
Hirano S, Ando M (1996) Apoptotic cell death following exposure to fluoride in rat alveolar macrophages. Arch Toxicol 70:249–251
Loweth AC, Williams GT, Scarpello JH, Morgan NG (1996) Heterotrimeric G proteins are implicated in the regulation of apoptosis in pancreatic b-cells. Exp. Cell Res 229:69–76
Conroy LA, Jenkinson EJ, Owen JT, Michel RJ (1995) Phosphatidylinositol 4,5-bisphosphate hydrolysis accompanies T-cell receptor-induced apoptosis of murine thymocytes within the thymus. Eur J Immunol 25:1828–1835
Liu YJ, Guan ZZ, Gao Q, Pei JJ (2011) Increased level of apoptosis in rat brains and SH-SY5Y cells exposed to excessive fluoride—a mechanism connected with activating JNK phosphorylation. Toxicol Lett 204(2):183–189
Yang S, Wang Z, Farquharson C, Alkasir R, Zahra M, Ren G, Han B (2011) Sodium fluoride induces apoptosis and alters bcl-2 family protein expression in MC3T3-E1 osteoblastic cells. Biochem Biophys Res Commun 410(4):910–915
Slamenˇova´ D, Ga´belova´ A, Ruppova´ K (1992) Cytotoxicity and genotoxicity testing of sodium fluoride on Chinese hamster V79 cells and human EUE cells. Mutat Res 279:109–115
Li P, Xue Y, Zhang W, Teng F, Sun Y, Qu T et al (2013) Sodium fluoride induces apoptosis in odontoblasts via a JNK-dependent mechanism. Toxicology 308:138–145
Lee JH, Jung JY, Jeong YJ, Park JH, Yang KH, Choi NK, Kim SH, Kim WJ (2008) Involvement of both mitochondrial- and death receptor-dependent apoptotic pathways regulated by Bcl-2 family in sodium fluoride-induced apoptosis of the human gingival fibroblasts. Toxicology 243:340–347
Danial NN (2007) BCL-2 family proteins: critical checkpoints of apoptotic cell death. Clin Cancer Res 13(24):7254–7263
Liu XL, Li CC, Liu KJ, Cui CY, Zhang YZ, Liu Y (2012) The influence of fluoride on the expression of inhibitors of Wnt/β-Catenin signaling pathway in rat skin fibroblast cells. Biol Trace Elem Res 148(1):117–121
Duan X, Xu H, Wang Y, Wang H, Li G, Jing L (2014) Expression of core-binding factor α1 and osteocalcin in fluoride-treated fibroblasts and osteoblasts. J Trace Elem Med Biol 28(3):278–283
Yan X, Feng C, Chen Q, Li W, Wang H, Lv L, Wang J et al (2009) Effects of sodium fluoride treatment in vitro on cell proliferation, apoptosis and caspase-3 and caspase-9 mRNA expression by neonatal rat osteoblasts. Arch Toxicol 83(5):451–458
Dangel NJ, Knoll A, Puchta H (2014) MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J 78(5):822–833
Acknowledgments
This research presented here was supported by the MEP-PRC Project 2012ZX07101-002&20110 9025. The authors are extremely grateful to the staff at the Department of Environment and Health and the Minzu University of China for their health statistics technical assistance. The findings and conclusions in this report are those of the authors.
Conflict of Interest
The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, H., Wang, H., Jiao, Y. et al. Effect of Sodium Fluoride on the Proliferation and Gene Differential Expression in Human RPMI8226 Cells. Biol Trace Elem Res 167, 11–17 (2015). https://doi.org/10.1007/s12011-015-0271-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-015-0271-1