Abstract
Fluoride and lead are two common pollutants in the environment. Previous investigations have found that high fluoride exposure can increase the lead burden. In this experiment, in order to study on the molecular mechanisms of central neural system injury induced by the above two elements, differently expressed protein spots in hippocampus of male mice treated with 150 mg sodium fluoride/L and/or 300 mg lead acetate/L in their drinking water were detected by two-dimensional electrophoresis (2-DE) and mass spectrometry (MS). The behavior tests showed that 56 days of fluoride and lead administration significantly reduced the vertical activity and lowered the memory ability of mice. In addition, results of 2-DE and MS revealed that nine spots demonstrated above a twofold change in the same trend in all treatment groups, which were mainly related with (1) energy metabolism, (2) cell stress response/chaperones, (3) cytoskeleton development, (4) protein metabolism, and (5) cell surface signal transduction. The findings could provide potential biomarkers for lesion in nervous system induced by fluoride and lead exposure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Excessive fluoride exposure presents a severe hazard to diverse soft organs such as the brain [1–3], thyroid [4], kidney [5], and testis [6]. Fluoride has been documented to easily distribute in the body through the circulatory system and penetrate the cellular membrane, inducing cellular damage [7]. Dysfunction of central nervous system occurs when fluoride passes the blood–brain barrier (BBB) and accumulates in different parts of the brain. It was reported by a number of epidemiological investigations from China [8, 9], India [10–13], Iran [14, 15], and Mexico that Children who lived in high-fluoride areas presented lower intelligence quotient (IQ) [16].
The behavior is the product of what occurs in the nervous system; therefore, behavioral analysis is considered to be an essential assay of neural function [17]. Experiments by Y-maze test [18], open field test [19], and forced swim test [20] from our and other lab showed that animals with adversely affected behavior had high fluoride concentration in the brain or blood. Previous studies showed that fluoride significantly changed the expressions of several functional proteins in animal brain like metabotropic glutamate receptor 5 (mGluR5) [21], N-methyl-D-aspartate receptor 1 (NMDAR1) [22], neuronal nicotinic acetylcholine receptors (nAChRs) [23], and nuclear transcription factor kappa-B r65 (NF-κB r65) [2], which closely related to the cognitive ability. However, the specific mechanisms still remain largely unknown.
Actually in the normal environment, humans and animals are exposed to more than one chemical. The interaction of multiple exposures may enhance or reduce the toxic effect of individual toxicants [24]. Coplan et al. [25] and Masters et al. [26] reported that in communities treated with silicofluoride in drinking water, the prevalence of children with elevated blood lead was nearly double than that in non-fluoridated areas. In China, Zhai et al. also found the elevated lead level in children who live in high fluoride area [27]. The findings of epidemiological investigations are supported by experimental studies which found that fluoride increased the blood lead concentrations in rats [28]. Our earlier behavioral experiment showed that lead and fluoride together significantly decreased the learning ability of rats [18]. But the toxicological similarity shared by both fluoride and lead lacks further study. Therefore, in the present study, to identify the specific proteins related to fluoride–lead-induced neurotoxicity, two-dimensional gel electrophoresis (2-DE) combined with mass spectrometric (MS) technologies were applied to explore the differentially expressed proteins in mouse hippocampus.
Materials and Methods
Animals and Treatment
Sixty healthy Kunming mice (female/male = 2:1), about 20 g in weight, were supplied by the experimental animal center of Shanxi Medical University. Animals were kept in plastic cages and allowed to acclimate for 1 week in our laboratory with their standard diets. Then each pair of two female and one male was placed together for mating. When the vaginal plug was observed, the females were separated from the male and kept individually. To produce fluoride and/or lead exposure pups, pregnant mice were divided into control and experimental groups as follows: (1) control group: received double-distilled water; (2) high fluoride (HiF) group: received sodium fluoride (150 mg/L); (3) high lead (HiPb) group: received lead acetate (300 mg/L); and (4) high fluoride plus high lead (HiF+HiPb) group: received sodium fluoride (150 mg/L) and lead acetate (300 mg/L). Before postnatal day 14, the pups derived their nutrients only from maternal milk. After day 14, they gradually began to eat feed and drink water, concomitantly with suckling maternal milk. At the age of postnatal day 21, the pups ate and drank entirely by themselves and given the same treatment as their parental generation until the postnatal day 56. The doses of sodium fluoride and lead acetate were selected according to our previous study [18]. To avoid the gender difference, only the male offspring mice were used in this experiment. Animals had free access to food and water under standard temperature (22–25 °C), 12/12-h light/dark cycle, ventilation, and hygienic conditions. The study design was approved by the Institutional Animal Care and Use Committee of China.
Spontaneous Activity Test
The spontaneous ability of male mice was detected in an activity chamber as previously described with minor modifications [18]. Briefly, the chamber consists of a clear glass box with two video cameras fixed on the top and one side of the chamber to monitor the mice behavior over 6 min. The floor of the chamber was divided into 25 checks. The frequency of walking across more than one check and the frequency of standing on two feet were recorded as the horizontal and vertical activities.
Novel Object Recognition Test
The novel object recognition (OR) test proceeded in three phases [29]: (1) adaptation period (AP): two novel objects (A and B) were placed equidistantly into the glass chamber, and the mice were allowed to investigate for 10 min and explore the objects. The exploring time was recorded; (2) short-term memory (STM): 90 min later, object B was replaced with the object C, and the time spent on exploring A and C was recorded; (3) long-term memory (LTM): 24 h later, object C was replaced with the novel D. The time spent on exploring A and D was recorded. Exploration was defined as sniffing and/or touching the objects. Finally, memory index (MI) was applied to reflect memory ability, which was calculated by the ratio of the exploring time on A to the total exploring time on two objects in the each phase.
Sample Preparation
After a 24-h food-and-water fast, mouse pups were anesthetized with 20 % urethane (ethyl carbamate, NH2COOC2H5) solution at the age of postnatal day 56. The hippocampus were quickly removed and stored at −80 °C until use. The frozen samples were suspended and homogenized in 1 mL ice-cold lysis buffer consisting of 30 mM Tris–HCl, 7 M urea, 2 M thiourea, 4 % w/v CHAPS, and a protease inhibitor mixture. The homogenate was centrifuged for 10 min at 3,000 rpm, incubated for 1 h at room temperature, and centrifuged again at 20,000g for 1 h at 4 °C. The supernatant was collected. Protein concentration was determined by the Bradford Protein Assay Kit following the manufacturer’s protocols (Beyotime Institute of Biotechnology, China).
2-DE Gel Electrophoresis
Proteins were first separated with isoelectric focusing (IEF), according to our previous report [30]. Each sample with 350 μL was pipetted into a rehydrating tray and covered by a 7-cm immobilized pH gradient (IPG) strip, pH 4–7 for 15 h of rehydration at room temperature. IEF was running with the conditions as follows: 250 V for 1 h linear gradient, 500 V for 1 h linear gradient, 4,000 V for 3 h linear gradient, 4,000 V rapid gradient until reaching total 20,000 Vh, and 500 V rapid gradient for 24 h. The strip was equilibrated for 15 min prior to second-dimension separation in solution A (50 mM Tris–HCl, pH 8.8 6 M urea, 2 % sodium dodecyl sulfate (SDS), 30 % glycerol, and 1 % DTT) followed by re-equilibration for 15 min in solution B (50 mM Tris–HCl pH 8.8, 6 M urea, 2 % SDS, 30 % glycerol, and 4.5 % iodoacetamide). Proteins were further separated using the 12 % SDS-PAGE gel for second-dimension electrophoresis. Electrophoresis was performed at 10 mA/gel for 30 min and then at 30 mA/gel until bromophenol blue reached the end of the gel. Following electrophoresis, gels were stained by Coomassie blue G-250 overnight at room temperature. Then the gels were destained with 1 % glacial acetic acid and washed with MilliQ H2O for three times. UMAX2100XL Image Scanner was applied to acquire the Coomassie blue-stained gel images. Image analysis including spot detection, spot editing, background subtraction, and spots matching were performed using PDQuest 8.0 image analysis software (Bio-Rad, Hercules, CA). Compared with those in the controls, the protein spots differently expressed up to twofold with the same differential trends in three treatment groups were chosen for mass spectrometry analysis.
In-Gel Digestion and Mass Spectrometry Analysis
Protein spot of interest was excised and removed from the gel. The gel piece was washed with destaining solution (0.1 M ammonium bicarbonate, 50 % acetonitrile) for three times of 30 min each at room temperature. After removing the liquid part, the gel pieces was allowed to dry and rehydrated with the addition of modified trypsin in 4 μL 20 mM NH4HCO3 and incubated for 13 h at 37 °C. Peptides were extracted by two steps with trifluoroacetic acid (TFA) solution. The first elution was performed with 8 μL 5 % TFA for 1 h at 37 °C and the second step with 8 μL 2.5 % TFA for 1 h at 37 °C. The peptide-containing solution was vacuum-dried and resuspended with 2 μL 0.5 % TFA, followed by reconstruction with saturated solution of 50 % acetonitrile and 0.1 % TFA. These reconstructed samples were loaded on a Scorce 384 target plate. All mass spectra were obtained on a Bruker Autoflex MALDI-TOF (matrix-assisted laser desorption-time of flight) mass spectrometer (MS, Bruker, Germany). Peptide mass fingerprints were generated by the reflectron mode with a 337-nm nitrogen laser, with an acceleration voltage of 20 kV and a reflected voltage of 23 kV.
MASCOT search engine (http://www.matrixscience.com) was used to search for mass spectra data in the entire National Center for Biotechnology Information (NCBI) mouse database, in order to identify the target proteins according to peptide mass fingerprints. During the searching, the max molecular weight error of peptide was set as 0.5 Da, with the assumption that the peptides were monoisotopic, oxidized at methionine residues, and carbamiodomethylated at cysteine residues. The functions of target proteins were annotated by using Molecule Annotation System 3.0 (MAS 3.0) (http://bioinfo.capitalbio.com/mas3/).
Statistical Analysis
GraphPad Prism5 software (GraphPad Software Inc., San Diego, USA) was applied in this study. All data were analyzed using one-way ANOVA analysis in which Dunnett’s Post hoc test was selected. Data was expressed as mean ± SEM, *p < 0.05, and **p < 0.01.
Results
Spontaneous Activity and Novel Object Recognition
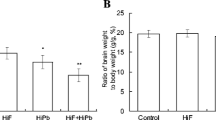
In Fig. 1, mice in HiF+HiPb group showed a significant low vertical activity (p < 0.01), indicating the inhibition of outside exploration. Although there was a decreasing trend in horizontal activity, no statistical difference was observed. The results of novel object recognition in Fig. 2 revealed that the HiF+HiPb significantly decreased the MIs during STM (p < 0.05) phase and LTM (p < 0.01) phase, compared with those in the control. HiF showed no effect on MI in STM and LTM, while HiPb significantly reduced in the LTM. Besides, no difference occurred in AP phase.
Effects of fluoride and/or lead on recognition memory activity of offspring male mice. MI in AP (adaptation period), STM (short-term memory), and LTM (long-term memory) was calculated by TA/(TA+TB), TA/(TA+TC), and TA/(TA+TD), respectively. Each bar represents the mean ± SEM (n = 15). **p < 0.01 (significant differences compared with the control); *p < 0.05. TA, TB, TC, and TD mean the exploring time of mouse on object A, B, C, and D, respectively
Protein Fractionation and Identification
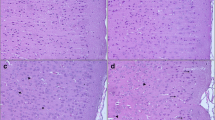
As shown in Fig. 3, the map of hippocampus protein spots from control and fluoride- and/or lead-treated mice was on the gels stained with Coomassie brilliant blue. After the establishment of reference gel, the spots in each group are well matched by PDQuest software package. Compared with those in the controls, the expressions of 92 protein spots in HiF, HiPb, and HiF+HiPb were significantly changed. Nine spots were altered with the same trend in all treatment groups, and the expression differences were up to twofold. We chose these nine protein spots for mass spectrometry analysis.
Two-dimensional electrophoresis representative gels of brain protein from the control (a), high fluoride (b), high lead (c), high fluoride plus high lead (d) groups. Proteins were separated on 7-cm pH 4–7 IEF strips followed by SDS-PAGE and Coomassie blue G-250 staining. The proteins were then detected and compared using the PDQuest software package. Selected spots represented the brain with different levels (twofold) compared with the controls
Among these spots, six of them were significantly upregulated and three were significantly downregulated. The differentially expressed proteins are identified as pyruvate dehydrogenase E1β subunit (Pdhb1), NADH dehydrogenase, ATPase, heat shock 70-kDa protein 8 (Hspa8), isoform 1 of 60 kDa heat shock protein/mitochondrial Hsp60 (Hspd1), dihydropyrimidinase-like 2 (Dpysl2), α-spectrin 2 (Spna2), growth factor receptor protein bound 2 (Grb2), and alanyl-tRNA synthetase (Aars). The pI and the molecular weight reported in the mass data for all the identified protein spots were consistent with the spot position on the gel, as seen in Table 1.
Discussion
Our previous study observed that 30 days of fluoride and lead exposure adversely affected spontaneous behavior and lowered the learning ability of rats before the occurrence of dental lesions [18]. In this study, the same doses of fluoride and lead resulted in a significant decrease in vertical activity and memory ability of mice, indicating a successful establishment of mouse model, and we also found that by 2-DE coupled with MS, nine proteins changing above twofold were identified in hippocampus, among which six were upregulated and three were downregulated. These proteins are involved in energy metabolism (Pdhb1, NADH dehydrogenase, ATPase), cell stress response/chaperones (Hspd1, Hspa8), cytoskeleton development (Dpysl2, Spna2), protein metabolism (Aars, Hspa8), and cell surface signal transduction (Grb2), as shown in Table 2.
Fluoride and Lead Induced Protein Changes in Energy Metabolism
Pyruvate dehydrogenase complex (PDC) is considered to be a gateway enzyme for carbohydrate oxidative metabolism in mitochondria [31], in which process pyruvate is catalyzed, followed by the formation of acetyl-CoA and NADH. The function of this complex largely depends on the heterotetramer (αβ/α′β′) of pyruvate dehydrogenase E1 α (Pdh1α) and β (Pdh1β) subunits [32]. Clinical consequences of PDC dysfunction mainly presented the nervous syndromes, including developmental delay, seizures, central hypotonia, peripheral neuropathy, and congenital brain malformations [33]. Pdh1α gene mutations are frequently reported to induce the PDC deficiency [34], and the over-expressed αβ/α′β′ heterotetramer can in turn enhance the Pdh1α mutations [35]. In the present study, with the comparison to controls, the protein expression of Pdh1β was significantly increased in fluoride and/or lead treatment groups, implying an abnormal αβ/α′β′ heterotetramer structure and may finally lead to low level of NADH or acetyl-CoA in hippocampus. It is worthy to note that the NADH dehydrogenase protein level in this study was downregulated by both fluoride and lead, which might be the results of the possibly low NADH induced by abnormal carbohydrate oxidation. Cells with low NADH commonly need for ATP to supply enough energy [36]. Although we have not detected the cellular ATP amount, enhanced ATPase level in mice exposed to fluoride and lead in this study hinted the energy requirement.
Fluoride and Lead Induced Protein Changes in Cell Stress Response
Under environmental stress conditions, the expression of heat shock proteins (HSPs) in stressed cells is increased and remains on elevated level for a prolonged period to prevent cellular damage [37]. HSPs, as molecular chaperones, play a major role in cytoprotection by inhibiting irreversible protein aggregation or misfolding [38]. According to the sequence homology and molecular weight, HSPs were classified into several families. Hspa8 and Hspd1, which were both upregulated by fluoride plus lead exposure in this study, are the members of Hsp70 and Hsp60, respectively. Hsp70 superfamily is the most abundant and the most highly conserved HSPs [39]. Previous study on zebrafish embryos pointed out that the stress-inducible HSP70 functions in attenuating the apoptotic-like process [40]. Mitochondrial Hsp60 (Hspd1) is essential for early embryonic development in mice [41]. In the staurosporin-induced apoptosis model, Hsp60 and Hsp10 (the co-chaperone of Hsp60) were observed to release the active caspase-3 [42]. Accumulated investigations have reported that fluoride or lead evokes cell apoptosis through different pathways, including Fas/FasL, Caspase, and bcl-2/c-fos. Therefore, the elevated Hspa8 and Hspd1 suggested that neurons in hippocampus may be in the stressed conditions induced by fluoride and lead exposure.
Fluoride and Lead Induced Protein Changes in Cytoskeleton
Dpysl2, which involves in neuronal differentiation and axonal guidance [43], is the most highly expressed isoform of dihydropyrimidinase (DRP) family in different brain areas such as the olfactory bulb, hippocampus, and cerebellum in adult nervous system [44]. Spna2 is a protein that functions in maintaining neuronal membrane integrity [45]. Dpysl2 and Spna2 play roles in the pathological processes in various nervous system diseases like vitamin A depletion [46], stroke [47], and alcohol-induced brain pathology [48]. By using proteomic analysis, Chen et al. found that Dpysl2, Spna2, and Hsp70 were upregulated in focal cerebral stroke in rats induced by ischemia [47]. The increased gene expressions of Dpysl2 and Spna2 were also detected in rat cortex with a significant apoptosis rate and injured swollen cells [49]. In this study, besides the elevated Hsp70 member mentioned above, the protein expressions of Dpysl2 and Spna2 were also increased in groups administrated with fluoride and/or lead. Morphological experiments by transmission electron microscope demonstrated that fluoride causes swelling of the mitochondria, granular endoplasmic reticulum, damages the membrane of nuclear and synapses, and decreases the number of synapses [50]. Together with the current results, we suggested that morphological alterations in hippocampus of mice exposed to fluoride and lead probably related to the upregulated Dpysl2 and Spna2.
Fluoride and Lead Induced Protein Changes in Cell Surface Signal Transduction
Grb2, as an adapter protein, can positively regulate the cell surface expression of FasL and mediate FasL reverse signaling via activation of Ras and the extracellular signal-regulated kinase (ERK) MAPK pathway 1, finally participating in cell proliferation or differentiation [51, 52]. The downregulated Grb2 in this study suggested that the toxicity of fluoride and lead combination may involve in the Ras-signaling pathway to adversely affect neuron proliferation. Further investigation can focus on selecting the target proteins in the downstream in this signal pathway.
In conclusion, this study presented the identification of proteomic pattern in hippocampus of mice exposed to fluoride and/or lead. Further studies are required to verify the differentially expressed proteins associated with important biological activities to clarify the pathological mechanisms underlying fluoride- and lead-induced neurotoxicity.
References
Adebayo OL, Shallie PD, Salau BA et al (2013) Comparative study on the influence of fluoride on lipid peroxidation and antioxidants levels in the different brain regions of well-fed and protein undernourished rats. J Trace Elem Med Biol 27(4):370–374
Zhang J, Zhu WJ, Xu XH et al (2011) Effect of fluoride on calcium ion concentration and expression of nuclear transcription factor kappa-B r65 in rat hippocampus. Exp Toxicol Pathol 67(5):407–411
Liu YJ, Guan ZZ, Gao Q et al (2011) Increased level of apoptosis in rat brains and SH-SY5Y cells exposed to excessive fluoride—a mechanism connected with activating JNK phosphorylation. Toxicol Lett 204(2–3):183–189
Zhou BH, Wang HW, Wang JM et al (2007) Effects of malnutrition and supplemented nutrition on non-specific immune function changes induced by fluoride in rabbits. Fluoride 40(3):169–177
Xu H, Hu LS, Chang M et al (2005) Proteomic analysis of kidney in fluoride-treated rat. Toxicol Lett 160(1):69–75
Liu HT, Niu RY, Wang JM et al (2008) Changes caused by fluoride and lead in energy metabolic enzyme activities in the reproductive system of male offspring rats. Fluoride 41(3):184–191
Bouaziz H, Ben Amara I, Essefi M et al (2010) Fluoride-induced brain damages in suckling mice. Pestic Biochem Physiol 96(1):24–29
Ding Y, Gao Y, Sun H et al (2011) The relationships between low levels of urine fluoride on children's intelligence, dental fluorosis in endemic fluorosis areas in Hulunbuir, Inner Mongolia, China. J Hazard Mater 186(2–3):1942–1946
Wang SX, Wang ZH, Cheng XT et al (2007) Arsenic and fluoride exposure in drinking water: children's IQ and growth in Shanyin county, Shanxi province, China. Environ Health Perspect 115(4):643–647
Eswar P, Nagesh L, Devarajc C (2011) Intelligence quotients of 12–14 years old school children in a high and a low fluoride village in India. Fluoride 44(3):168–172
Saxena S, Sahay A, Goel P (2012) Effect of fluoride exposure on the intelligence of school children in Madhya Pradesh, India. J Neurosci Rural Pract 3(2):144–149
Trivedi M, Sangai N, Patel R et al (2012) Assessment of groundwater quality with special reference to fluoride and its impact on IQ of schoolchildren in six villages of the Mundra region, Kachchh, Gujarat, India. Fluoride 45(4):377–383
Shivaprakash PK, Ohri K, Noorani H (2011) Relation between dental fluorosis and intelligence quotient in school children of Bagalkot district. J Indian Soc Pedod Prev Dent 29(2):117–120
Poureslami HR, Horri A, Garrusib B (2011) IQ of children age 7–9 in a high and a low F water city in Iran. Fluoride 44(3):163–167
Seraj B, Shahrabi M, Shadfar M et al (2012) Effect of high water fluoride concentration on the intellectual development of children in makoo/iran. J Dent (Tehran) 9(3):221–229
Rocha-Amador D, Navarro ME, Carrizales L et al (2007) Decreased intelligence in children and exposure to fluoride and arsenic in drinking water. Cad Saude Publ 23(Suppl 4):579–587
Whishaw IQ, Haun F, Kolb BL (1999) Analysis of behavior in laboratory rodents. IN: Windhorst U, Johansson H, editors. Modern techniques in neuroscience research. Secaucus: Springer; 1243–1244
Niu RY, Sun ZL, Wang JM et al (2008) Effects of fluoride and lead on locomotor behavior and expression of Nissl body in brain of adult rats. Fluoride 41(4):276–282
Ekambaram P, Paul V (2001) Calcium preventing locomotor behavioral and dental toxicities of fluoride by decreasing serum fluoride level in rats. Environ Toxicol Pharmacol 9(4):141–146
Balaji B, Kumar EP, Kumar A (2012) Evaluation of standardized Bacopa monniera extract in sodium fluoride-induced behavioural, biochemical, and histopathological alterations in mice. Toxicol Ind Health. doi:10.1177/0748233712468018
Jiang S, Su J, Yao S et al (2014) Fluoride and arsenic exposure impairs learning and memory and decreases mGluR5 expression in the hippocampus and cortex in rats. PLoS One 9(4):e96041
Niu RY, Sun ZL, Cheng ZT et al (2008) Effects of fluoride and lead on n-methyl-d-aspartate receptor 1 expression in the hippocampus of offspring rat pups. Fluoride 41(2):101–110
Gao Q, Liu YJ, Guan ZZ (2008) Oxidative stress might be a mechanism connected with the decreased alpha 7 nicotinic receptor influenced by high-concentration of fluoride in SH-SY5Y neuroblastoma cells. Toxicol In Vitro 22(4):837–843
Inkielewicz-Stepniak I, Czarnowski W (2010) Oxidative stress parameters in rats exposed to fluoride and caffeine. Food Chem Toxicol 48(6):1607–1611
Coplan MJ, Patch SC, Masters RD et al (2007) Confirmation of and explanations for elevated blood lead and other disorders in children exposed to water disinfection and fluoridation chemicals. Neurotoxicology 28(5):1032–1042
Masters RD, Coplan MJ, Hone BT et al (2000) Association of silicofluoride treated water with elevated blood lead. Neurotoxicology 21(6):1091–1100
Zhai Y, Dong J, Cao XZ et al (2006) Epidemiological investigation about the relation between blood lead level and high fluorine drinking water in children. Child Health Care China 21(8):1088–1090 [in Chinese]
Sawan RM, Leite GA, Saraiva MC et al (2010) Fluoride increases lead concentrations in whole blood and in calcified tissues from lead-exposed rats. Toxicology 271(1–2):21–26
Bevins RA, Besheer J (2006) Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc 1(3):1306–1311
Ge Y, Niu R, Zhang J et al (2011) Proteomic analysis of brain proteins of rats exposed to high fluoride and low iodine. Arch Toxicol 85(1):27–33
Okajima K, Korotchkina LG, Prasad C et al (2008) Mutations of the E1beta subunit gene (PDHB) in four families with pyruvate dehydrogenase deficiency. Mol Genet Metab 93(4):371–380
Patel MS, Korotchkina LG (2006) Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans 34:217–222
Kerr DS, Schmotzer C (2004) Variability of human pyruvate dehydrogenase complex deficiency. In: Patel MS, Jordan F (eds) Thiamine: catalytic mechanisms and role in normal and disease states. Marcel Dekker, New York, pp 471–483
Lissens W, De Meirleir L, Seneca S et al (2000) Mutations in the X-linked pyruvate dehydrogenase (E1) alpha subunit gene (PDHA1) in patients with a pyruvate dehydrogenase complex deficiency. Hum Mutat 15(3):209–219
Korotchkina LG, Ciszak EM, Patel MS (2004) Function of several critical amino acids in human pyruvate dehydrogenase revealed by its structure. Arch Biochem Biophys 429(2):171–179
Soole KL, Menz RI (1995) Functional molecular aspects of the NADH dehydrogenases of plant mitochondria. J Bioenerg Biomembr 27(4):397–406
Rupik W, Jasik K, Bembenek J et al (2011) The expression patterns of heat shock genes and proteins and their role during vertebrate's development. Comp Biochem Physiol A 159(4):349–366
Heikkila JJ (2010) Heat shock protein gene expression and function in amphibian model systems. Comp Biochem Physiol A 156(1):19–33
Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62(6):670–684
Elicker KS, Hutson LD (2007) Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene 403(1–2):60–69
Christensen JH, Nielsen MN, Hansen J et al (2010) Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress Chaperones 15(6):851–863
Samali A, Cai J, Zhivotovsky B et al (1999) Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of Jurkat cells. EMBO J 18(8):2040–2048
Quinn CC, Gray GE, Hockfield S (1999) A family of proteins implicated in axon guidance and outgrowth. J Neurobiol 41(1):158–164
Wang LH, Strittmatter SM (1996) A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci 16(19):6197–6207
Voas MG, Lyons DA, Naylor SG et al (2007) AlphaII-spectrin is essential for assembly of the nodes of Ranvier in myelinated axons. Curr Biol 17(6):562–568
Zhang M, Huang K, Zhang Z et al (2011) Proteome alterations of cortex and hippocampus tissues in mice subjected to vitamin A depletion. J Nutr Biochem 22(11):1003–1008
Chen A, Liao WP, Lu Q et al (2007) Upregulation of dihydropyrimidinase-related protein 2, spectrin α II chain, heat shock cognate protein 70 pseudogene 1 and tropomodulin 2 after focal cerebral ischemia in rats—a proteomics approach. Neurochem Int 50(7–8):1078–1086
Matsuda-Matsumoto H, Iwazaki T, Kashem MA et al (2007) Differential protein expression profiles in the hippocampus of human alcoholics. Neurochem Int 51(6–7):370–376
Indraswari F, Wong PT, Yap E et al (2009) Upregulation of Dpysl2 and Spna2 gene expression in the rat brain after ischemic stroke. Neurochem Int 55(4):235–242
Valdez-Jiménez L, Soria Fregozo C, Miranda Beltrán ML et al (2011) Effects of the fluoride on the central nervous system. Neurologia 26(5):297–300
Thornhill PB, Cohn JB, Stanford WL et al (2008) The adaptor protein Grb2 regulates cell surface Fas ligand in Schwann cells. Biochem Biophys Res Commun 376(2):341–346
Anteby EY, Ayesh S, Shochina M et al (2005) Growth factor receptor-protein bound 2 (GRB2) up-regulation in the placenta in preeclampsia implies a possible role for ras-signalling. J Obstet Gynecol Reprod Biol 118(2):174–181
Acknowledgments
This research was supported by the China National Natural Science Foundation (Grant Nos. 31101869, 31201965, and 31172376), Shanxi Province Natural Science Foundation (Grant No. 2012021027–5), and Program for the Top Young Academic Leaders of Higher Learning Institutions of Shanxi.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ruiyan Niu and Shuangling Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Niu, R., Liu, S., Wang, J. et al. Proteomic Analysis of Hippocampus in Offspring Male Mice Exposed to Fluoride and Lead. Biol Trace Elem Res 162, 227–233 (2014). https://doi.org/10.1007/s12011-014-0117-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-014-0117-2