Abstract
Copper is a toxic heavy metal widely used to microbial control especially in agriculture. Consequently, high concentrations of copper residues remain in soils selecting copper-resistant organisms. In vineyards, copper is routinely used for fungi control. This work was undertaken to study copper resistance by rhizosphere microorganisms from two plants (Avena sativa L. and Plantago lanceolata L.) common in vineyard soils. Eleven rhizosphere microorganisms were isolated, and four displayed high resistance to copper. The isolates were identified by 16S rRNA gene sequence analysis as Pseudomonas putida (A1), Stenotrophomonas maltophilia (A2) and Acinetobacter sp. (A6), isolated from Avena sativa rhizosphere, and Acinetobacter sp. (T5), isolated from Plantago lanceolata rhizosphere. The isolates displayed high copper resistance in the temperature range from 25°C to 35°C and pH in the range from 5.0 to 9.0. Pseudomonas putida A1 resisted as much as 1,000 mg L−1 of copper. The isolates showed similar behavior on copper removal from liquid medium, with a bioremoval rate of 30% at 500 mg L−1 after 24 h of growth. Speciation of copper revealed high copper biotransformation, reducing Cu(II) to Cu(I), capacity. Results indicate that our isolates are potential agents for copper bioremoval and bacterial stimulation of copper biosorption by Avena sativa and Plantago lanceolata.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Copper (Cu(II)) is an essential micronutrient to living organisms. However, at high concentrations in the environment, copper is hazardous to living systems [1]. Vineyard soils generally contain high levels of copper. This occurrence is due to frequent sprays used to control leave diseases in horticulture [2]. Such contamination results in the selection of copper-resistant microorganisms and plants.
In vineyard areas of South of Brazil, oatmeal crops are widely used to control soil erosion and to improve physical and chemical soil properties. Oatmeal plants however display signs of copper toxicity [3]. Phytoremediation is one approach to bioremediation of metals. The efficiency of phytoremediation is however not very high [4]. Nonetheless, rhizosphere microorganisms from contaminated areas appear to be more efficient in plant growth-promoting under contaminated sites [5–7]. Variation of speciation of copper in the maize rhizosphere was reported by Tao et al. [8], and it was related to microbial activities in the rhizosphere zone.
Copper-resistant microorganisms can play a major role in copper removal from the environment [9]. Bioreduction of Cu(II) to Cu(I) increases its bioavailability to microbial cells [10, 11] as well as plants [9, 12]. Thus, copper-resistant microorganisms can be used in phytoremediation to promote phytoaccumulation of heavy metals. Moreover, copper-resistant microorganisms adapted in the rhizosphere environment can be better suited for bioaugmentation and can promote phytoremediation, such as the Avena sativa plants exhibited after inoculation of the rhizosphere bacteria isolated and characterized [3].
Selection of metal-resistant microorganisms with high bioremoval capacity is a major step in heavy metal bioremediation and phytoremediation. However, environmental parameters such as pH, temperature and contaminant concentration can interfere in the microbial activity [13]. In this study, it was isolated, and selected highly copper-resistant bacteria from Avena sativa and Plantago lanceolata rhizosphere soils are collected from vineyard areas; it was characterized as the isolates for copper bioreduction and biosorption. Selected isolates were then subjected to molecular characterization by 16S rRNA gene sequence analysis. The copper speciation by rhizosphere microorganisms was also examined.
Materials and Methods
Soil Samples
Avena sativa and Plantago lanceolata soil samples were collected from two copper-contaminated vineyard areas from Southern Brazil, in the EMBRAPA experimental station, Bento Gonçalves, RS, Brazil (29°09'53.92'' S and 51°31'39.40'' W). These soils were classified as Inceptisol and Mollisol. Table 1 presents the physical–chemical parameters: pH, cation exchange capacity, organic matter, clay, copper dissolved in acid (Cu) and copper soluble in water (Cu(H2O)). Copper soluble in water was extracted from 10 g of soil in 20 mL of deionized water for 60 min, with shaking. Copper was determined by atomic absorption spectrophotometer.
Enrichment and Isolation of Cu(II)-Resistant Bacteria
Enrichment of Cu(II)-resistant bacteria was set up using 100 mL of nutrient broth in 250-mL Erlenmeyer flasks to which we added 500-mg mL−1 Cu(II) using CuSO4.5H2O, subsequently mentioned as Cu(II). The medium was adjusted to pH 7.0 using 1-M NaOH or 0.1-M HCl. The nutrient broth medium with copper was sterilized by autoclaving at 121°C for 20 min. The two different rhizosphere soils were independently used to inoculate (1% w/v) sterile nutrient broth amended with Cu(II) and incubated for 24 h, with orbital shaking (150 rpm) at 30°C. Subsequently, 1 mL of the enrichment culture was transferred to 99 mL of sterile nutrient broth amended with Cu(II) and incubated for 24 h, with shaking (150 rpm) at 30°C. The enrichment procedure was repeated for a third round. Cu(II)-resistant bacteria were thereafter purified by repeated streaking of the third-round cultures on nutrient agar plates containing Cu(II) (500 mg L−1). Cu(II)-resistant bacterial strains isolated from enrichment consortia were evaluated to Cu(II) resistance and biosorption.
Analysis of Copper and Bacterial Mass
Total copper was analyzed using an atomic absorption spectrometer. Aliquots of culture supernatant (200-μL aliquots) were diluted 20 times and injected into the atomic absorption spectrometer. Copper remaining was calculated as the difference of the total copper added to the medium and the total copper removed from the medium after different microbial treatments. Copper reduction was quantified by measuring monovalent copper complex with 1-mM neocuproine hydrochloride [14]. Cultures were centrifuged (2,500 rpm, 10 min). For copper reduction, 1-mL aliquot of the cell-free culture supernatant was mixed with 2 mL of 1-mM neocuproine to complex Cu(I) and incubated for 30 min at 30°C in a 13 × 100 mm glass culture tubes. Thereafter, the absorbance was read using a spectrophotometer at λ454 [14]. Copper remaining and copper reduction were calculated from copper (I or II) standard curves (25–500 mg L−1 of copper) prepared in the nutrient broth medium. Copper resistance was determined by measuring bacterial biomass development using at λ600.
Effect of Copper Concentration
The effects of different concentrations of Cu(II) were determined using nutrient broth amended with 250, 500, 750, 1,000 and 1,250 mg L−1 of Cu(II). The inoculum was grown at 30°C for 24 h with shaking (150 rpm) and thereafter adjusted to 0.85 OD600 units with sterile salt solution (0.85%). The sterile Cu(II) media were independently inoculated with 100-μL aliquot of each inoculum and incubated at 30°C for 24 h with shaking (150 rpm).
Effect of Temperature and pH
Temperature and pH effects on bacterial growth were examined using nutrient broth amended with Cu(II) (500 mg L−1). For the temperature effect, cultures were incubated at 25, 30, 35 and 40°C. In the pH experiment, sterilized nutrient broth amended with Cu(II) (500 mg L−1) was adjusted to pH 3.0, 5.0, 7.0, 9.0 and 11.0 by addition of predetermined amounts of sterilized 1-M NaOH or 0.1-M HCl. Cultures were inoculated with 100 μL of inoculum (OD600 = 0.85). Inoculum preparation and biomass determination were as described previously.
Time Course of Copper Biosorption and Reduction
The time course of divalent copper removal by biosorption and bioreduction was examined as follows. Nutrient broth amended with 500 mg L−1 of Cu(II) was used. The medium was inoculated with 100 μL of each isolate (OD600 = 0.85). Cultures were incubated at 30°C with shaking (150 rpm) and analyzed at different time intervals (0, 6, 24, 30 and 48 h). Biomass, copper reduced and copper removed were then analyzed, and copper bioremoval was calculated as discussed previously.
DNA-Based Identification of Isolates
Isolates were identified by 16S ribosomal RNA gene sequence analysis as follows. The isolates were grown by streaking on nutrient agar with incubation at 30°C for 24 h. DNA of isolates was extracted from colony-forming units pooled from the nutrient agar plate using Promega Wizard Genomic DNA Purification Kit (Promega, Madison, WI) with slight modification. Briefly, cells were re-suspended in 300 μL of nucleic acid lyses solution, incubated at 80°C for 15 min and allowed to cool at room temperature. RNase solution (1.5 μL) was added and incubated at 37°C for 60 min. Protein precipitation solution (100 μL) was added and incubated on ice for 5 min. Following centrifugation, the supernatant was transferred to a tube, and cold ice 95% ethanol was added. The precipitate was recovered by centrifugation. The pellet was washed with 70% ethanol at ambient temperature and re-suspended in sterile nuclease-free distilled water. Two primers corresponding to E. coli positions 27 F (5′-AGATTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGAC TT-3′) were used for PCR amplification of the 16S ribosomal RNA [15]. The PCR reaction mixture consisted of 12.5 μL of PCR master mix (Promega, Madison, WI), genomic DNA template (0.5 μL), primer 27 F (2.5 μL = 12.5 pmol) and primer 1492R (2.5 μL = 12.5 pmol) and made up to 25-μL final volume with nuclease-free water. The 16S rRNA gene was amplified using a 35-cycle PCR (initial denaturation, 95°C for 5 min; subsequent denaturation, 95°C for 0.5 min; annealing temperature, 50°C for 1 min; extension temperature, 72°C for 1 min and final extension, 72°C for 5 min). The PCR amplification products were analyzed by electrophoresis on a 1% agarose gel. Millipore Montage PCR filter units (Millipore, Billerica, MA) were used to remove primers, salts and unincorporated dNTPs according to the manufacturer’s instructions. DNA cycle sequencing was performed using BigDye terminator kit (Applied Biosystems, Foster City, CA) with sequencing primer 519r (5′-GWATTACCGCGGCKGCTG-3′) in independent reactions at the Institute of Integrative Genome Biology (IIGB) of the University of California, Riverside, CA.
DNA Sequence Similarity and Phylogenetic Analysis
GenBank BLAST (N) was used for homology searches. The 16S rRNA gene sequence was deposited to the GenBank database. Phylogenetic analysis was conducted using RDP release 10 software [16].
Results
Isolate Identification
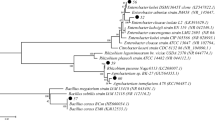
A total of nine copper-resistant bacteria, six from Avena sativa (A) rhizosphere and three from Plantago lanceolata (T) rhizosphere, were isolated. These isolates resistant to high copper concentration (500 mg L−1) were further studied. The four selected isolates were identified by 16S rRNA, similarity searches were analyzed with my RDP software (Fig. 1), and the sources, the number of the 16S rDNA nucleotides, the GenBank Submission and the GenBank Match are presented in Table 2. The isolates were identified as Pseudomonas putida (A1) (BO–1 in the tree), Stenotrophomonas maltophilia (A2) (BO–2 in the tree) and Acinetobacter sp. (A6) (BO–3 in the tree) and were isolated from Avena sativa rhizosphere soil zone, whereas one isolate, Acinetobacter sp. (T5) (BO–6 in the tree), was isolated from Plantago lanceolata rhizosphere (Fig. 1). Isolates A1 and A2 were 98% similar to the GenBank BLAST analysis match, while isolates A6 and T5 were 96% and 94% similar to GenBank BLAST analysis match, respectively (Table 2).
Phylogenetic tree showing evolutionary distance of the selected isolates from the rhizosphere zone of two plants grown in the copper-contaminated areas based on 16S rRNA gene sequence. The identification of the isolates in the tree are isolate A1 (Pseudomonas putida) is BO–1, isolate A2 (Stenotrophomonas maltophilia) is BO–2, isolate A6 (Acinetobacter sp.) is BO–3, and isolate T5 (Acinetobacter sp.) is BO–6. The numbers in the nodes are the bootstrap of 100 replicates. The scale is the evolutionary distance value
Effect of Copper Concentration
The effect of copper concentration on the resistance profile of the four bacteria isolated from rhizosphere of Avena sativa and Plantago lanceolata plants was measured (Fig. 2). In general, all isolates showed growth decrease with increase in copper concentration. Isolates A1 and A6 showed similar resistance pattern at different copper concentrations and resisted up to 750 mg L−1 in liquid medium. On the other hand, isolate A2 showed superior growth at all copper concentrations followed by T5, isolated from Avena sativa and Plantago lanceolata rhizosphere zone, respectively. Furthermore, A2 and T5 were the only isolates that tolerated 1,000 mg L−1 of copper in liquid medium. However, none of the isolates survived at 1250 mg L−1 of copper concentration.
Effect of Temperature
The effect of temperature on the copper resistance profile of bacteria isolated from rhizospheres of Avena sativa and Plantago lanceolata plants was studied (Fig. 3). Isolates A2, A6 and T5 were grown maximally between 25°C and 30°C in copper-amended medium. Contrarily, growth of isolate A1 in copper-amended medium was optimal at 35°C, with as high as 1.4 OD600 units recorded. None of the isolates survived at 40°C in copper-amended medium.
Effect of pH
The effects of pH on Cu(II)-resistant bacterial isolates are presented in Fig. 4. The optimum initial pH for biomass production by all isolates was at pH 7.0. There was no growth of the isolates at pH 3.0. Isolate A1 grew substantially at initial pH 11.0 (1.1 OD600 units), but pH 11.0 was generally inhibitory to all the isolates. Slight growth (0.2 OD600 units) of isolate A6 was observed at initial pH 11.0. Isolate A2 was sensitive to pH changes, with high biomass production only at pH 7.0 (1.4 OD600 units). Isolates A6 and T5 were similar in their growth profiles at different pH (Fig. 4a). Growth of the most of the isolates induced significant changes in pH (Fig. 4b).
Time Course of Copper Resistance and Copper Bioremoval
Time course of Cu(II) resistance and Cu(II) bioremoval by selected rhizosphere bacterial isolates was evaluated (Fig. 5). Isolates A6 and T5 showed the highest biomass production after 9 h of incubation (1.1 and 1.2 OD600 units, respectively). Isolates A2 and A1 showed the highest biomass production after 24 h of incubation (1.2 and 0.9 OD600 units, respectively). In both groups, the exponential phase of biomass production occurred after 9 h of incubation at 30°C with orbital shaking (150 rpm) (Fig. 5a). Copper bioremoval capacities of the four isolates were similar. The highest rate of Cu(II) bioremoval in liquid medium was in the first 9 h of incubation (25% removal, which is 125 mg L−1 of copper removed). However, the isolates showed high Cu(II) bioremoval capacity (30% of the total amended copper or 150 mg L−1 removed) from liquid medium with high copper concentration (500 mg L−1) in 24 h of incubation (Fig. 5b).
Dynamics of Copper Speciation
Figure 6a presents the dynamics of Cu(II) biosorption and bioreduction in medium amended with 500 mg L−1 of copper. Pseudomonas species A1 displayed peak biomass production at 6 h of incubation (0.7 OD600 units), and it was stable until 48 h of incubation. Cu(II) bioremoval by isolate A1 increased until 48 h of incubation, with more than 150 mg L−1 of copper removed. Consequently, copper remaining in the medium was less than 350 mg L−1, and more than 240 mg L−1 was reduced to Cu(I). This indicates that only 110 mg L−1 of the total Cu(II) added (500 mg L−1) remained after 48 h of incubation.
Time course and relationship of biomass development, copper biosorption and bioreduction in culture of Pseudomonas putida A1 (a), Stenotrophomonas maltophilia A2 (b), Acinetobacter calcoaceticus A6 (c) and Acinetobacter calcoaceticus T5 (d) at high copper concentration (500 mg L−1). Cultures were incubated for 48 h at 30°C with orbital shaking (150 rpm). Biomass (circle), Cu(II) removed (filled circle), Cu(II) remaining (filled square) and Cu(II) reduced (square) were evaluated. The error bars are standard errors of the mean
Isolate A2 (Stenotrophomonas maltophilia) culture reached 0.65 OD600 units after 6 h of incubation, and biomass production thereafter remained stable after 48 h of incubation. Cu(II) bioremoval increased, and more than 165 mg L−1 of copper removal was observed after 48 h of incubation. Cu(II) remaining was less than 335 mg L−1, where more than 100 mg L−1 of Cu(II) was reduced to Cu(I). This indicates that 235 mg L−1 of the 500-mg L−1 Cu(II) added remained after 48 h of incubation of isolate A2 (Fig. 6b).
In isolate A6 (Acinetobacter sp.), culture biomass production rose to 0.65 OD600 units after 6 h and increased to 0.75 OD600 units after 48 h of incubation (Fig. 6c). Cu(II) removal increased and was greater than 160 mg L−1 of copper from the liquid medium amended with 500 mg L−1 of copper after 48 h of incubation. Consequently, copper remaining in the medium was 340 mg L−1, of which 160 mg L−1 of Cu(II) was reduced to Cu(I). Thus, Cu(II) remaining after 48 h of incubation was 265 mg L−1 of Cu(II), originally added to the culture medium.
Acinetobacter sp. T5 isolated from Plantago lanceolata rhizosphere grew exponentially, reaching 0.40 OD600 units after 6 h, which increased to 0.45 OD600 units after 36 h (Fig. 6d). Copper bioremoval increased until 36 h of incubation, and more than 190 mg L−1 of copper bioremoval from liquid medium amended with 500 mg L−1 of copper was recorded at 36 h of incubation. Accordingly, copper remaining in the medium was less than 310 mg L−1 after 36 h of incubation. About this concentration, more than 190 mg L−1 of Cu(II) was reduced to Cu(I), indicating that 120 mg L−1 of Cu(II) originally added remained after 36 h of incubation.
Discussion
Copper pollution is widespread in the environment, and intensive research on the use of metal-resistant bacteria to remove metals from polluted sites is necessary. This study characterized four highly copper-resistant bacteria from the Avena sativa and Plantago lanceolata rhizospheres. However, some species of rhizosphere bacteria such as Pseudomonas and Acinetobacter were found in copper-resistant plants grown in copper-contaminated soils [5], although the copper uptake was not evaluated. Stenotrophomonas maltophilia was reported to be resistant to heavy metals [17], and Clausen [18], Chen et al. [19], Uslu and Tanyol [20], Chen et al. [21] reported metal resistance in Pseudomonas putida. Acinetobacter calcoaceticus strains were explored for the bioremediation of sites contaminated with metals such as chromium [18, 22], copper and arsenic [18] and sites contaminated with aromatic compounds [23]. However, copper resistance, bioreduction and biosorption capacity displayed by two of our rhizosphere bacterial isolates T5 and A6 are much higher than previously reported.
The development of biomass by bacteria was substantial in liquid medium containing copper concentrations greater than 500 mg mL−1 of Cu (II). Tolerance and sorption of divalent copper by our isolates were higher than observed in Pseudomonas putida CZ1 tolerant to only low concentrations of copper and zinc, in a range between 20 and 25 mg L−1 [19]. Interestingly, our isolate Pseudomonas putida A1 removed approximately 150 mg L−1 of copper after 24 h of growth in medium containing 500 mg L−1 of copper.
Factors such as pH, temperature and concentrations of pollutants influence microbial metabolism as enzyme activity [24]. Such factors either promote or inhibit enzyme activity [11, 25] and biosorption [13, 19, 26, 27]. In the culture of Pseudomonas fluorescens, the presence of 20 mg L−1 of copper reduced the level of O2, which may interfere with the aerobic metabolism of the cell [28]. Temperature is an important factor, especially in relation to the speed of chemical and biochemical reactions. Optimal temperatures for growth of our isolates were between 25°C and 30°C. A similar optimal temperature for removal of copper was observed in a study with Pseudomonas putida [20]. In our study, isolate A1 identified as Pseudomonas putida optimally removed copper at 35°C. The genera Pseudomonas are important for bioremediation of contaminated sites [9, 18, 20, 21, 29].
The operon cop has a system for genetic regulation that is expressed in function of the copper concentration in the cell [24]. The gene copA captures the copper when the concentration is low and transport it to the cytoplasm, and copB removes the copper from inside cells when there is toxic concentrations; the gene copY is the repressor, and copZ is the activator, which regulate the copper concentration in the cells [30]. Also, the environment conditions can increase and/or decrease the copper bioavailability for the microorganisms [29]. The optimum pH for enzyme activities is important for any metabolism; any pH lower than the optimum would protonate the ionized residue, while any pH higher than the optimum would ionize the protonated residue, leading to decreased activity [24]. Most enzymes have an optimum temperature, which may be related to the type of organism from which the enzyme was isolated. Some organisms grow well near the room temperature, and so, their enzymes are most active at a temperature around 30–40°C [24]. Besides, the copper reductase activity is involved in copper reduction that has been exhibited as one of the most important pathways for copper biosorption [29].
Understanding copper speciation is important to predict copper availability in the environment [10]. Matrix pH is considered as a key factor in the variability of metal species in the environment. Thus, it can be used to determine copper species found in the environment, revealing copper bioavailability and potential toxicity in contaminated sites [31]. Changing pH from 5.0 to 3.0 decreased biosorption of Cu(II) and Mn(II) in a study with Pseudomonas species and Bacillus species [32]. All isolates in our study increased culture pH values to 7.5 and 8.5 after 24 or 48 h of incubation. Culture pH influenced the growth of the isolates, and it was high at neutral pH. Nonetheless, isolates T5 as well as A6 grew significantly at pH 5.0. This might be related to the genetic similarity of the isolates because both microorganisms were identified as Acinetobacter sp. The pH increase can be attributed to the exchange of Cu(II) complexed or bound with other organic compounds (COO-) existing in the medium to the H+ ion, decreasing the concentration of free hydrogen, thus increasing the pH [33].
The highest sorption of copper by the isolates in this study was 190 mg L−1, achieved by isolate T5. The quantities removed in this study are much higher than 32 mg L−1 removed by Pseudomonas putida [21] and 16 mg L−1 of copper removed by Acinetobacter calcoaceticus [18]. Bacillus licheniformis removed 59 mg L−1 of copper [18]. Copper bioremoval capacities of our isolates are hence remarkable.
In summary, our isolates displayed high capacity for copper resistance, reduction and uptake in aqueous media. The capacity of the rhizosphere bacteria to reduce copper as well as copper uptake are important, where the reduction of Cu(II) to Cu(I) increases copper bioavailability in the rhizosphere zone. Cu(II)-resistant bacteria isolated from the rhizosphere region of Avena sativa and Plantago lanceolata were characterized. The results indicate that our isolates are potential agents for Cu(II) bioremoval and bacterial stimulation of Cu(II) biosorption by Avena sativa and Plantago lanceolata. Furthermore, our isolates demonstrated high ability to copper biotransformation in the liquid medium, being important agents to copper reactions in the rhizosphere zone, when it was bioaugmented by the inoculation of the Avena sativa plants.
References
Atlas RM, Bartha R (1997) Microbial ecology: fundamentals and applications, 4th edn. Benjamin/Cummings Science Publishing, Menlo Park
Nóvoa-Muñoz JC, Queijeiro JMG, Blanco-Ward D et al (2007) Total copper content and its distribution in acid vineyards soils developed from granitic rocks. Sci Total Environ 378:23–27
Andreazza R, Okeke BC, Lambais MR et al (2010) Bacterial stimulation of copper phytoaccumulation by bioaugmentation with rhizosphere bacteria. Chemosphere 81:1149–1154
McCutcheon SC, Schnoor JL (2003) Phytoremediation: transformation and control of contaminants. Wiley-Interscience, Hoboken
He LY, Zhang YF, Ma HY (2010) Characterization of copper-resistant bacteria and assessment of bacterial communities in rhizosphere soils of copper-tolerant plants. App Soil Ecol 44:49–55
Sachdev D, Nema P, Dhakephalkar P (2010) Assessment of 16 S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol Res 165:627–638
Sorkhoh NA, Ali N, Salamah S et al (2010) Enrichment of rhizospheres of crop plants raised in oily sand with hydrocarbonutilizing bacteria capable of hydrocarbon consumption in nitrogen free media. Inter Biodeter Biodegr 64:659–664
Tao S, Chen YJ, Xu FL et al (2003) Changes of copper speciation in maize rhizosphere soil. Environ Pollut 122:447–454
Andreazza R, Pieniz P, Wolf L et al (2010) Characterization of copper biosorption and bioreduction by a highly copper resistant bacterium isolated from copper-contaminated vineyard soil. Sci Total Environ 408:1501–1507
Cuppett JD, Duncan SE, Dietrich AM (2006) Evaluation of copper speciation and water quality factors that affect aqueous copper tasting response. Chem Senses 31:689–697
Whiteley CG, Lee DJ (2006) Enzyme technology and biological remediation. Enz Microb Technol 38:291–316
Römkens PFAM, Bouwman LA, Boon GT (1999) Effect of plant growth on copper solubility and speciation in soil solution samples. Environ Pollut 106:315–321
Umrania VV (2006) Bioremediation of toxic heavy metals using acidothermophilic autotrophes. Bioresource Technolol 97:1237–1242
Smith CF, Mccurdy WH Jr (1952) 2,9-Dimethyl-1,10-phenanthroline—new specific in spectrophotometric determination of copper. Anal Chem 24:371–373
Lane D (1991) 16S/23S sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York
Cole JR, Wang Q, Cardenas E et al (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Pages D, Rose J, Conrod S (2008) Heavy metal tolerance in Stenotrophomonas maltophilia. PLoS One 2:1–6
Clausen CA (2000) Isolating metal-tolerant bacteria capable of removing copper, chromium, and arsenic from treated wood. Waste Manag Res 18:264–268
Chen XC, Wang YP, Lin Q (2005) Biosorption of copper (II) and zinc (II) from aqueous solution by Pseudomonas putida CZ1. Colloid Surface B 46:101–107
Uslu G, Tanyol M (2006) Equilibrium and thermodynamic parameters of single and binary mixture biosorption of lead (II) and copper (II) ions onto Pseudomonas putida: effect of temperature. J Hazard Mater 135:87–93
Chen X, Shi J, Chen Y et al (2007) Determination of copper binding in Pseudomonas putida CZ1 by chemical modifications and X-ray absorption spectroscopy. App Microbiol Biotech 74:881–889
Zakaria ZA, Zakaria Z, Surif S et al (2007) Hexavalent chromium reduction by Acinetobacter haemolyticus isolated from heavy-metal contaminated wastewater. J Hazard Mater 146:30–38
Thangaraj K, Kapley A, Purohit HJ (2008) Characterization of diverse Acinetobacter isolates for utilization of multiple aromatic compounds. Bioresource Technol 99:2488–2494
Whiteley CG, Lee DJ (2006) Enzyme technology and biological remediation. Enzyme Microb Tech 38:291–316
Rodriguez-Montelongo LR, Volentini SI, Farías RN et al (2006) The Cu(II)-reductase NADH dehydrogenase-2 of Escherichia coli improves the bacterial growth in extreme copper concentrations and increases the resistance to the damage caused by copper and hydroperoxide. Arch Biochem Bioph 451:1–7
Tunali S, Çabuk A, Akar T (2006) Removal of lead and copper ions from aqueous solutions by bacterial strain isolated from soil. Chem Eng J 115:203–211
Özer A, Gürbüz G, Çalimli A et al (2009) Biosorption of copper(II) ions on Enteromorpha prolifera: application of response surface methodology (RSM). Chem Eng J 146:377–387
Poirier I, Jean N, Guary JC et al (2009) Responses of the marine bacterium Pseudomonas fluorescens to an excess of heavy metals: physiological and biochemical aspects. Sci Total Environ 406:76–87
Andreazza R, Okeke BC, Pieniz P et al (2011) Bioreduction of Cu(II) by cell-free copper reductase from a copper resistant Pseudomonas sp. NA. Biol Trace Elem Res. doi:10.1007/s12011-010-8899-3
Solioz M, Stoyanov JV (2003) Copper homeostasis in Enterococcus hirae. FEMS Microb Rev 27:183–195
Twiss MR, Errécalde O, Fortim C (2001) Coupling the use of computer chemical speciation models and culture techniques in laboratory investigations of trace metal toxicity. Chem Spec Bioavailab 13:9–24
Voss M, Thomas RWSP (2001) Sorção de cobre e manganês por bactérias rizosféricas do trigo. Ciênc Rural 31:947–951
Vilar VJP, Botelho CMS, Boaventura RAR (2007) Copper desorption from Gelidium algal biomass. Water Res 41:1569–1579
Acknowledgements
The authors would like to thank CNPq and CAPES for a scholarship to Robson Andreazza and Auburn University for the opportunity given to Robson to conduct part of his Ph.D. research. An equipment grant to Benedict Okeke to purchase a thermal cycler was received from Auburn University at Montgomery.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Andreazza, R., Okeke, B.C., Pieniz, S. et al. Characterization of Copper-Resistant Rhizosphere Bacteria from Avena sativa and Plantago lanceolata for Copper Bioreduction and Biosorption. Biol Trace Elem Res 146, 107–115 (2012). https://doi.org/10.1007/s12011-011-9228-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-011-9228-1