Abstract
Photorhabdus luminescens is an entomopathogenic rod-shaped bacterium infected with insect nematodes of the Heterorhabditidae family. It kills insects through the secretion of high molecular weight toxin complexes. In this study, Plutella xylostella larvae were orally administered P. luminescens for bioassay. After incubation in Luria-Bertani (LB) medium for a sufficiently long period, the mortality rates of P. xylostella observed after diluting the fermentation broth 50 times and diluting the supernatant 5 times were 18.89% and 91.11%, respectively. Retentates measuring more than 70 kDa showed 88% mortality after ultrafiltration (UF) membrane treatment. Thus, the supernatant of P. luminescens had insecticidal activity, and the main insecticidal toxin complexes had a molecular weight exceeding 70 kDa. The L9 (34) Taguchi orthogonal experimental optimized medium mode-predicted insecticidal activity levels were 84% and 119% in the 50-fold diluted fermentation broth and 5-fold diluted supernatant, respectively. Moreover, the insecticidal activity was improved to 92.2% in the 100-fold diluted fermentation broth and to 97.8% in the 10-fold diluted supernatant in the experiments. All combinations tested showed clear indications of lethality, including swelling, vesicle formation, cytoplasm vacuolization, and brush border membrane lysis. Thus, these results promote the use of P. luminescens 0805-P2R as a potent biopesticide to effectively control P. xylostella.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Photorhabdus luminescens is a symbiotic bacterium isolated from entomopathogenic nematodes Heterorhabditidae. It was adopted as a biopesticide because of different pathogenic factors in insect hosts through either bloodstream injection or oral ingestion [1]. P. luminescens is a non-spore-forming, motile, bioluminescent, gram-negative bacterium that belongs to the family Enterobacteriaceae and has four toxin groups. It is pathogenic to numerous insects and has a complex symbiotic relationship with nematodes. Among all sequenced bacterial genomes, the P. luminescens genome comprises the highest number of toxin genes. Additionally, an analysis of the complete genome sequence of P. luminescens revealed numerous adhesins, proteases, and lipases, which may be expressed during the pathogenic phase of its complex lifecycle. Many “toxin complex” (tc) loci were also revealed on the chromosome of P. luminescens strains [1], and all of these strains code for different high molecular weight insecticidal tc toxins. Some of the tc proteins may destroy the insect midgut, similar to Bacillus thuringiensis delta endotoxins. Moreover, P. luminescens can kill insect hosts through chromosomal insertion of a single gene called makes caterpillars floppy (mcf) [2].
The diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae), is a worldwide pest of Brassicaceae crops [2, 3]. However, information regarding the toxicity or histopathological effects of P. luminescens 0805-P2R or of its toxin, bacterial cell, and supernatant against this insect pest is not available. Accordingly, the aims of the current study were to (1) investigate the toxicity of P. luminescens 0805-P2R against P. xylostella and (2) identify the main insecticidal factor and its histopathological effects. Furthermore, by determining the insecticidal activity and histopathological effects of P. luminescens 0805-P2R, we proposed a mode of action of biological control agents.
Materials and Methods
Strains and Medium Culture Conditions
A P. luminescens 0805-P2R strain was used in this study. The strain was maintained on nutrient bromothymol blue agar (NBTA) plates [2.3% nutrient agar (Difco™, USA), 0.0025% bromothymol blue (Merck), and 0.004% 2,3,5-triphenyltetrazolium (Sigma-Aldrich)] at 30 °C and subcultured weekly.
Culture media was used in this study: 2.5% Luria-Bertani (LB) medium [4] [1% tryptone, 0.5% yeast extract (YE), and 1% NaCl at pH 7] and cultivation medium in this study [1.25% molasses, 0.3% YE, 0.5% yeast extract powder (YP), 0.2% isolated soy protein (ISP), 0.5% amino acid (AA), 1.5% sucrose, 0.5% maltodextrin, 0.75 mM K2HPO4, and 0.75 mM MgSO4 at pH 5.5], where the amino acid (lactalbumin enzymatic hydrolysate) in this study was purchased from the Sigma-Aldrich Corporation (St. Louis, MO, USA).
Pathogenicity tests were performed from 0 to 120 h (0, 12, 24, 48, 72, 96, and 120 h) after culture. P. luminescens 0805-P2R was cultured at 30 °C in LB agar plates. A single colony was selected to prepare a cell suspension in sterile saline solution (0.85% NaCl); for the culture, the optical density (OD) at 600 nm was 0.2, equivalent to 1 × 107 log (colony-forming unit (CFU)/mL). Colony counter 270 (SUNTEX, Taiwan) was used to quantify the cells. OD and luminescence were monitored at 0, 12, 24, 48, 72, 96, and 120 h using a V-730 ultraviolet-visible/near-infrared spectrophotometer (Japan) and a Centro luminometer (Model 4900, Awareness Technology), respectively. For monitoring the relative luminescence unit (RLU), the cell-free cultivation medium was used to calibration as zero point and the RLU of cultivation broth was read directly by the luminometer after proper dilution with distilled water. The extracellular protein secreted by P. luminescens 0805-P2R cultivated in LB broth at different incubation times and dilution ratios was analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
L9 (34) Taguchi Design for Medium Optimization
The L9 (34) Taguchi orthogonal experimental design was used to determine the optimal culture medium for insecticidal activity. The factors considered for culture medium optimization were as follows: (A) nitrogen source, (B) carbon source, (C) dissolved oxygen, and (D) cultivation time. The optimization was conducted using a four-factor (X1–X3) three-level system (namely 1, 2, and 3), where Latin square array and 9 experimental runs were performed. The values derived for the factors at the three levels are outlined as follows: (A) nitrogen source—(1) 0.1% YE, 0.5% YP, 0.25% AA, 0.2% ISP, and 0.25% peptone; (2) 0.3% YE, 0.5% YP, 0.5% AA, 0.2% ISP, and 0.5% peptone; and (3) 0.5% YE, 0.5% YP, 0.75% AA, 0.2% ISP, and 0.75% peptone; (B) carbon source—(1) 0.5% molasses and 0.5% sucrose; (2) 1% molasses and 1% sucrose; and (3) 1.5% molasses and 1.5% sucrose; (C) dissolved oxygen—(1) silicone plug; (2) rubber plug; (3) and cotton plug; (D) cultivation time—(1) 20 h; (2) 72 h; and (3) 120 h. Table 1 lists the design matrix for nutrient concentrations and their levels. All experiments included 0.75 mm K2HPO4, 0.75 mm MgSO4, and 0.1% Tween 80. The current study adopted whole broth and supernatant as different treatments.
Bioassays
An oral bioassay was performed using laboratory-reared P. xylostella under constant conditions (temperature 25 °C; relative humidity 60–70%; and environment 15-h/9-h light/dark photoperiod). The strain was provided by Syngenta (Wilmington, DE, USA) Ltd. Thirty-third-instar P. xylostella larvae were reared separately in 9-cm sterile Petri dishes containing 1-cm3 artificial diet. To test the efficiency of P. luminescens 0805-P2R against P. xylostella, 1.5-g bacterial culture was poured on the surface of a cube of fresh artificial diet at a concentration of 107 cells/mL after different culture incubation times. Mortality was defined as the number of deaths observed within 72 h of feeding larvae with thirty-third-instar P. xylostella larvae per treatment.
Preparation and Sectioning of P. xylostella Midgut Tissues
Third-instar dead larvae of P. xylostella were placed on ice for 15 min after exposure to the P. luminescens 0805-P2R whole broth with a cell concentration of 107 cells/mL or supernatant for 2 days. Larvae were rinsed with 70% ethanol. Midgut tissue samples were fixed with Bouin Hollande 10% sublimate [5]. The samples were incubated in fixative for 30 min, moved to fresh fixative solution and incubated for 24 h, and then washed for 12 h in distilled water. Subsequently, the samples were dehydrated using ethanol at different concentrations: (1) 70% ethanol for 1 h, (2) 96% ethanol for 1 h, and (3) 100% ethanol for 2 h. After being infiltrated using a 50% ethanol–50% xylol mixture for 1 h, 100% xylol solution for 1 h, and 50% xylol–50% Paraplast (Oxford Labware) mixture for 12 h at 58 °C, gut tissues were embedded in Paraplast twice for 24 h at 58 °C. Finally, the applied Paraplast was hardened with ice. Sections measuring 5 μm were placed in carriers coated with a mixture of 1.5% egg albumin and 3% glycerol in distilled water [5]. After deparaffinization using 100% toluene, the obtained sections were stained with hematoxylin and eosin [6] for histopathological localization of toxin effects.
Statistical Analysis
P. luminescens 0805-P2R cell toxicity levels represent the average of three replicates of each executed experiment. All statistical analyses were performed using R software [7]. The median lethal concentration and 90% lethal concentration were derived using GraphPad Prism software, version 7.0 (GraphPad Software, San Diego, CA). The statistical significance of the model equation was evaluated using Fisher’s test, with the significance level being set at p < 0.05. The quality of model fit is expressed as R2.
Results
Bacterial Pathogenicity
Regarding the number of bacterial colonies, the current study revealed the first 2 h to represent the lag phase, 2–24 h to represent the exponential phase, and 24–48 h to represent the stable phase (stationary phase); the period after 72 h represented the death phase (decline phase). The highest colony number of P. luminescens 0805-P2R culture in LB medium was 9.35 ± 0.19 log (CFU/mL) and was observed at 24 h; two bioluminescence expression peaks were observed. At 12 and 48 h, no cold light activity was observed in the supernatant. According to the relationship between the bioluminescence expression peaks and number of bacterial colonies, the light-emitting protein expressed by photobacterium under cold light may not have been secreted extracellularly but intracellularly (Fig. 1).
For the extracellular protein secreted by P. luminescens 0805-P2R cultivated in LB broth, after 48 h of fermentation, a clear band appeared when the molecular weight was greater than 210 kDa, as revealed by SDS-PAGE results, indicating that this substance increased significantly in the metabolite. This band was maintained until 72 h, after which it became less apparent at 120 h (Fig. 2).
L9 (34) Taguchi Design and Medium Optimization
The experiment revealed that whole broth optimized using the seventh formula (i.e., 0.5% YE, 0.5% YP, 0.75% AA, 0.2% ISP, 0.75% peptone, 0.5% molasses, 0.5% sucrose, cotton plug, and culture time 72 h) engendered the highest insect mortality rate (up to 83.33%). The bacterial concentration was the highest in the ninth formula, namely 8.17 ± 0.1 [log (CFU/mL). The best factor combinations for whole broth and supernatant treatments were A3B2C3D2 and A2B2C3D2, respectively (Fig. 3).
Effect factor for insecticidal activity of a whole broth and b supernatant of P. luminescens 0805-P2R incubated in medium optimized using L9 (34) Taguchi experimental design method. (A1) 0.1% YE, 0.5% YP, 0.25% AA, 0.2% ISP, and 0.25% peptone. (A2) 0.3% YE, 0.5% YP, 0.5% AA, 0.2% ISP, and 0.5% peptone. (A3) 0.5% YE, 0.5% YP, 0.75% AA, 0.2% ISP, and 0.75% peptone. (B1) 0.5% molasses and 0.5% sucrose. (B2) 1% molasses and 1% sucrose. (B3) 1.5% molasses and 1.5% sucrose. (C1) 1, silicone plug; 2, rubber plug; and 3, cotton plug. (D1) 1, 20 h; 2, 72 h; and 3, 120 h
Bioassay for Determining Insecticidal Activity of P. luminescens 0805-P2R Against P. xylostella As Well As Histopathological Effects of P. luminescens 0805-P2R on Midgut Tissues
The cells were removed by centrifugation at 10,000 rpm for 10 min, and the supernatant was filtered through a 0.45-μm membrane. Then, the filtrate was ultrafiltered by a tangential flow filtration system (MAP-TFF, Lefo Science, Taipei, Taiwan) equipped with a hollow fiber filter module with a molecular weight cutoff 70 kDa (MicroKros, Spectrum Labs, Rancho Dominguez, CA, USA). The retentate was concentrated to one tenth of the original volume. The whole broth, supernatant, retentate, and permeate were subjected to bioassay.
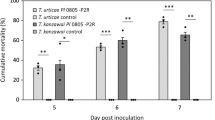
A 50-fold dilution of the whole fermentation broth has a significant insecticidal activity against the third instar larvae. On the other hand, a 5-fold dilution of the supernatant has a higher insecticidal activity. In the Taguchi experiment design, treatments 2, 5, 6, and 7 all have more than 95% lethal activity. After the supernatant was separated and treated with a hollow fiber column, a five-fold dilution of the retentate showed high insecticidal activity, and treatments 5 and 7 both had a lethal activity of more than 95% (Table 2). According to factor response analysis, the optimum conditions for the insecticidal activity of the whole fermentation broth were A3B2C3D2, and the optimum conditions for the insecticidal activities of the supernatant and the retentate were A2B2C3D2, which were different from the whole fermentation broth. The optimum condition for the insecticidal activity of the permeate was A2B1C2D3, which was obviously different from the above treatments (Table 3). Even without dilution, its insecticidal activity was lower than above treatments (Table 3). In this experiment, the insecticidal active substance in the fermentation broth of P. luminescens was a substance having a molecular weight of more than 70 kDa, with the band of SDS-PAGE that showed that the insecticidal active substance may be 210 kDa. The mode-predicted insecticidal activity was calculated by equations of UWB = ū + [(A3 − ū) + (B2 − ū) + (C3 − ū) + (D2 − ū)] and USP = ū + [(A2 − ū) + (B2 − ū) + (C3 − ū) + (D2 − ū)] for the whole fermentation broth and the supernatant, respectively. The predicted insecticidal activity was 84% for the 50-fold diluted fermentation broth obtained using A3B2C3D2 and was 119% for the 5-fold diluted supernatant (Table 3).
A histopathological study of P. luminescens 0805-P2R bacterial cells (whole broth) alone was conducted on the third-instar larvae of P. xylostella (Fig. 4b, c). Moreover, the histopathological effect of the supernatant mixture on P. xylostella third-instar larvae was studied (Fig. 4d). Midgut samples obtained from noninfected P. xylostella (i.e., controls) exhibited an intact morphology and a well-preserved layer of epithelial cells with an unaffected apical microvilli membrane (Fig. 4a). By contrast, samples obtained from P. xylostella third-instar larvae infected with P. luminescens 0805-P2R cells for 72 h demonstrated intensive vacuolization, without brush border membrane destruction (Fig. 4b). Conversely, midgut samples obtained from P. xylostella larvae exposed to the supernatant exhibited swollen and vacuolated cells with a regularly disposed brush border and constricted lumen (Fig. 4d). However, the midgut epithelium showed histopathological alterations, including cytoplasmic vacuolization, brush border membrane destruction, and apical membrane fragmentation (Fig. 4c). This structural disorganization of the P. xylostella intestinal epithelium was similar to that observed when insect larvae were treated with a mixture of TT01 cells (5 cells/mL) at a concentration of 20 ng/cm2 and strong altered columnar cells with many vesicles (Fig. 4d).
Histopathological effect of a non-treatment on midgut tissue of the third-instar P. luminescens larvae (control check, CK); P. luminescens 0805-P2R whole broth after b 24 h and after c 72 h. d Histopathological effects of supernatant on midgut tissue of the third-instar P. luminescens larvae. Am apical membrane, Bm basal membrane, Lu lumen, V vacuole formation. Image is × 40 magnified. Red bar = 0.1 mm
Discussion
The current study revealed that the insecticidal activity of a photobacterium bacterial solution and supernatant of the bacterial solution was positively correlated with treatment time. For the extracellular protein, molecular weight greater than 210 kDa increased significantly in the metabolite and maintained 3 days. Moreover, retentates measuring more than 70 kDa also showed insecticidal activity.
Photobacterium, a new insecticidal biological pesticide, is a multifunctional biological agent with insecticidal and antibacterial properties. Accordingly, this study primarily investigated the insecticidal activity of photobacterium against P. xylostella. Taguchi’s experimental design method was used for culture medium optimization, thus enabling rapid determination of the most suitable medium for the strain 0805-P2R. The best factor combination for medium optimization was A3B2C3D2 (nitrogen source: 0.5 YE, 0.5% YP, 0.75% AA, 0.2 ISP, and 0.75 peptone; carbon source: 0.1% molasses and 1% sucrose; dissolved oxygen: cotton plug; incubation time: 72 h). This optimized medium provided the best culture condition for polished rod bacterial strains for the investigation of insecticidal activity.
Histopathological investigations reveal that after P. xylostella infection with P. luminescens 0805-P2R bacterial cells and the supernatant, most of the columnar cells of the gut epithelium exhibited alterations, demonstrating the potential of the P. luminescens 0805-P2R toxin; the midgut tissue is the primary site of action of the toxin [8]. P. xylostella mixture treatment caused intensive damage to all columnar cells as well as completes apical membrane destruction. This explains the synergy between P. luminescens 0805-P2R cells and supernatant toxin. A similar finding was reported by previous research [9]. Benfarhat-Touzri et al. [10] indicated that the Cry1Ac toxin of B. thuringiensis and P. luminescens 0805-P2R cells exhibited synergistic effects against Spodoptera littoralis.
In conclusion, our study demonstrated the lethal effect of supernatant with P. luminescens 0805-P2R against P. xylostella; P. luminescens 0805-P2R caused 100% mortality 3 days after treatment. This result was confirmed by the histopathological alteration of the P. xylostella midgut epithelium. These findings support the feasibility of P. luminescens 0805-P2R as a novel biopesticide agent against polyphagous lepidopteran pests.
References
Ffrench-Constant, R. H., Dowling, A., & Waterfield, N. R. (2007). Insecticidal toxins from Photorhabdus bacteria and their potential use in agricultureToxicon, 49, 436–451.
Zalucki, M. P., Shabbir, A., Silva, R., Adamson, D., Shu-Sheng, L., & Furlong, M. J. (2012). Estimating the economic cost of one of the world’s major insect pests, Plutella xylostella (Lepidoptera: Plutellidae): just how long is a piece of string? Journal of Economic Entomology, 105(4), 1115–1129.
Furlong, M. J., Wright, D. J., & Dosdall, L. M. (2013). Diamondback moth ecology and management: problems, progress, and prospects. Annu Rev Entomol, 58, 517–541.
Sambrook, J., Frisch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual (2nd ed.). Cold New York: Spring Harbor Laboratory, Cold Spring Harbor.
Denolf, P., Jansens, S., Van Houdt, S., Peferoen, M., Degheele, D., & Van Rie, J. (1993). Biotinylation of Bacillus thuringiensis insecticidal crystal proteins. Applied and Environmental Microbiology, 59(6), 1821–1827.
Tyagi, R. (2005). Cell biotechnology. New Delhi: Discovery Publishing Pvt. Ltd.
R Core Team, (2014) R foundation for statistical computing, Vienna. ISBN 3-900051-07-0.
Ben Hamadou, D., Boukedi, H., Abdelkefi-Mesrati, L., Tounsi, S., & Jaoua, S. (2013). Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. Journal of Invertebrate Pathology, 114, 139–143.
Jallouli, W., Boukedi, H., Sellami, S., Frikha, F., Abdelkefi-Mesrati, L., Tounsi, S. (2018). Combinatorial effect of Photorhabdus luminescens TT01 and Bacillus thuringiensis Vip3Aa16 toxin against Agrotis segetum. Toxicon 142, 52–57.
Benfarhat-Touzri, D., Ben Amira, A., Ben Khedher, S., Givaudan, A., Jaoua, S., & Tounsi, S. (2013). Combinatorial effect of Bacillus thuringiensis kurstaki and Photorhabdus luminescens against Spodoptera littoralis (Lepidoptera: Noctuidae). Journal of Basic Microbiology, 54, 1160–1165.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, LH., Wang, YT., Hsieh, FC. et al. Insecticidal Activity of Photorhabdus luminescens 0805-P2R Against Plutella xylostella. Appl Biochem Biotechnol 191, 191–200 (2020). https://doi.org/10.1007/s12010-020-03289-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03289-8