Abstract
Endoglucanase secreted by the fungus Trichoderma atroviride is a kind of cellulase. An endoglucanase gene egII was cloned from T. atroviride AS3.3013 and expressed in Saccharomyces cerevisiae INVScI. The open reading frame of the egII gene was composed of 1257 bp, encoding 418 amino acids with a molecular weight of 44.23 kDa plus a signal peptide of 21 amino acids. Based on sequence similarity, TaEGII belonged to the glycosyl hydrolase family 5. Expression of the egII gene in T. atroviride AS3.3013 can be induced by microcrystalline cellulose (MCC), bran, carboxymethyl cellulose (CMC), rice straw, and corn stalk but is inhibited by glucose. A highly efficient integrated expression vector (pYPIGH-B includes a sequence of the α-mating factor signal peptide (MF-α)) was constructed. The enzymatic activity of the supernatant of recombinant yeast YPIGH-B3 was 1.29 times higher than that of YES2-egII, demonstrating that the MF-α can significantly improve the expression of the recombinant EGII in S. cerevisiae. The recombinant endoglucanase TaEGII produced by S. cerevisiae showed maximum activity at pH 5.0 and temperature 60 °C. Under these conditions, the Km and Kcat values for Avicel and raffinose hydrolysis were 1.22 × 10–2 mg ml−1, 9.09 × 10−2 s−1 and 1.06 × 10–2 mg ml−1 , 9.18 × 10–2 s−1, respectively. The enzymatic activity of recombinant TaEGII was stable when incubated from 40 to 60 °C for 1 h. It was stable in a wide range of pH (4.0–7.0) and sensitive to various metal ions. Transgenic yeast strain YPIGH-B3 might be applied to cellulosic ethanol production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Due to increased energy needs and a need to decrease fossil energy consumption, fuel ethanol is becoming an important new energy worldwide [1]. Cellulose is the most abundant and renewable biomass and can be used as a substitute for fossil energy [2, 3]. For bioethanol production, the depolymerization of cellulose into reducing glucose is a prerequisite for microbial fermentation [4], and enzyme digestion is an attractive strategy because it lacks harmful effects on the environment. Cellulases are produced by a wide spectrum of microbes in nature [5] and include endoglucanases (E.C.3.2.1.4), β-glucosidases (E.C.3.2.1.21), and exoglucanases (E.C.3.2.1.91) [6]. The filamentous fungus Trichoderma atroviride produces cellulases with high enzyme activity. However, cost-effective production of these enzymes on a large scale is challenging [7]. As an approach to solve this problem, efforts have been made to engineer the heterologous expression of cellulases. The most commonly employed yeast strain for recombinant protein expression is Saccharomyces cerevisiae [8]. However, yeast expression levels may be low, and the activities of yeast-expressed enzymes are generally lower than that of native enzymes [9]. This low expression may be because the heterologous gene encodes a signal peptide that is not accurately identified in the yeast host or if the copy number of the integrating vector is low [4]. To address this issue, a high copy integrating vector including a signal peptide is often constructed to express exogenous protein. The ribosomal DNA of S. cerevisiae encompasses 100–200 tandemly repeated units and is considered an attractive target for such multiple integration. In this study, a new endoglucanasegene egII was cloned from T. atroviride AS3.3013 and a high copy integration vector (pYPIGH) was constructed. Five recombinant endoglucanase yeast strains (YPIGH-H1, YPIGH-H2, YPIGH-B2, YPIGH-B3, and YPIGH-D) were obtained. An episomal vector pYES2-EGII was also constructed. The yield, secretion, and activity of the EGII expressed from S. cerevisiae were studied.
Materials and Methods
Strains, Media, and Plasmids
T. atroviride AS3.3013 was cultured on potato dextrose agar medium (PDA: glucose 20.0 g, potato leachate 1.0 L, agar 15.0 g; pH 7.0) at 28 °C for 4 days until spores were formed. Seed liquid preparation was performed in accordance with the method described by Huang et al. [10]. The expression host was S. cerevisiae INVScI and was cultivated in yeast peptone dextrose (YPD) medium (1% yeast extract, 2% peptone, and 2% glucose (w/v)). To study the transcription of the recombinant gene, S. cerevisiae INVScI was cultured on synthetic complete lacking uracil (SC-U) medium [4]. Escherichia coli DH5α and E. coli JM109 strains were grown in Luria-Bertani broth (LB) at 37 °C or agar supplemented with the appropriate antibiotic. The pYES2, pUC18, pPIC9K, and pCSN43 plasmids were used for the construction of a integrating vector with high copy.
Cloning of the Endoglucanase Gene egII from T. atroviride
The hyphostoma of T. atroviride AS3.3013 were collected and ground into powder in a mortar using liquid nitrogen. The mycelia were lysed, and total RNA was isolated using Trizol reagent (Invitrogen, USA) and treated with DNase I (Roche, Switzerland). The complementary DNA (cDNA) was generated by reverse transcription of total RNA with Revertaid First-Strand cDNA Synthesis Kit (Fermentas, Canada) and used as the polymerase chain reaction (PCR) template.
The egII gene was amplified using primers MEGRF/MEGRR1/MEGRR (Table 1). The primers were designed according to the gene sequence of egII (DQ178347) from Hypocrea jecorina. The full-length endoglucanase gene egII was amplified under defined PCR conditions of initial denaturation at 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 55.6 °C for 45 s, and 72 °C for 90 s in 25 μl reaction with final extension of 7 min at 72 °C. The PCR fragment (1.3 kb) was purified, cloned into the pMD18-T vector (TaKaRa, Japan), and introduced into E. coli DH5α by CaCl2 transformation.
The transformants were grown onto LB plates containing ampicillin (100 μg ml−1). Colonies were identified by PCR and restriction digestion analysis, and the cDNA sequence was analyzed.
Bioinformatic Analysis of egII
Homology of endoglucanase gene egII was identified using the BlastX database. The ORF database (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to determine the open reading frame (ORF). The Protparam tool (http://web.expasy.org/protparam/) was used to calculate the theoretical molecular mass and isoelectric point (pI) of the protein. The Signal P 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) was used to predict the signal peptide of the gene. InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) identified the conserved domain of the EGII protein. The ClustalX program aligned the EGIIamino acid sequences from different fungi.
Expression Response of Native egII to Different Induced Substrates
The gene expression of egII from T. atroviride AS3.3013 was studied using Mandels’ medium (MM including (NH4)2SO4 1.4 g l−1, arbamide 0.3 g l−1, KH2PO4 2.0 g l−1, CaCl2 0.3 g l−1, MgSO4·7H2O 0.3 g l−1, peptone 1.0 g l−1, Tween 80 1.0 g l−1, FeSO4·7H2O 5.0 mg l−1, ZnSO4·7H2O 1.4 mg l−1, MnSO4·H2O 1.6 mg l−1, and CoCl2 2.0 mg l−1, pH 5–6) [11] supplemented with different substrates. Substrates were added to Mandels’ medium at 2% and included glucose, sucrose, carboxymethyl cellulose (CMC), microcrystalline cellulose (MCC), corn straw, rice straw, and bran. One hundred milliliters of medium with 106 conidia per milliliter was injected into 500-ml conical flasks, then cultivated at 200 rpm and 28 °C for 48 h as seed liquid. The seed liquid (10% (v/v) (described above) was added to the MM with different substrates and incubated at 28 °C in a rotary shaker at 200 rpm. Samples were harvested from different cultured times for 1, 2, 3, 4, and 5 days. The hyphostoma were gathered and ground into powder in liquid nitrogen for RNA isolation. Total RNA was stored at −80 °C and used for RT-PCR. The egII and 18S ribosomal DNA (rDNA) genes were amplified using primers LF/LR and 18sF/18sR, respectively. The PCR procedure and reaction conditions were performed according to the manufacturer’s instructions (TaKaRa, Japan).
Construction of a High Copy Integrating Recombinant Vector
To clone the sequence of the rDNA, the genomic DNA of S. cerevisiae was extracted by traditional method. Then, the fragment of rDNA S1 sequence was amplified from the isolated genomic DNA using primers seq1F and seq1R. The pUC18 plasmid was isolated by plasmid extraction kit (Fermentas, #K0503) and digested with EcoRI and KpnI. The amplified rDNA S1 sequence was digested with EcoRI and KpnI (Thermo, USA) and then inserted into the pUC18 vector, yielding pUCTY1. The PCR fragment rDNA S2 sequence was amplified with primers seq2F and seq2R from the genomic DNA, digested with KpnI and HindIII, and then inserted into the pUCTY1 vector, yielding pUCTY12. The hygromycin-resistant gene (hyg) that was used to identify the transformants of S. cerevisiae was amplified by primers hygF and hygR using the pCSN43 (Invitrogen, USA) vector as template. The hyg fragment and vector pUCTY12 were digested with KpnI, and the digested vector pUCTY12 was dephosphorylated with alkaline phosphatase (CIP). Then, the fragments were ligated with T4 ligase to form plasmid pUCTY12-hyg. The sequence of the multiple clone site (dcs) was cloned from the vector pYES2 (Invitrogen, USA), digested with NheI and PacI, and then inserted into vector pUCTY12-hyg, yielding the integrating vector pYPIGH.
To conveniently purify the recombinant protein, a His tag was added to the egII coding sequence. To increase the secretion of protein, the sequence of the α-mating factor strong signal peptide (MF-α) was amplified from plasmid pPIC9k by primers M1 and M2 and inserted into pUC18, yielding pUC18-M. The fragment of the whole gene (H1) and the fragment of the gene egII not including the signal peptide (H2) was amplified with primers H1F/H1R and H2F/H2R, respectively. Then, fragments H1 and H2 were separately inserted into pUC18-M, yielding pUC18-MH1 and pUC18-MH2. The fragment of MF-α was added to the plasmid with egII but no natural signal peptide (B), and the fragment of MF-α and egII (D) was amplified using primers BF/BR and DF/DR, respectively. The four fragments (H1, H2, B, and D) were separately inserted into pYPIGH, yielding pYPIGH-H1, pYPIGH-H2 pYPIGH-B, and pYPIGH-D.
To compare the expression efficiency, the episomal vector pYES2-EGII was constructed in this study. First, the egII gene was amplified using the primers MEGRF/MEGRR. Then, the PCR fragment and the plasmid pYES2 were digested with XbaI and NotI and ligated together, yielding pYES2-EGII.
Recombinant plasmids pYPIGH-B, pYPIGH-D, pYPIGH-H1, and pYPIGH-H2, pYES2-EGII, and the empty vector plasmid YPIGH were transformed into S. cerevisiae INVScI using the electricity conversion method [12]. Yeast transformants were designated S. cerevisiae strains YPIGH-B, YPIGH-D, YPIGH-H1, YPIGH-H2, YES2-EGII, and YPIGH and stored in SC-U glycerol medium at −80 °C.
Real-Time RT-PCR Expression Analysis of egII in Yeast
To study expression of egII, the yeast transformants were cultured in SC-U medium containing 2% β-D-galactose and were collected after induction for 2, 3, 4, 5, or 6 days. RNA samples were reverse transcribed with the Revert Aid™ First Strand cDNA Synthesis Kit (Fermentas, Canada). First, 12 μl reactions containing 5 μl RNA template, 1 μl oligo(dT)18 primer, and 6 μl water nuclease-free were mixed according to the instructions and incubated at 65 °C for 5 min. Second, the above reactants were added to 4 μl 5× reaction buffer, 1 μl RiboLock™ RNase Inhibitor, 2 μl 10 mM dNTP mix, and 1 μl Revert Aid™M-MuLV Reverse Transcriptase, incubated at 42 °C for 60 min, followed by termination of the reaction by heating at 70 °C for 5 min. The cDNA product was tested with the SYBR® Premix Ex Taq™II (TaKaRa, Japan) using gene-specific primers (LF and LR). Part of the 18s RNA gene was amplified with primers (18sF and 18sR) as a control. Twenty-five microliter reactions containing 2 μl cDNA template, 0.4 μmol l−1 PCR primers (10 μmol), 12.5 μl SYBR® Premix Ex Taq™II (2×), and 8.5 μl ddH2O were prepared according to the instructions. The real-time RT-PCR amplification program consisted of an initial denaturation step at 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The threshold cycle (Ct) values were adjusted artificially. Samples with a Ct value <40 were considered positive.
Protein Purification
The supernatant from yeast cultures expressing the plasmid was used to purify the protein with the Ni-Agarose His protein purification kit (CWBIO, China). The purified proteins were identified by SDS-PAGE electrophoresis.
The molecular weight of the purified recombinant TaEGII was determined by SDS-PAGE using 12% sodium dodecyl sulfate-polyacrylamide gels according to the method of Laemmli [13] with some modifications. Coomassie brilliant blue R-250 was used to stain the gels. The purified enzyme activity was identified by the method of transparent circle [14].
Assay of Endoglucanase Activity
The CMCase activity was measured according to the method described by Meinke et al. [15]. S. cerevisiae transformants were grown on SC-U medium at 30 °C, and the expression of egII was induced by 2% β-D-galactose. Yeast cells were collected and centrifuged after inducing for 24, 36, 48, 60, and 72 h. The supernatants containing crude enzyme were purified to measure CMCase activity. First, the supernatants were salted out by (NH4)2SO4 for one night at 4 °C, then centrifuged at 4 °C, 12,000 rpm, and the precipitates were gathered. The pellets were dissolved into citric acid-sodium citrate buffer (pH 5.0) for 30 min, and the CMCase activity was measured.
Properties of Recombinant EGII
The yeast transformant S. cerevisiae YPIGH-B3 was induced by 2% β-D-galactose at 30 °C for 4 days. The preparation of crude enzyme was performed as above. Reaction mixtures (pH = 5.0) were incubated at 30 to 80 °C for 1 h at 10 °C intervals, chilled on ice, and then assayed for activity to determine the effect of temperature. The pH value of the reaction mixture was adjusted between 2.0 and 8.0 at an interval of 1.0 pH unit using Na2HPO4-citrate buffer (pH 2.0 to 8.0). After incubating at 4 °C for 48 h, the solution was adjusted to the optimum pH for enzyme activity and measured.
For examination of the metal ion effect on the enzyme activity, various titers of metal salt (FeSO4, MnSO4, ZnSO4 MgSO4, CoCl2, NaCl, KCl, and BaCl2) were added in the reaction mixture at a final concentration of 0.75 mM each. Activity was examined at 60 °C and pH 5.0.
To analyze the kinetic parameters of recombinant TaEGII, the Michaelis constant (Km) and maximum velocity (Vmax) of purified recombinant enzyme were determined by measuring the rates of CMC-Na, Avicel, cellulose, cellobiose, and raffinose hydrolysis under standard assay conditions. The reaction was carried out by incubating these five substrates at concentrations ranging from 0.5 to 2.5 mg ml−1 at 60 °C and pH 5.0. Km and Vmax values were determined using the Lineweaver-Burk plot (LBP). The concentration of the purified protein was calculated by Bradford method [16], using bovine serum albumin (BSA) as the standard.
Results
Construction of a High Copy of the Integration of Vectors
An integrated plasmid pYPIGH (Supplementary Fig. S1) for S. cerevisiae was successfully constructed for protein expression. Four integrated expression vectors (pYPIGH-B, pYPIGH-D, pYPIGH-H1, and pYPIGH-H2) and one episomal recombinant vector (pYES2-EGII) were successfully constructed. The pYPIGH-H1 expressed the egII gene without modification, and pYPIGH-D expressed the egII gene with MF-α. The pYPIGH-H2 plasmid encodes the egII gene without the natural signal peptide, and the pYPIGH-B plasmid encodes the egII gene without the natural signal peptide with the MF-α. To target pYPIGH efficiently to the chromosomal rDNA locus, the integrated plasmids were linearized using BglII restriction enzyme before transforming into S. cerevisiae INVScI.
Sequence Analysis of egII
The length of the egII gene coding region was 1257 bp. The mature TaEGII protein could encode 418 amino acids with a theoretical pI of 4.96 and a molecular mass of 44.23 kDa. The DNA sequence of egII has been submitted to the GenBank nucleotide sequence database under the accession number of KP098496.
The putative signal peptide was predicted with Signal P 3.0 server, and there was a signal peptide (21 amino acids) in the N-terminal of the TaEGII amino acid sequence (AJP16798). The signal peptide cleavage site was between positions A21 and Q22, indicating a predicted extracellular protein. Analysis with InterProScan revealed that the T. atroviride AS3.3013 TaEGII protein sequence has a catalytic domain of the GH family 5 (InterProAcc. No. IPR001547), indicating that it is an endoglucanase.
Multiple sequence alignment of amino acid sequence of EGII with sequences of related fungal endoglucanase was carried out using ClustalX. The amino acid sequence identities between AJP16798 from T. atroviride AS3.3013 and other fungal species ranged from 54 to 99%, showing that the sequences were not highly conserved for all fungi. However, the catalytic domains were highly conserved, with a common region of IIGQGGPTN[D] (Fig. 1).
Multiple sequence alignment of catalytic domains of endoglucanase from 27 fungal species/strains. All these endoglucanase are members of glycoside hydrolase family 5; their catalyzing domains are highly conserved. The asterisks refer to the conserved amino acid residues. ESZ90854.1: Sclerotinia borealis F-4157; XP_001552808: Botrytis cinerea B05.10; XP_001598802: Sclerotinia sclerotiorum 1980; KKO98175.1: Trichoderma harzianum; XP_013960589: Trichoderma virens Gv29-8; KUF02176.1: Trichoderma gamsii; XP_013943634: Trichoderma atroviride IMI206040; AHW57398.1: Trichoderma asperellum; XP_013948379: Trichoderma atroviride IMI206040; KUE96135.1: Trichoderma gamsii; AHW57399.1: Trichoderma asperellum; KKP03485.1: Trichoderma harzianum; AAR29981.1: Trichoderma sp. C-4; XP_013952028: Trichoderma virens Gv29-8; ABA64553.1: Trichoderma reesei; P07982.1: Trichoderma reesei; XP_006962583: Trichoderma reesei QM6a; ADJ10627.1: Trichoderma viride; AFK10489.1: Clonostachysrosea f. catenulate; AFD50195.1: Trichoderma orientale; ACH92572.1: Trichoderma sp. SSL; AJP16798.1: Trichoderma atroviride; BAA36216.1: Trichoderma viride; XP_003656246: Thielavia terrestris NRRL8126; XP_007914601: Phaeoacremonium minimum UCRPA7; EMD34838.1: Gelatoporia subvermispora B; BAF75943.1: Polyporus arcularius
Native Expression of egII
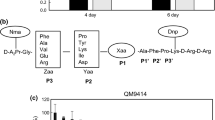
The native expression of egII in T. atroviride AS3.3013 was analyzed for different substrates (Fig. 2). No transcript signal was detected for glucose (2%), and the highest expression was seen in MCC, indicating that MCC was the optimal induced substrate. When induced by CMC, MCC, or bran, expression was highest at 3 days. When induced by sucrose, corn straw, and rice straw, expression was highest at 4 days.
Transcript of Gene egII in Yeast Recombinant
We next examined expression of the different egII constructs in recombinant yeast using real-time RT-PCR analyses (Fig. 3). The transcript properties were similar between YPIGH-B and YPIGH-D, and the highest expression was observed from transformant YPIGH-B3 at 4 days. For all constructs, the messenger RNA (mRNA) level of egII detected 2 days after inoculation was low, increased rapidly between 3 and 4 days, and then decreased after 4 days.
Real-time RT-PCR analysis of egII transcripts in the yeast transformants. Yeast transformants were cultured in SC-U medium containing 2% β-D-galactose and were collected at inducing for 2, 3, 4, 5, and 6 days, respectively. The above showed different transformant expression phenomena. In addition, control and YES2-EGII indicated S. cerevisiae INVScI and the non-egIIgene yeast transformant including episomal recombinant vector pYES2-EGII, respectively
Assay of CMCase Activity
The CMCase activities of YPIGH-H1, YPIGH-H2, YPIGH-B3, and YPIGH-D showed a peak value at 48 h under 2% β-D-galactose induction (Fig. 4). The recombinant EGII activity of the transgenic yeast YPIGH-B3 (117.79 U g−1) was higher than that of YPIGH-H, YPIGH-H2, and YPIGH-D, indicating that the MF-α increased secretion of EGII. No CMCase activity was detected for the YPIGH control strain lacking the EGII contruct, confirming that the observed enzyme activity resulted from the expression of the EGII gene. The CMCase activity of transformant YES2-EGII was lower than transformant YPIGH-B3, indicating that expression of the integrated vector was higher than that of the free vector.
Yeast recombinant enzymatic activity of EGII. The crude enzymes of recombinant yeast with the egII gene were used for measuring endoglucanase activity. The transformants YPIGH and YES2-EGII were used as control. The transformants were induced by β-D-galactose at 30 °C for 24 to 72 h, at 12-h intervals. The enzyme activity at different induced times was measured. EGII activity of yeast YPIGH (dot), EGII activity of yeast YPIGH-H1 (square), EGII activity of yeast YPIGH-H2 (regular triangle), EGII activity of yeast YPIGH-B (del triangle), EGII activity in yeast YPIGH-D (diamond), EGII activity in yeast YES2-EGII (circle)
Properties of Recombinant EGII
We next determined the properties of TaEGII expressed in transgenic yeast YPIGH-B3 by assaying culture supernatants for CMCase activity. The CMCase activity increased slowly from 50 to 60 °C and peaked at 60 °C (110.75 U g−1), then decreased when the temperature was higher than 60 °C (Supplementary Fig. S2a), suggesting that the 60 °C was the optimal reaction temperature for CMCase in the yeast transformant. The enzyme activity was highest at pH 5.0 with the activity value of 119.87 U g−1, and enzyme activity decreased rapidly at pH values greater than 6.0 or less than 3.0 and was low (55.19 U g−1) at pH 7.0 (Supplementary Fig. S2b). The enzyme activity was stable between 40 and 60 °C (88.5–94.41 U g−1) and decreased rapidly above 70 °C (Supplementary Fig. S2c). The activity was stable at pH 4.0–7.0 (97.24–106.1 U g−1) and decreased rapidly at pH greater than 7.0 (Supplementary Fig. S2d). Additionally, the examination of metal ion effect on the enzyme activity was carried out by measuring the activity in the presence of 0.75 mM of each metal ion. The CMCase activity of TaEGII was significantly different for the different metal ions (F = 119.43, df = 8, P < 0.01). The enzyme activity was stimulated by Mn2+, Fe2+, and Mg2+ (168.89 U g−1 when activated by Fe2+) but was inhibited by Zn2+, K+, Na+, and Co2+ (Supplementary Fig. S3).
The kinetic constants for CMC-Na, Avicel, cellulose, cellobiose, and raffinose hydrolysis were determined as shown in Table 2. The Km values were 2.46 × 10−2 and mg 1.06×10−2 mg ml−1 for cellobiose and raffinose, respectively. The enzyme showed the highest affinity for raffinose and the lowest affinity for cellobiose. The values of Kcat for Avicel and raffinose were lower than that for CMC-Na, cellulose, and cellobiose. The values of Kcat/Km for raffinose and cellobiose were 8.64 and 3.87 s−1 ml mg−1, respectively. The enzyme for raffinose had the highest catalytic efficiency.
Confirmation of Expression and Active Protein
To confirm expression of the correct protein, cellular supernatants were separated by SDS-PAGE, showing β-D-galactose-induced expression of 44.3 kDa proteins after 2–6 days (Supplementary Fig. S4). The protein could be purified using the His tag as shown in Supplementary Fig. S5. The activities of the purified enzymes were confirmed using a transparent circle assay (Supplementary Fig. S6).
Discussion
Studies of the fungi Trichoderma spp. have attracted increased attention because these microbes can efficiently produce cellulases that can depolymerize cellulose to glucose. T. atroviride produces many cell lysis enzymes, such as glycoside hydrolases, chitinase, and proteases, and is commonly used in biological control [17]. However, individual genes have not been studied as sources of potential cellulose enzymes, and the sequences of these enzymes have not been published. The cDNA sequence of T. atroviride AS3.3013 egII isolated here represents a novel gene encoding a 44.23 kDa endoglucanase. BlastX analysis showed a relatively high degree of similarity to endoglucanase (EC.3.2.1.4) belonging to the GH family 5. The highest sequence identity (99%) was to the EGII from the fungus Trichoderma orientale strain EU7-22, but there was low sequence similarity to Cel5A from Bacterium, perhaps due to phylogenetic evolution.
The expression patterns of T. atroviride egII revealed that the expression was induced by different substrates, and optimal expression was seen for microcrystalline cellulose (Fig. 2). The expression of egII was completely inhibited by glucose. This result was similar to the observed expression of the egIV gene from Trichoderma reesei [18], but the putative membrane-bound endoglucanase cel5b from H. jecorina was moderately expressed during growth on glucose, glycerol, sophorose, and lactose and only slightly induced over this level by cellulose [19]. However, nanocellulose prepared by controlled microbial hydrolysis (NCm) was an ideal inducer of cellulase [20]. Different cellulases can be regulated by different substrates.
The signal peptide helps the protein cross the cell membrane and strongly determines the efficiency of protein secretion. Huang et al. [10] constructed S. cerevisiae expression plasmids IpYEMα-xegVIII and IpYES2-egVIII, that included the native S. cerevisiae secretion signal peptide (MF-α), and found 0.86-fold enhancement of expression. In our study, five plasmids were constructed to increase the secretion and expression of recombinant protein. The native signal peptide (21 amino acids) of EGII from T. atroviride was recognized by yeast, but the S. cerevisiae MF-α enhanced EGII secretion and activity 1.29-fold. This enhancement may facilitate protein purification.
Although the enzymatic characterization of EGII (Cel5A) from T. reesei and Trichoderma viride was studied, this is the first report of the functional expression of T. atroviride EGII in S. cerevisiae and the characterization of the recombinant enzyme. The yeast-expressed EGII was stable at temperatures up to 70 °C with an optimal temperature of 60 °C (Supplementary Fig. S2c) and over a wide range of pH (4.0–7.0) (Supplementary Fig. S2d), indicating the potential application of the enzyme for cellulose hydrolysis at moderate temperatures and pH levels. The optimal temperature of 60 °C (Supplementary Fig. S2a) was higher than that reported previously for EG5A from Phanerochaete chrysosporium (55 °C) [21] and lower than that of EGI from Bacillus subtilis (65 °C) [22]. The inhibition of the activity of the recombinant endoglucanase from P. chrysosporium by Fe2+, Mg2+, Ca2+, and Fe3+ was reported Huy et al. [21], but the activity of recombinant enzyme from Trichoderma viride was stimulated by Fe2+, Na+, and Mg2+ [4]. Here, we found that the enzyme activity of recombinant TaEGII was highly activated by Fe2+, Mn2+, and Mg2+ and inhibited by Zn2+, K+, Na+, and Co2+ (Supplementary Fig. S3). Thus, endoglucanases can be activated or inhibited by different metal ions, and endoglucanases from different microbes may be not similarly regulated by metal ions. The recombinant TaEGII for CMC-Na has lower affinity (1.44 × 10−2 mg ml−1) (Table 2) than the endoglucanases from T. reesei [23], T. viride [10], and Actinomyces sp. [24] but higher affinity than the endoglucanase from Rhizopus stolonifer [25]. The affinity for MCC and the catalytic efficiency for cellobiose of recombinant TaEGII were lower than those of EGVII from T. viride [10].
In summary, the novel egII gene (GenBank Acc. No. KP098496) from T. atroviride AS3.3013 has successfully been cloned, and recombinant yeast strain YPIGH-B3 with high expression using the S. cerevisiae MF-α secretion signal sequence was obtained. The recombinant TaEGII from yeast strain YPIGH-B3 exhibited the highest activity (168.89 U g−1) at 60 °C, pH 5.0, and 0.75 mM Fe2+ and showed a high specificity and hydrolysis capacity towards raffinose and Avicel. Coupled with its broad range of pH, all these features make the enzyme very useful for the biomass degradation and bioethanol production.
References
Zhou, J., Wang, Y. H., Chu, J., Zhuang, Y. P., Zhang, S. L., & Yin, P. (2008). Identification and purification of the main components of cellulases from a mutant strain of Trichoderma viride T100-14. Bioresource Technology, 99, 6826–6833.
Bayram Akcapinar, G., Gul, O., & Sezerman, U. O. (2012). From in silico to in vitro: modelling and production of Trichoderma reesei endoglucanaseIand its mutant in Pichia pastoris. Journal of Biotechnology, 159, 61–68.
Kawai, T., Nakazawa, H., Ida, N., Okada, H., Ogasawara, W., Morikawa, Y., & Kobayashi, Y. (2013). A comprehensive analysis of the effects of the main component enzymes of cellulase derived from Trichoderma reesei on biomass saccharification. Journal of Industrial Microbiology & Biotechnology, 8, 805–810.
Huang, X. M., Fan, J. X., Yang, Q., Chen, X. L., Liu, Z. H., Wang, Y., & Wang, D. (2012). Cloning, expression, and characterization of endoglucanase gene egIV from Trichoderma viride AS 3.3711. Journal Microbiology Biotechnology, 22, 390–399.
Kuhad, R. C., Gupta, R., & Singh, A. (2011). Microbial cellulases and their industrial applications. Enzyme Research, 2011, 1–10.
Karmakar, M., & Ray, R. R. (2011). Current trends in research and application of microbial cellulases. Rearch Journal of Microbiology, 6, 41–53.
Narasimha, G., Sridevi, A., Viswanath, B., Subhosh, C. M., & Reddy, R. (2006). Nutrient effects on production of cellulolytic enzymes by Aspergillus niger. African Journal of Biotechnology, 5, 472–476.
Li, X.-H., Wang, D., Zhou, F., Yang, H.-J., Bhaskar, R., Hu, J.-B., Sun, C.-G., & Miao, Y.-G. (2010). Cloning and expression of a cellulase gene in the silkworm, Bombyx mori by improved Bac-to-Bac/BmNPV baculovirus expression system. Molecular Biology Reports, 37, 3721–3728.
Penttilä, M. E., André, L., Lehtovaara, P., Bailey, M., Teeri, T. T., & Knowles, J. K. (1988). Efficient secretion of two fungal cellobiohydrolases by Saccharomyces cerevisiae. Gene, 63, 103–112.
Huang, X. M., Yang, Q., Liu, Z. H., Fan, J. X., Chen, X. L., Song, J. Z., & Wang, Y. (2010). Cloning and heterologous expression of a novel endoglucanase gene egVIII from Trichoderma viride in Saccharomyces cerevisiae. Applied Biochemitry Biotechnology, 162, 103–115.
Van Wyk, J. P., & Mohulatsi, M. (2003). Biodegradation of wastepaper by cellulase from Trichoderma viride. Bioresource Technology, 86, 21–23.
Wu, S., & Letchworth, G. J. (2004). High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. BioTechniques, 36, 152–154.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Li, F., Zhu, X.-W., Li, N.-Y., Zhang, S., Zhao, X., Li, P. G., Zhu, Q. C., & Lin, H. P. (2014). Screening of lignocellulose-degrading superior mushroom strains and determination of their CMCase and laccase activity. The Scientific World Journal, 2014, 1–6.
Meinke, A., Damude, H. G., Tomme, P., Kwan, E., Kilburn, D. G., Miller Jr., R. C., Warren, R. A., & Gilkes, N. R. (1995). Enhancement of the endo-β-1,4-glucanase activity of an exocellobiohydrolase by deletion of a surface loop. Journal of Biological Chemistry, 270, 4383–4386.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Lima, F. B. D., Félix, C., Osório, N., Alves, A., Rui, V., & Domingues, P. (2016). Secretome analysis of Trichoderma atroviride T17 biocontrol of Guignardia citricarpa. Biological Control, 99, 38–46.
Saloheimo, M., Nakari-Setälä, T., Tenkanen, M., & Penttilä, M. (1997). cDNA cloning of a Trichoderma reesei cellulase and demonstration of endoglucanase activity by expression in yeast. European Journal of Biochemistry, 249, 584–591.
Kubicek, C. P., Mikus, M., Schuster, A., Schmoll, M., & Seiboth, B. (2009). Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnology for Biofuels, 2, 323–330.
Satyamurthy, P., Jain, P., Karande, V. S., & Nadanathangam, V. (2016). Nanocellulose induces cellulase production in Trichoderma reesei. Process Biochemistry, 51, 1452–1457.
Huy, N. D., Nguyen, C. L., Park, H. S., Loc, N. H., Choi, M. S., & Kim, D. H. (2015). Characterization of a novel manganese dependent endoglucanase belongs in GH family 5 from Phanerochaete chrysosporium. Journal of Bioscience & Bioengineering, 121, 154–159.
Zi-Zhong, T., Zhen-Fang, W., Hui, C., Xin, L., Xue-Yi, H., & Qi, W. (2013). Characterization of novel EGs reconstructed from Bacillus subtilis endoglucanase. Applied Biochemistry and Biotechnology, 169, 1764–1773.
Qin, Y., Wei, X., Song, X., & Qu, Y. (2008). Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. Journal of Biotechnology, 135, 190–195.
Kim, S. C., Kang, S. H., Choi, E. Y., Hong, Y. H., Bok, J. D., & Kim, J. Y. (2016). Cloning and characterization of an endoglucanase gene from Actinomyces sp Korean native goat 40. FEMS Microbiology Letters, 158, 25–31.
Tang, B., Wei, L., Tang, W., Li, S., & Zhou, R. (2015). Effect of linker flexibility on the catalytic features of a glycoside hydrolase family 45 endoglucanase from Rhizopus stolonifer. Applied Biochemistry and Biotechnology, 176, 2242–2252.
Acknowledgements
This research was funded by the Natural Science Foundation of Heilongjiang Province (C2015002), the National Natural Science Foundation of China (31372351 and 31272484), Northeast Agriculture University Innovation Foundation for Postgraduate (yjscx14082), and Doctoral Starting Funds Program of Northeast Agricultural University (2012RCB95).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Supplementary Fig. S1
Structure of plasmid pYPIGH. The TYI and TY12 boxes are the fragment of ribosomal DNA of yeast. The DCS and hyg boxes indicate multiple cloning site which is derived from the plasmid pYES2 and hygromycin gene that roots in pasmid pCSN43, respectively. To target the integration of plasmid pYPIGH to the ribosomal DNA, the plasmid was linearized with BglII before transformation to yeast. (DOCX 19 kb)
Supplementary Fig. S2
Activity and properties of recombinant endoglucanase EGII. a. Effects of temperature on endoglucanase EGII activity. The enzyme activity was measured at 30 °C to 80 °C for 1 h at 10 °C intervals (pH 5.0). b. Effects of pH on endoglucanase EGII activity. The enzyme activity was measured in the reaction buffer at different pH values between 2.0 and 8.0 at intervals of 1.0 pH units. c. Effects of temperature on endoglucanase EGII stability. The dialyzed fraction (pH 5.0) was incubated at 30 °C to 90 °C for 1 h at 10 °C intervals, and then the residual activity was measured by incubation at 60 °C for 1 h. d. Effects of pH on endoglucanase EGII stability. The dialyzed fraction was incubated in different pH buffers (pH 1.0–8.0) for 48 h at 4 °C, and then the residual activity was measured under standard conditions (pH = 5.0). All the experiments were performed three times. (DOCX 59 kb)
Supplementary Fig. S3
Effects of exposure to different metal ions on the activity of endoglucanase EGII. Several different reaction buffers were prepared, each spiked with 0.75 mM of a metal ion. One unit of CMCase activity was defined as the amount of enzyme required for releasing total reduced sugar equivalent to 1 mmol glucose per minute. Con: control, the activity of EGII was measured under the normal reaction condition without any additional ions. The experiments were performed three times. Different letters above the columns indicate a significant difference determined by Duncan’s multiple comparisons test (P < 0.01) (DOCX 137 kb)
Supplementary Fig. S4
SDS-PAGE analysis of crude recombinant EGII. Lane M: Premixed Protein Marker (Low); Lane 1: blank control; Lane2–6: yeast transformant YPIGH-B3 induced 2–6 days crude enzyme. (DOCX 94 kb)
Supplementary Fig. S5
SDS-PAGE analysis of purified recombinant EGII. Lane M: Premixed Protein Marker (Low); lane 1:10 μl purified recombinant protein; lane 2:15 μl purified recombinant protein; lane 3:20 μl purified recombinant protein. (DOCX 101 kb)
Supplementary Fig. S6
Transparent Circle analysis of recombinant EG II. a: the back of the plate; b:the front of the plate. NO.1: 5 μl of purified enzyme; NO.2: 10 μl of purified enzyme; NO.3: 15 μl of purified enzyme; NO.4: 20 μl of purified enzyme. (DOCX 84 kb)
Rights and permissions
About this article
Cite this article
Huang, Xm., Li, Qq., Chen, Xl. et al. Expression and Characteristics of an Endoglucanase from Trichoderma atroviride (TaEGII) in Saccharomyces cerevisiae . Appl Biochem Biotechnol 182, 1158–1170 (2017). https://doi.org/10.1007/s12010-016-2389-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2389-9