Abstract
Toll-like receptors (TLRs) are a class of innate immune receptors that sense pathogens or their molecular signatures and activate signaling cascades to induce a quick and non-specific immune response in the host. Among various types of TLRs, TLR22 is exclusively present in teleosts and amphibians and is expected to play the distinctive role in innate immunity. This report describes molecular cloning, three-dimensional (3D) modeling, and expression analysis of TLR22 in rohu (Labeo rohita), the most commercially important freshwater fish species in the Indian subcontinent. The open reading frame (ORF) of rohu TLR22 (LrTLR22) comprised of 2,838 nucleotides (nt), encoding 946 amino acid (aa) residues with the molecular mass of ∼107.6 kDa. The secondary structure of deduced LrTLR22 exhibited the presence of signal peptide (1–22 aa), 18 leucine-rich repeat (LRR) regions (79–736 aa), and TIR domain (792–935 aa). The 3D model of LrTLR22-LRR regions together elucidated the horse-shoe-shaped structure having parallel β-strands at the concave surface and few α-helices at the convex surface. The TIR domain structure revealed alternate presence of five α-helices and β-sheets. Phylogenetically, LrTLR22 was closely related to common carp and exhibited significant similarity (92.2 %) and identity (86.1 %) in their amino acids. In rohu, TLR22 was constitutively expressed in all embryonic developmental stages, and tissue-specific analysis illustrated its expression in all examined tissues, highest was in liver and lowest in brain. In vivo modulation of TLR22 gene expression was analyzed by quantitative real-time PCR (qRT-PCR) assay following stimulation with lipopolysaccharide (LPS), synthetic double stranded RNA (polyinosinic-polycytidylic acid), and bacterial (Aeromonas hydrophila) RNA. Among these ligands, bacterial RNA most significantly (p < 0.05) induced TLR22 gene expression in most of the tested tissues. In A. hydrophila infection, induction of TLR22 gene expression was also observed in majority of the tested tissues. Together, these data suggested that in addition to sensing other microbial signatures, TLR22 can recognize bacterial RNA and may play the important role in augmenting innate immunity in fish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Innate immune system is the first line of defense of the host that plays essential role in defending pathogenic invasion. It is the only defense mechanism in invertebrates and the major component of defense in vertebrates. It is regulated by various pattern recognition receptors (PRRs) that are distributed in cell surface, intracellular compartments, or secreted into the blood stream and tissue fluids. Toll-like receptors (TLRs) are one class of PRRs that play an important role in innate immunity in clearing pathogens by sensing microorganisms or their specific and conserved signatures called pathogen-associated molecular patterns (PAMPs). Well-characterized PAMPs include various bacterial components such as peptidoglycan (PGN), lipoteichoic acid (LTA), flagellin, lipopolysaccharides (LPS), lipoproteins, heat shock protein (hsp), CpG-DNA, nucleic acids, yeast derivative like zymosan and double stranded RNA (dsRNA) of viruses etc. Structurally, TLRs are type 1 integral membrane glycoproteins, having leucine-rich repeat (LRR) domain in their extracellular regions for binding PAMPs and a Toll/interleukin-1 receptor domain (TIR) that transmits downstream signals into the cytosol by recruiting and activating a cascade of adaptor molecules [1, 2]. TLR-induced signals are largely divided into MyD88-dependent and MyD88-independent signaling pathways. MyD88-dependent signaling pathways that are involved in mediating TLR-induced responses include nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) pathways and are responsible for the expression of pro-inflammatory cytokines. MyD88-independent or TRIF (TIR-domain-containing adapter-inducing interferon-β)-dependent signaling pathway is involved in the induction of type I IFNs.
Depending upon their primary amino acid sequence similarity and specificity of ligand (PAMPs) recognition, TLRs are classified into various types. Among them, TLR22 is unique because of its exclusive presence in aquatic animals (teleosts and amphibians). It was reported in goldfish (Carassius auratus) [3], Atlantic salmon (Salmo salar) [4], fugu (Takifugu rubripes) [5], Japanese flounder (Paralichthys olivaceus) [6], rainbow trout (Oncorhynchus mykiss) [7], zebrafish (Danio rerio) [8], large yellow croaker (Pseudosciaena crocea) [9], and in grass carp (Ctenopharyngodon idella) [10]. The fugu TLR22 (fgTLR22) recognized long-sized (∼1,000 bp) dsRNA on the cell surface, whereas TLR3 (fgTLR3) resided in endoplasmic reticulum and recognized relatively short-sized (∼200 bp) dsRNA [11]. However, both of them linked the IFN-inducing pathway via the Toll-IL-1R homology domain-containing adaptor protein 1 (TICAM-1/TRIF). In large yellow croaker, TLR22 was highly expressed in anterior kidney, and its expression was upregulated by poly I:C in the anterior kidney and spleen [9]. In Japanese flounder, it was mainly expressed in peripheral blood leucocytes (PBL)-rich organs such as kidney, spleen and gill and was induced by both peptidoglycan and poly I:C [6]. In rainbow trout, basal expressions of TLR22 transcripts were highest in spleen and lowest in liver. In response to inactivated Aeromonas salmonicida, the expression of TLR22 was induced in PBLs, spleen, and head kidney [7]. In grass carp, TLR22 expression was highest in gill and lowest in spleen and liver. Several single nucleotide polymorphisms (SNPs) were also reported in grass carp TLR22 [10].
In the world, India ranks third in aquaculture [12]. The freshwater fish production in India is mostly contributed by the Indian major carps (IMC), and among them, rohu (Labeo rohita) is the most widely cultured and commercially important fish species. Occurrences of various diseases are one of the limiting factors in aquaculture. To protect fish against diseases, innate immunity contributed by various PRRs is expected to be very important. In this context, TLR22 being unique PRR in aquatic animals draws more attention than its counterparts. Keeping these in view, this work was undertaken to clone and characterize TLR22, to investigate its structural insights, and to find out its role in recognizing various PAMPs.
In this article, we reported the coding sequence of TLR22 in rohu, predicted its three-dimensional structure, and investigated the temporal expression following various ligands exposure and bacterial infection. For the first time, we report that bacterial RNA is recognized by TLR22.
Materials and Methods
Fish
Rohu (L. rohita) fingerlings were obtained from the Central Institute of Freshwater Aquaculture (CIFA), Bhubaneswar, India, with an average weight ∼50 g and were stocked at 50 fingerlings in a 500-l fiberglass reinforced plastic (FRP) tank with aeration facility. Before the start of the experiment, fish were acclimatized for 8 weeks with daily two-third water exchanges and feeding once with commercial carp diet. During the experiment, pH of the tank water varied from 7.4 to 7.6 and the temperature ranged from 25 to 28 °C.
Bacteria
Aeromonas hydrophila (ATCC-35654) was cultured in LB broth (USB, USA) at 37 °C for 16 h with constant shaking. Viable count was determined as colony-forming unit (CFU) following 10-fold serial dilutions and plating on nutrient agar.
Cloning of Rohu TLR22
To clone TLR22 complementary (c)DNA, total RNA was extracted from rohu gill, cDNA was prepared following the protocol as described later, and PCR was carried out with three sets of degenerate primers (Table 1(A)) derived from the conserved TLR22 nucleotide sequences of D. rerio (GenBank ID NM_001128675), C. auratus (GenBank ID AY162178), and T. rubripes (GenBank ID NM_001113193). PCR was performed using 1 μl cDNA in a 50-μl reaction volume under the conditions of 1 cycle of initial denaturation at 94 °C for 2 min followed by 45 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min, and a final extension at 72 °C for 5 min. Ten microliters of the PCR product was analyzed in 2 % agarose gel, and a single specific band that matched with the expected size was purified with agarose gel DNA extraction kit (Roche, Germany). The purified DNA was cloned in pGEM-T Easy vector (Promega, Madison, USA), and both strands DNA sequencing was carried out with T7 and SP6 primer (ABI prism 3000). Obtained sequences were analyzed by BLAST search to confirm it as TLR22 of L. rohita (LrTLR22). From this partial LrTLR22 sequence, two sets of primers (TLR22-(iv) Fw/TLR22-(iv) Rv and TLR22-(v) Fw, TLR22-(v) Rv; Table 1(A)) were designed, PCR was carried out and the amplified PCR products were cloned, and DNA sequencing was carried out. After removing the overlapping bases, the sequences were joined together. To obtain full-length LrTLR22 cDNA sequence, 3′- and 5′ rapid amplification of cDNA ends (RACE ) were conducted with SMARTer RACE cDNA amplification kit (Clontech, USA) following manufacturer’s protocol. For amplification of LrTLR22 cDNA 3′ and 5′ end regions, gene-specific forward (T22-GS-Fw-3′) and reverse (T22-GS-Rv-5′) primers were designed (Table 1(A)) from the partial LrTLR22 cDNA sequence. Touchdown PCR was carried out using TLR22-GS-FW-3′/universal primer mix (UPM) and T22-GS-NP-Rv-5′/UPM primer sets (Table 1(A)) respectively under these conditions: initial denaturation for 1 cycle at 94 °C for 2 min, followed by 5 cycles of 94 °C/30 s and 72 °C/3 min; next, 5 cycles at 94 °C/30 s, 62 °C/30 s, and 72 °C/3 min; next, 26 cycles at 94 °C/30 s, 55 °C/30 s, and 72 °C/3 min; and 1 cycle at 72 °C/10 min. Nested PCR for 3′ and 5′ RACE was performed using gene-specific nested primer (GS-NP)-FW-3′/nested universal primer (NUP) for 3′ and T22-GS-NP-Rv-5′/NUP for 5′ nested primer sets (Table 1(A)). Using 1 μl of primary RACE-PCR product as template, nested PCR was carried out with the following conditions: initial 1 cycle of 94 °C/5 min, then 35 cycles of 94 °C/30 s, 56 °C/30 s, and 72 °C/90 s followed by 1 cycle of 72 °C/10 min. The PCR products were cloned in pGEM-T Easy vector, sequenced, and validated through BLAST search, and overlapping sequences from the fragments were combined together to obtain the complete LrTLR22.
Analysis of LrTLR22 cDNA Sequence
The deduced LrTLR22 protein was analyzed by SMART (http://smart.embl-heidelberg.de) to identify various structural features, viz., signal peptide, leucine-rich repeat (LRR) regions, transmembrane (TM), and toll-interleukin 1 receptor (TIR) domains. The similarity and identity of TLR22 amino acids between rohu and other fish species were analyzed in MatGAT program [13]. The phylogenetic tree of TLR22 among various fish species was constructed by the neighbor-joining method of MEGA4 program [14], and the branches were validated by bootstrap analysis from 1,000 replications using default parameter.
3D Modeling
In LrTLR22, the ecto-domain (ECD) comprising LRR motifs (79–736 aa) and the TIR domain (792–935) were scanned against the PDB database to identify suitable templates for 3D-model building. The respective suitable templates with the highest sequence identities with the target sequences were set to generate 3D coordinates of LrTLR22-ECD and LrTLR22-TIR domain in Modeller9v12 [15]. Among different sets of models, the model with the lowest discrete optimized protein energy (DOPE) score was chosen for further refinement and studies. The loop regions of the lowest DOPE score models of LrTLR22-ECD and LrTLR22-TIR were refined at ModLoop server (http://modbase.compbio.ucsf.edu/modloop/) and were submitted to 3D refine server (http://sysbio.rnet.missouri.edu/3Drefine/) to minimize the structural ambiguities. The refined models were thus validated at SAVes server (http://nihserver.mbi.ucla.edu/SAVES/).
Molecular Dynamics Simulation of Homology Models
Molecular dynamics (MD) simulations were carried out using the GROMACS program (version 4.5.5) [16] with GROMOS43a1 force field in a computer with Intel Core i3 processor running at 3.07 GHz well equipped with CentOS 5.6 (http://www.centos.org/). The modeled structures (LrTLR22-ECD and LrTLR22-TIR) were immersed in a cubic water box (1.0 nm thickness), and the net charges were neutralized by the addition of Cl− and Na+ counterions. Long-range electrostatics were handled using the particle mesh Ewald method. The steepest descent energy minimization was used to remove possible bad contacts until energy convergence reached 1,000 kJ mol−1 nm−1). The systems were subjected to equilibration at 300 K and normal pressure constant (1 bar) for 200 ps under NPT condition. A production run of 5 and 4 ns was performed for LrTLR22-ECD and TIR domains, respectively. The time step of the simulation was set to 2 fs, and the coordinates were saved for analysis at every 1 ps. GROMACS and VMD 1.9.1 (www.ks.uiuc.edu/Research/-vmd/) routines were utilized to analyze the MD trajectories and the quality of the simulations. The graphs of trajectory analysis were created using Xmgr 4.1.2 (http://plasma-gate.weizmann.ac.il/Xmgr/). All visualizations were illustrated using PyMOL (http://www.pymol.org/) and VMD.
Model Validation
The final snapshot obtained at the end of MD simulation was considered to represent the structures of the LrTLR22-ECD and LrTLR22-TIR domains. The final 3D structures were validated at SAVes, WHAT IF [17], and MolProbity [18] servers.
Ontogenic Expression
To investigate ontogenic expression of TLR22 gene, fertilized eggs of rohu were collected from CIFA hatchery and maintained at ambient temperature (28–29 °C). Confirmation of ongoing developmental stages were carried out at different time intervals under microscopic observation, and samples were collected separately at 0 h (fertilized egg), 2, 4, 7, 9, 12, 16, 18, 20, 24, and 48 h in TRIzol reagent (Invitrogen, USA). RNA was isolated from each sample, quantified in UV-spectrophotometry, and using 1 μg of total RNA, cDNA was prepared with oligo-dT primer. Semi-quantitative reverse transcriptase (RT)-PCR was employed to analyze TLR22 gene expression among the samples keeping β-actin as housekeeping gene. The condition of semi-quantitative PCR was as follows: One cycle of initial denaturation at 94 °C for 2 min followed by 30 cycles (β-actin)/42 cycles (TLR22) of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 5 min. Ten microliters of the PCR product was analyzed in 2 % agarose gel.
Basal Expression
To study basal expression of TLR22 gene in various organs/tissues, brain, intestine, skin, blood, muscle, eye, heart, gill, liver, kidney, and spleen were collected separately from three healthy rohu (n = 3) fingerlings (weight ∼50 g each) in TRIzol reagent. From each sample, total RNA was isolated, and cDNA was prepared with oligo-dT primer following the protocol as described later. TLR22 and β-actin gene expressions in each sample were analyzed by quantitative real-time PCR (qRT-PCR) assay, and the mean value was represented graphically.
Purification of Bacterial RNA
In a 200-ml conical flask, A. hydrophila was cultured in 50 ml LB broth (USB, USA) at 37 °C with constant shaking. After 16 h of growth, the bacterial culture was centrifuged at 1,600×g for 5 min at 4 °C. The pellet was washed thrice with chilled phosphate buffered saline (PBS), and total cellular RNA was extracted with TRIzol reagent (Invitrogen, USA) following manufacturer’s protocol. Extracted RNA was treated with 5 U of DNase I (MBI, Fermentas) at 37 °C. After 30 min, 2 μl of 0.5 M EDTA was added to stop the reaction, and RNA was purified following the protocol of phenol-chloroform precipitation. Precipitated RNA was air dried and was dissolved in 30 μl of diethylpyrocarbonate (DEPC)-treated water. To check the purity of RNA, 10 μl of RNA sample was treated with 2 μl of RNase A at room temperature for 1 h and was loaded in 1 % agarose gel.
In Vivo Bacterial Challenge
Healthy rohu fingerlings (∼50 g) were divided into control and treated groups keeping three fish in each group. For bacterial infection, fish were intra-peritoneally (i.p.) injected with 100 μl of PBS containing A. hydrophila (1 × 106 CFU/fish) and were designated as treated fish group. The control group was injected with 100 μl of PBS only and was kept separately in the aerated tank. After 24 and 48 h of bacterial infection, control and treated groups of fish were sacrificed and tissues were collected separately in TRIzol reagent and were kept at −80 °C till further analysis.
Ligands Stimulation
Rohu fingerlings with an average weight of ∼50 g were divided into two groups keeping three fish in each group. To stimulate fish with LPS, purified LPS of Escherichia coli (serotype O55: B5) (SIGMA, Germany) was reconstituted in endotoxin-free water at 5 mg ml−1, and PBS (100 μl) containing 20 μg of diluted LPS was injected to the fingerlings (treated group) through intra-venous (i.v.) route. The control fish group was i.v. injected with 100 μl of PBS. For dsRNA stimulation, poly I:C (Sigma, USA) was diluted in DEPC-treated water at 10 mg ml−1, and 100 μl of DEPC-treated water containing 300 μg of diluted poly I:C was injected through i.v. route into each of the three fish (treated fish group). To stimulate with bacterial RNA, 10 μg purified RNA of A. hydrophila diluted in 100 μl of DEPC-treated water was i.v. injected. Control fish group received only 100 μl of DEPC-treated water. After 4 h of ligands exposure, control and treated fish groups were sacrificed and organs/tissues were separately collected in TRIzol reagent for RNA extraction.
RNA Isolation from Tissue and cDNA Synthesis
Total RNA from tissue samples were extracted with TRIzol reagent (Invitrogen, USA) following manufacturer’s protocol. The concentration of RNA was measured by UV-spectrophotometer (Eppendorf, Germany), and the quality was assessed by visualizing the band intensity of 28 and 18S ribosomal RNA in 1 % agarose gel. For cDNA synthesis, 1 μg of total RNA was treated with 1 U of DNase I (MBI, Fermentas, USA) and reverse transcription was carried out using oligo-dT primer and RevertAid 1st strand cDNA synthesis kit (MBI, Fermentas, USA).
Quantitative Real-Time PCR Analysis
The target (TLR22) and reference gene (β-actin) expressions were analyzed by quantitative real-time RT-PCR (qRT-PCR) assay in LightCycler®480 II-real-time PCR detection system (Roche, Germany). The qRT-PCR reactions were carried out in triplicate wells of a 96-well plate, and each of the well contained 10 μl reaction mixture with the following compositions: cDNA 1 μl, FW and RV primers (2.5 μM each; Table 1(B)) 0.25 μl each, 2X LightCycler®480 SYBR Green I master mix (Roche, Germany) 5 μl and PCR grade H2O 3.5 μl. The conditions of PCR were as follows: 1 cycle of initial denaturation at 95 °C for 10 min followed by 45 cycles of 94 °C/10 s, 58 °C (for TLR22 and β-actin) for 10 s, and 72 °C/10 s. For negative control, PCR was carried out without template (cDNA). The qRT-PCR products were checked in 2 % agarose gel to confirm the correct size and single band amplification. The ratio of relative gene expression was derived by normalizing the expression of target gene, as determined by mean crossing point (cp) deviation by that of a housekeeping reference gene, β-actin following 2-ΔΔCT method [19]. The data obtained from qRT-PCR analysis was represented as mean of three individual experiments ± standard error (s.e.). The significant difference of gene expression between the control and treated fish groups was determined by the Student’s t test using Microsoft Excel 2010 with p < 0.05 as significance level.
Results
Cloning and Characterization of LrTLR22 cDNA
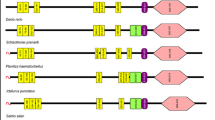
The cDNA sequence that encoded open reading frame (ORF) of LrTLR22 was derived from the gill and was submitted to the GenBank (KJ187303). The LrTLR22-ORF consisted of 2,838 nucleotides encoding a polypeptide of 946 amino acid (aa) residues with an estimated molecular mass of ∼107.6 kDa. Prediction of structural organization by SMART revealed signal peptide (1–22 aa), leucine-rich repeat (LRR) regions (79–736 aa), and TIR domains (792–935 aa). The comparison of domain organization among various fish species TLR22 were shown in Fig. 1.
Schematic representation of domains in various fish species TLR22. The secondary structures of TLR22 were predicted by SMART program. Signal peptide (small solid box), leucine-rich repeat (LRR) domains (rectangular box), C-terminal LRR domain (oval shaded box), transmembrane domain (elongated box), and the TIR domain (rectangular shaded box) were shown with their respective positions
3D Structure Analysis
Template search at BLAST revealed that the ligand-recognizing LRR regions in LrTLR22-ECD shared the close structural relationship with human toll-like receptor 3 (TLR3) extracellular domain and crystal structure of mouse TLR3 ecto-domain (PDB ID: 1ZIW and 3CIG). The TLR22-TIR domain shared close structural similarity with crystal structure of the TIR domain of human TLR1 (PDB ID: 1FYV). The LrTLR22-ECD formed a horse-shoe-shaped structure, where most of the LRR domains were comprised of parallel β-strands and connected by long loop and with few α-helices (Fig. 2a). The β-strands faced towards the concave surface, and the α-helices were present at the convex surface. The LrTLR22-TIR domain comprised of five α-helices and five β-sheets and was depicted in alternate fashion (Fig. 2b).
The Ramachandran plot analysis at SAVes for the homology models showed that the phi-psi angles of most of the residues were in the allowed regions (Fig. S1). In LrTLR22-ECD, the plot showed 72.3, 24.6, 2.5, and 0.7 % of residues remained in core, allowed, generously allowed, and disallowed regions, respectively. Analysis of LrTLR22-TIR showed 81.4, 14.7, 1.6, and 2.3 % of residues were in core, allowed, generously allowed, and disallowed regions, respectively. The Verify3D score of LrTLR22-ECD and LrTLR22-TIR domains showed 90.14 and 100 % residues had an average 3D-1D score >0.2, respectively. The overall G-factor of LrTLR22-ECD and LrTLR22-TIR domains were calculated to be −0.48 and −0.23, respectively (> −0.5 represented good models). The average coarse packing quality, planarity, and the collision with symmetry axis, bond lengths, and bond angles obtained by the WHAT IF server of both models were within the cutoff score. The results of MolProbity server of both models were also within the range and supported the feasibility of our proposed models.
Analysis of the MD trajectories of LrTLR22-ECD showed the RMSD trajectory rose for the initial 2 ns with an average RMSD of 2.921 Å and attained a stable plateau at the last 3 ns with an average RMSD of 4.591 Å (Fig. S2a). The MD trajectory of LrTLR22-TIR analysis showed the RMSD graph fluctuated slightly for the initial 2 ns and thereafter stabilized with an average RMSD of 2.124 Å (Fig. S2b). The RMSF of Cα atoms of each amino acid in LrTLR22-ECD and LrTLR22-TIR domains identified LRR11-13 in LrTLR22-ECD (Fig. S2c) and AB, BB, and DD loops in LrTLR22-TIR as the most flipped regions (Fig. S2d).
Phylogenetic Relationship of TLR22 Among Various Fish Species
To identify the similarity and identity, the deduced amino acids sequence of LrTLR22 was compared with other fish species in MatGAT program. As shown in Table 2, LrTLR22 exhibited the highest amino acid similarity (92.2 %) and identity (86.1 %) with common carp. With other fish species, the similarity and identity of LrTLR22 amino acids were ∼60–90 % and ∼40–80 %, respectively. To identify the conserved motifs, LrTLR22 amino acid sequence was compared with other species TLR22 by CLUSTALW and several highly conserved regions (identical amino acids in black box) and well-conserved regions (>50 % consensus) were identified in LRR (Fig. S3) and TIR domains (Fig. S4) across the species. To identify the evolutionary relationship, a molecular phylogenetic tree was constructed based on pairwise alignments of LrTLR22 amino acid sequence among various fish species. The result (Fig. 3) showed two clearly distinct clusters: one for freshwater fish species (cluster-I) and the other one for marine fish (cluster-II). Among the freshwater fish species, LrTLR22 was closely related to common carp TLR22. The close evolutionary relationship of TLR22 between rohu and common carp may possibly indicate their functional similarity.
Phylogenetic relationship of TLR22 among various fish species. TLR22 amino acid sequences of various fish species were aligned using CLUSTALW program, and the un-rooted phylogenetic tree was generated by the neighbor-joining method within the MEGA4 program. The branches were validated by bootstrap analysis from 1,000 replications, which were represented by percentages in branch nodes. Full-length TLR22 amino acid sequences used for tree construction were retrieved from the following database: common carp ADR66025.1, goldfish AAO19474.1, grass carp ADX97523.2, zebrafish NP_001122147.1, catfish AEI59679.1, salmon CAJ80696.1, rainbow trout NP_001117884.1, fugu AAW69372.1, large yellow croaker ADK77870.1, mandarin fish AFC95889.1, and rohu KJ187303
TLR22 Gene Expression During Embryogenesis
In rohu, TLR22 gene expressions in various developmental stages were analyzed by semi-quantitative RT-PCR assay. The results (Fig. 4a) showed constitutive expression of TLR22 gene in all embryonic developmental stages, including fertilized eggs (0 h), hatchlings (18 h), and larvae (24–48 h).
a Ontogenic expression of TLR22 gene. Total RNA was extracted from various developmental stages (denoted by hours) of rohu, and semi-quantitative RT-PCR was carried out to analyze the expression of TLR22 gene, keeping β-actin as an internal control. b Tissue-specific expression of TLR22. Total RNA was extracted from gill, liver, kidney, intestine, spleen, heart, blood, skin, muscle, brain, and eye. The cDNA was prepared and quantitative real-time PCR (qRT-PCR) was carried out to examine the expression of TLR22 gene among these tissues. Expression of TLR22 gene was represented as a ratio relative to β-actin (internal control) levels in the same samples. Among the tissues examined, brain expressed the lowest TLR22 and was chosen as calibrator (1). TLR22 expressions in other tissues were represented as fold changes from the calibrator. The results were expressed as mean ± standard error (showing bars in the graph) from three fish (n = 3)
Tissue-Specific Expression of TLR22
Basal expression of TLR22 gene in various organs, viz., brain, eye, gill, liver, kidney, heart, intestine, blood, muscle, and skin, was analyzed by quantitative real-time PCR (qRT-PCR) assay. As shown in Fig. 4b, TLR22 expression was observed in all examined tissues but the level of expression varied considerably amid the tissues. Among the analyzed tissues, the lowest TLR22 expression was observed in brain, and in comparison to brain (calibrator tissue; 1-fold), the highest expression was recorded in liver (∼30-fold) followed by kidney (∼25-fold), gill (∼15-fold), heart (∼10-fold), eye (∼6-fold), muscle (∼5-fold), blood (∼5-fold), skin (∼3-fold), and intestine (∼1.5-fold).
LPS-Mediated TLR22 Gene Expression
In response to LPS stimulation, TLR22 gene expression in rohu was enhanced in liver, spleen, skin, and blood. The data of qRT-PCR (Fig. 5) revealed significant (p < 0.05) induction of TLR22 in spleen (∼2.7-fold) followed by skin (∼2.5-fold) and liver (∼1.8-fold) and a moderate increase in blood. In LPS-stimulated gill, kidney, and intestine, TLR22 gene expression was downregulated as compared to control.
LPS-mediated TLR 22 gene expression. In rohu fingerlings, LPS (20 μg/fish) was i.v. injected. At 4-h post treatment, total RNA was extracted from various tissues, and cDNA was analyzed by real-time PCR. The relative expression of TLR22 gene was normalized to the expression of β-actin (internal control) and was expressed as fold changes relative to the un-treated control group. The mean value of three fish (n = 3) is shown and the bars indicate standard error. Significant difference (p < 0.05) between control and treated fish group is indicated with asterisks (*)
Modulation of TLR22 Expression by Synthetic dsRNA
The structure of poly I:C resembles viral double stranded RNA (dsRNA). Modulation of TLR22 gene transcripts by poly I:C was analyzed by qRT-PCR assay following i.v. injection of poly I:C. As shown in Fig. 6, among all tested tissues, the highest induction of TLR22 gene expression was noticed in liver (∼1.8-fold), and moderate induction in spleen and intestine. In the treated fish gill, TLR22 expression remained unchanged but in kidney, skin, and blood, TLR22 expression was downregulated as compared to controls.
Modulation of TLR22 gene expressions by synthetic dsRNA. In rohu fingerlings, poly I:C (300 μg/fish) was i.v. injected, and after 4 h, various tissues were collected and were used for total RNA extraction, cDNA preparation, and real-time PCR analysis. The relative expression of TLR22 gene was normalized to the expression of β-actin (internal control) and was expressed as fold changes relative to the un-treated control group. The mean value of three fish (n = 3) is shown and the bars indicate standard error. Significant difference (p < 0.05) between control and treated fish group is indicated with asterisks (*)
Modulation of TLR22 Expression by Bacterial RNA
To investigate the role of TLR22 in recognizing bacterial RNA, total RNA was purified from A. hydrophila and i.v. injected into rohu fingerlings. Purity of the RNA was checked after treating it with DNase I and RNase A. The result showed that the RNA remained intact after treating with DNase I (Fig. 7a left panel) and was completely digested with RNase A (Fig. 7a right panel) confirming the purity of the extracted RNA. Following bacterial RNA stimulation, significant (p < 0.05) induction of TLR22 gene expression was observed in all tested organs of the treated fish as compared to control, except in blood (Fig. 7b). Among the tested tissues, liver showed the highest induction of TLR22 (∼36-fold) followed by spleen (∼13-fold), kidney (∼9-fold), intestine (∼5.5-fold), skin (∼2.7-fold) and gill (∼1.7-fold). However, TLR22 gene expression in blood was downregulated in treated fish as compared to control.
Modulation of TLR 22 gene expressions by bacterial RNA. Total RNA was extracted from A. hydrophila, and 10 μg of the purified RNA was i.v. injected into rohu fingerlings. At 4-h post treatment, TLR22 gene expression in various tissues was analyzed by qRT-PCR. The relative expression of TLR22 gene was normalized to the expression of β-actin (internal control) and was expressed as fold changes relative to the un-treated control group. The mean value of three fish (n = 3) is shown and the bars indicate standard error. Significant difference (p < 0.05) between control and treated fish group is indicated with asterisks (*). a Photograph of agarose gel analysis confirming the purity of the A. hydrophila-derived RNA. b TLR22 gene expression in various tissues following A. hydrophila-derived RNA treatment
Response of TLR22 in Bacterial Infection
To investigate the response of TLR22 gene in bacterial infection, we infected rohu fingerlings with A. hydrophila and analyzed temporal expression of TLR22 gene in gill, liver, kidney, intestine, spleen, skin, and in blood by qRT-PCR. The data revealed that except in gill, TLR22 gene expression was significantly (p < 0.05) induced in all tested tissues of A. hydrophila-infected fish as compared to the control (Fig. 8). At 24 h, the highest induction of TLR22 was noted in blood (∼3-fold), and at 48 h, in kidney (∼9-fold), followed by skin (∼7-fold) and liver and spleen (∼4-fold). However, in contrast to all other tissues, gill remained unresponsive both at 24- and 48-h post infection.
Response of TLR22 gene following bacterial infection. Aeromonas hydrophila (1 × 106 CFU/fish) was injected into rohu fingerlings by i.p. route. After the designated time course, total RNA was extracted from gill, liver, kidney, intestine, spleen, skin, and blood. Real-time PCR was conducted to analyze TLR22 and β-actin expression in various organs/tissues. The results were expressed as mean ± standard errors (bars) from three fish (n = 3) after normalizing the values with β-actin (internal control). Significant difference (p < 0.05) between the control and treated group is indicated with asterisks (*)
Discussion
This report describes the identification of TLR22 in rohu, a member of the Indian major carp (IMC), and a freshwater fish species of highest commercial importance in the Indian subcontinent. The ORF of LrTLR22 gene consisted of 2,838 nucleotides that encoded a putative protein of 946 amino acid (aa) residues. In TLR, LRR motifs were known to be involved in various PAMPs recognition. In LrTLR22, the spans of LRR regions were almost equivalent with other fish species. The SMART analysis did not show the transmembrane domain in LrTLR22. This was in contrast to common carp but similar to fugu TLR22 (Fig. 1). Absence of transmembrane domain in TLR22 of rohu and fugu might suggest its intracellular localization. In LrTLR22, the ecto-domain (ECD) comprising LRR motifs was predicted to form a horse-shoe-shaped structure and was similar to rohu TLR3-ECD [20] and human TLR3-ECD [21]. The structural homology between TLR22 and TLR3 might suggest their functional similarity in recognizing dsRNA.
In fish, wide variation in TLR22 genomic structure was reported. The number of intron(s) in TLR22 gene was one in grass carp [10], two in fugu [11], and three in Japanese flounder [6] and large yellow croaker [9]. PCR amplification of LrTLR22 using genomic DNA or cDNA as template resulted in same product size (Fig. S5) suggesting it as an intronless gene. Wide variations in TLR22 genomic structure among various fish species might suggest differential regulation in TLR22 gene transcription. Rohu, common carp, grass carp, gold fish, and zebrafish belonged to the Cyprinidae family. Therefore, as expected, they formed a separate sub-cluster and were far apart from catfish. The evolutionary close relationship of TLR22 among the cyprinid fish species might suggest the similarity in their PAMPs recognition, downstream signaling pathway, and induction of innate immunity.
Ontogenic expression of innate immune receptors was previously been reported in zebrafish [22] and channel catfish [23] embryos. In rohu, ontogenic expression of TLR2 [24], TLR3 [25], NOD1 [26], and NOD2 [27] gene was reported previously. In contrast to other TLR orthologs, TLR22 exists only in teleosts and amphibians and is unique in nature. Previously, TLR22 gene expression at various stages of embryonic development was reported in grass carp [28]. Therefore, expression of LR22 gene (Fig. 4a.) during ontogenesis of rohu was expected. Embryonic expression of TLR22 in grass carp and rohu might suggest the important role of TLR22 in protecting eggs, hatched embryos, and larvae from various pathogenic invasions.
In healthy (un-treated) rohu fingerlings, TLR22 expression was detected in all tested organs, and among them, liver expressed the highest TLR22. In Japanese flounder, TLR22 gene transcripts were mainly detected in peripheral blood leukocytes (PBL) [6] but it was ubiquitously expressed in fugu [5, 9]. In goldfish and rainbow trout, TLR22 gene expression was highest in spleen and lowest in liver [3, 7]. The grass carp was phylogenetically closely related to rohu. But the TLR22 gene expression in various organs/tissues of grass carp [10] significantly differed from rohu. These findings indicated that TLR22 gene expression in various organs/tissues greatly differed among teleosts. In spite of these variations, prominent expression of TLR22 in many immune-related organs in wide variety of fish species strongly suggested its important role in immunity.
LPS is one of the major cell wall components of the Gram-negative bacteria and is specifically recognized by TLR4 [29, 30]. In rohu, LPS induced TLR22, and the induction was similar to orange spotted grouper [31]. Till date, specific ligand(s) of TLR22 is not well established. Therefore, the data in rohu and orange spotted grouper may indicate LPS as one of the ligands of TLR22. Fish are aquatic animals, and are continuously exposed to different types of RNA liberated by various organisms [32, 33]. During evolutions, aquatic animals perhaps developed many RNA-sensing TLRs and other receptors to recognize these non-self RNAs, which are likely to be distinct from land animals. To investigate these RNA-sensing TLRs in rohu, we stimulated rohu fingerlings with poly I:C, a synthetic dsRNA that mimicked the architecture of viral dsRNA, and also with bacterial (A. hydrophila) RNA.
In response to dsRNA (poly I:C) stimulation, varied expression of LrTLR22 gene was noted in the tested tissues. Most significant induction of LrTLR22 was noted in liver, followed by spleen, suggesting liver as an important immune-related organ in fish. However, downregulation of TLR22 was observed in the kidney following poly I:C as well as LPS stimulation. In Argulus siamensis infection, downregulation of TLR22 gene expression was also reported in kidney [34]. These observations together might suggest a different role of LrTLR22 in kidney in contrast to other tissues.
In response to poly I:C, TLR22 gene expression was significantly low (Fig. 6) as compared to TLR3 [25]. On the other hand, significantly high induction of TLR22 gene expression was noted in majority of the bacterial RNA-treated fish tissues (Fig. 7) as compared to poly I:C. In fugu, short-sized (∼200 bp) dsRNA was reported to be recognized by TLR3, whereas long-sized (∼1,000 bp) dsRNA by TLR22 [11]. Bacterial RNA is mostly (80 %) comprised of ribosomal RNA [35] and is larger than poly I:C. Therefore, preferential recognition of poly I:C by TLR3 and long-sized bacterial RNA by TLR22 may likely to occur in rohu. Furthermore, LRR11-13 motifs in LrTLR22-ECD exhibited maximum fluctuation during MD simulation (Fig. S2c). The prominent motions accomplished in these regions are suggestive of ligand recognition and binding. In rohu TLR3, the involvement of the LRR 13–14 regions were shown to bind dsRNA of aquatic reovirus [20]. Therefore, conservation of these central LRR regions in both rohu TLR3 and 22 suggest that LRR11-13 might be the crucial domain of viral/bacterial dsRNA recognition in TLR3/22 subgroups. The flipping of AB, BB, and DD loops in the TIR domain (Fig. S2d) may also indicate their critical role in protein-protein interactions and downstream signal transduction. To confirm these observations, further studies employing site-directed mutagenesis in LrTLR22 are required.
In A. hydrophila-infected fish, TLR22 gene expression pattern in majority of the tested tissues (Fig. 8) showed significant similarity with the data of purified bacterial RNA-treated fish tissues (Fig. 7), suggesting the important role of TLR22 in recognizing bacterial RNA during infection.
In conclusion, this article describes molecular cloning, characterization, and 3D-structure analysis of TLR22 (the unique TLR in fish) in the Indian major carp (IMC) rohu and its function as an important innate immune receptor in recognizing LPS, poly I:C, and bacterial RNA. The data in this article is expected to help in understanding the critical role of TLR22 in inducing innate immunity in fish and warrant further investigation to understand the possible interplay between microbial signatures and TLR22 in fish.
References
Akira, S. (2009). Pathogen recognition by innate immunity and its signaling. Proceedings of the Japan Academy Series B, 85, 143–156.
Rauta, P. R., Samanta, M., Dash, H. R., Nayak, B., & Das, S. (2014). Toll-like receptors (TLRs) in aquatic animals: signaling pathways, expressions and immune responses. Immunology Letters, 158, 14–24.
Stafford, J., Stafford, J. L., Ellestad, K. K., Magor, K. E., Belosevic, M., & Magor, B. G. (2003). A toll-like receptor (TLR) gene that is up-regulated in activated goldfish macrophages. Developmental and Comparative Immunology, 27, 685–698.
Franch, R., Cardazzo, B., Antonello, J., Castagnaro, M., Patarnello, T., & Bargelloni, L. (2006). Full-length sequence and expression analysis of Toll-like receptor 9 in the gilthead seabream (Sparus aurata L.). Gene, 378, 42–51.
Oshiumi, H., Tsujita, T., Shida, K., Matsumoto, M., Ikeo, K., & Seya, T. (2003). Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics, 54, 791–800.
Hirono, I., Takami, M., Miyata, M., Miyazaki, T., Han, H. J., Takano, T., Endo, M., & Aoki, T. (2004). Characterization of gene structure and expression of two toll-like receptors from Japanese flounder, Paralichthys olivaceus. Immunogenetics, 56, 38–46.
Rebl, A., Siegl, E., Koller, B., Fischer, U., & Seyfert, H. M. (2007). Characterization of twin toll-like receptors from rainbow trout (Oncorhynchus mykiss): evolutionary relationship and induced expression by Aeromonas salmonicida. Developmental and Comparative Immunology, 31, 499–510.
Jault, C., Pichon, L., & Chluba, J. (2004). Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Molecular Immunology, 40, 759–771.
Xiao, X., Qin, Q., & Chen, X. (2011). Molecular characterization of a toll-like receptor 22 homologue in large yellow croaker (Pseudosciaena crocea) and promoter activity analysis of its 5′-flanking sequence. Fish & Shellfish Immunology, 30, 224–233.
Su, J., Heng, J., Huang, T., Peng, L., Yang, C., & Li, Q. (2012). Identification, mRNA expression and genomic structure of TLR22 and its association with GCRV susceptibility/resistance in grass carp (Ctenopharyngodon idella). Developmental and Comparative Immunology, 36, 450–462.
Matsuo, A., Oshiumi, H., Tsujita, T., Mitani, H., Kasai, H., Yoshimizu, M., et al. (2008). TLR22 recognizes RNA duplex to induce IFN and protect cells from Birnaviruses. Journal of Immunology, 181, 3474–3485.
FAO, NACA. (2003). Quarterly aquatic animal disease report (Asia and Pacific Region).
Campanella, J. J., Bitincka, L., & Smalley, J. (2003). MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics, 4, 29.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Sali, A., & Blundell, T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology, 234, 779–815.
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: fast, flexible, and free. Journal of Computational Chemistry, 26, 1701–1718.
Vriend, G. (1990). WHAT IF: a molecular modeling and drug design program. Journal of Molecular Graphics, 8, 52–56.
Chen, V. B., Arendall, W. B., 3rd, Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., et al. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica, Section D: Biological Crystallography, 66, 12–21.
Livak, K. J., & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) Method. Methods, 25, 402–408.
Sahoo, B. R., Basu, M., Swain, B., Maharana, J., Dikhit, M. R., Jayasankar, P., & Samanta, M. (2012). Structural insights of rohu TLR3, its binding site analysis with fish reovirus dsRNA, poly I:C and zebrafish TRIF. International Journal of Biological Macromolecules, 51, 531–543.
Botos, I., Liu, L., Wang, Y., Segal, D. M., & Davies, D. R. (2009). The Toll-like Receptor 3: dsRNA signaling complex. Biochimica et Biophysica Acta, 1789, 667–674.
Kanther, M., & Rawls, J. F. (2010). Host-microbe interactions in the developing zebrafish. Current Opinion in Immunology, 22, 10–19.
Peterson, B. C., Bosworth, B. G., & Bilodeau, A. L. (2005). Differential gene expression of IGF-I, IGF-II, and Toll-like receptors 3 and 5 during embryogenesis in hybrid (channel × blue) and channel catfish. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 141, 42–47.
Samanta, M., Swain, B., Basu, M., Panda, P., Mohapatra, G. B., Sahoo, B. R., & Maiti, N. K. (2012). Molecular characterization of toll-like receptor 2 (TLR2), analysis of its inductive expression and associated down-stream signaling molecules following ligands exposure and bacterial infection in the Indian major carp, rohu (Labeo rohita). Fish & Shellfish Immunology, 32, 411–425.
Samanta, M., Basu, M., Swain, B., Panda, P., & Jayasankar, P. (2013). Molecular cloning and characterization of Toll-like receptor 3, and inductive expression analysis of type I IFN, Mx and pro-inflammatory cytokines in the Indian carp, rohu (Labeo rohita). Molecular Biology Reports, 40, 225–235.
Swain, B., Basu, M., & Samanta, M. (2012). Molecular cloning and characterization of nucleotide binding and oligomerization domain-1 (NOD1) receptor in the Indian major carp, rohu (Labeo rohita), and analysis of its inductive expression and down-stream signalling molecules following ligands exposure and Gram-negative bacterial infections. Fish & Shellfish Immunology, 32, 899–908.
Swain, B., Basu, M., Sahoo, B. R., Maiti, N. K., Routray, P., Eknath, A. E., & Samanta, M. (2012). Molecular characterization of nucleotide binding and oligomerization domain (NOD)-2, analysis of its inductive expression and down-stream signalling following ligands exposure and bacterial infection in rohu (L. rohita). Developmental and Comparative Immunology, 36, 93–103.
Lv, J., Huang, R., Li, H., Luo, D., Liao, L., Zhu, Z., & Wang, Y. (2012). Cloning and characterization of the grass carp (Ctenopharyngodon idella) Toll-like receptor 22 gene, a fish-specific gene. Fish & Shellfish Immunology, 32, 1022–1031.
Hoshino, K., Takeuchi, O., Kawai, T., Sanjo, H., Ogawa, T., Takeda, Y., Takeda, K., & Akira, S. (1999). Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hypo-responsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. Journal of Immunology, 162, 3749–3752.
Basu, M., Maiti, N. K., & Samanta, M. (2013). Toll-like receptor (TLR) 4 in mrigal (Cirrhinus mrigala): response to lipopolysaccharide treatment and Aeromonas hydrophila infection. International Research Journal of Biological Sciences, 2, 20–27.
Ding, X., Lu, D. Q., Hou, Q. H., Li, S. S., Liu, X. C., Zhang, Y., et al. (2012). Orange-spotted grouper (Epinephelus coioides) toll-like receptor 22: molecular characterization, expression pattern and pertinent signaling pathways. Fish & Shellfish Immunology, 33, 494–503.
Seya, T., Matsumoto, M., Ebihara, T., & Oshiumi, H. (2009). Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunological Reviews, 227, 44–53.
Ando, T., Suzuki, H., Nishimura, S., Tanaka, T., Hiraishi, A., & Kikuchi, K. (2006). Characterization of extracellular RNAs produced by the marine photosynthetic bacterium Rhodovulum sulfidophilum. Journal of Biochemistry, 139, 805–811.
Saurabh, S., Mohanty, B. R., & Sahoo, P. K. (2011). Expression of immune-related genes in rohu Labeo rohita (Hamilton) by experimental freshwater lice Argulus siamensis (Wilson) infection. Veterinary Parasitology, 175, 119–128.
Eberle, F., Sirin, M., Binder, M., & Dalpke, A. H. (2009). Bacterial RNA is recognized by different sets of immune receptors. European Journal of Immunology, 39, 2537–2547.
Acknowledgments
The work was supported by the grant of National Fund for Basic Strategic and Frontier Application Research in Agriculture (NFBSFARA/BS-4003) and National Agricultural Innovation Project (NAIP/ C4-C30018) of the Indian Council of Agricultural Research (ICAR), Govt. of India. We express our sincere gratitude to Dr. A. Bandyopadhyay and Dr. S. Kochar, National Coordinators, NFBSFARA and NAIP-Comp-4, for meticulous suggestions and help. We thank Dr. P. Routray, Principal Scientist, Aquaculture Production and Environment Division, CIFA, for hatchery facility.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Fig. S1
Procheck analysis of rohu TLR22 domains. Ramachandran plot analysis of (a) LrTLR22-ECD and (b) LrTLR22-TIR domain. (GIF 198 kb)
Fig. S2
MD trajectory analysis of LrTLR22-ECD and TIR domains. RMSD graph of (a) LrTLR22-ECD and (b) LrTLR22-TIR domain. RMSF graph of (c) LrTLR22-ECD and (d) LrTLR22-TIR domain. Flipping regions are shown within the arc. (GIF 79 kb)
Fig. S3
Multiple sequence alignment among LRR motifs of TLR22. The LRR motifs of various species including rohu were predicted by SMART and were aligned by CLUSTALW. Aligned file was submitted in ESPript 3. to build blocks. The black background boxes represented strictly conserved regions and other boxes represented well-conserved (> 50 %) regions. (GIF 333 kb)
(GIF 321 kb)
Fig. S4
Multiple sequence alignment among TIR domains of TLR22. The TIR domains of various species including rohu were predicted by SMART and were aligned by CLUSTALW. Aligned file was submitted in ESPript 3.0 to build blocks. The black background boxes represented strictly conserved regions and other boxes represented well-conserved (> 50 %) regions. (GIF 147 kb)
Fig. S5
PCR amplification of TLR22 from genomic and cDNA template. PCR was carried out in a 50 μl reaction mixture using rohu genomic DNA (lane:1) or cDNA (lane: 2) as template, and 10 μl of the PCR product was analyzed in 1 % agarose gel. (GIF 45 kb)
Rights and permissions
About this article
Cite this article
Samanta, M., Swain, B., Basu, M. et al. Toll-Like Receptor 22 in Labeo rohita: Molecular Cloning, Characterization, 3D Modeling, and Expression Analysis Following Ligands Stimulation and Bacterial Infection. Appl Biochem Biotechnol 174, 309–327 (2014). https://doi.org/10.1007/s12010-014-1058-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1058-0