Abstract
Present investigation deals with the study, to quantify the antibacterial property of silver nanoparticles (SNPs), synthesized by pulsed laser ablation (PLA) in aqueous media, on some human enteropathogenic gram-positive and gram-negative bacterial strains. Antibacterial property was studied by measuring the zone of inhibition using agar cup double-diffusion method, minimum inhibitory concentration by serial dilution method, and growth curve for 24 h. The results clearly show the potency of antibacterial property of PLA-synthesized SNPs and suggest that it can be used as an effective growth inhibitor against various pathogenic bacterial strains in various medical devices and antibacterial control systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The resistivity of different bacterial strains against various antibiotics has been increasing day by day, and in this scenario, many researchers are trying to develop new nonresistant, economical, and effective antibacterial agents. Such problems have led to resurgence in the use of silver (Ag)-based antiseptics that may be linked to broad-spectrum activities and lower propensity to induce microbial resistance than antibiotics [1]. Since long, silver has been known to have disinfecting effects and thus found applications in traditional medicines and culinary items. Several salts of silver and there derivatives are commercially employed as antimicrobial agents [2]. The antibacterial effects of Ag salts have been noticed since antiquity [3], and Ag is currently used to control bacterial growth in a variety of applications, including dental work, catheters, and burn wounds [4, 5]. Antibacterial property of silver nanoparticles (SNPs), synthesized by various chemical precipitation methods, has been investigated by several groups on several bacterial strains [6–18]. SNPs have good antibacterial property in pure [6] and composite forms [7, 8]. Nanoparticles synthesized by biological systems also show good antibacterial property [9, 10]. Pal et al. [11] demonstrated that SNPs undergo a shape-dependent interaction with Escherichia coli, while Cristóbal et al. [6] showed that the antibacterial property of SNPs is size dependent. SNPs show antibacterial property even when synthesized by chemical reduction method [8, 12] or sol–gel method [13]. Biodegradable poly(l-lactide) ultrafine fibers containing SNPs prepared via electrospinning show antibacterial property [14]. Antibacterial property of SNPs can be enhanced by using various capping agents [15, 16]. SNPs loaded on poly(acrylamide-co-itaconic acid)-grafted cotton fabric can be used as an antibacterial agent [17]. Travan et al. [18] discovered a novel finding that chitlac hydrogels containing SNPs display a very effective bactericidal activity towards both gram-positive and gram-negative bacteria; however, it does not show any cytotoxic effect towards three different eukaryotic cell lines. In general for biological applications, purity of nanoparticles is required. In this context, pulsed laser ablation (PLA) of metal in liquid medium is a novel top-down technique to produce pure nanoparticles [19]. PLA have several advantages such as follows: it produces chemical contaminant-free nanoparticles, and it takes very less time for preparing nanoparticles in comparison to other conventional methods [20]. Nath et al. [21] have synthesized Cu/Cu2O nanocomposites by laser ablation method and observed effective reduction in bacterial colonies by these nanocomposites.
Silver-based antimicrobial agents receive much attention, because of the low toxicity of the active Ag+ ion to human cells [22, 23]. The mechanism of the bactericidal effect of silver nanocolloids against bacterial strain is still not very well known. Metal nanoparticles exhibit remarkable physical, chemical, and biological properties due to their large surface area and high reactivity compared with the bulk-sized particles [24]. SNPs may either interact at the cell wall/cell membrane/DNA levels or may even interrupt various metabolic pathways by interaction with various metabolic enzymes. SNPs release silver ions, which make an additional contribution to the bactericidal effect [25]. Morones et al. [26] showed that Ag+ and Ag0 both contribute to the antibacterial activity. They also showed that SNPs (where silver is present in the Ag0 form) also contain micromolar concentrations of Ag+. There are some reports that show that electrostatic attraction between negatively charged bacterial cells and positively charged NPs is crucial for the activity of NPs as bactericidal materials [27, 28]. In the review of Jones and Hoek [29], they summarized three most common mechanisms of toxicity for SNPs: (1) uptake of free silver ions followed by disruption of ATP production and DNA replication, (2) SNPs and silver ion-induced generation of reactive oxygen species (ROS), and (3) SNPs directly damage to the cell membranes, whereas Guzman et al. [30] assumed four different ways for antibacterial activity of SNPs: (1) interference with cell wall synthesis, (2) inhibition of protein synthesis, (3) interference with nucleic acid synthesis, and (4) inhibition of metabolic pathways.
In the present study, we used SNPs synthesized by PLA in aqueous medium for testing the antibacterial property against several human pathogenic gram-positive and gram-negative bacterial strains.
Materials and Methods
Chemicals and Bacterial Strains
Silver rod of 99.9 % purity was purchased from Johnson Matthey, and double-distilled water was purchased from Merck, India, for synthesis of nanoparticles by laser ablation method. The antibacterial experiment was carried out with two gram-positive (Bacillus subtilis and Staphylococcus aureus) and six gram-negative (E. coli, Enterobacter aerogenes, Klebsiella pneumonia, Pseudomonas aeruginosa, Salmonella typhi, and Salmonella typhimurium) bacterial strains. The bacterial strains were obtained from the National Collection of Industrial Microorganisms (NCIM), National Chemical Laboratory, Pune, India, and were maintained on nutrient agar in the culture lab of Department of Biotechnology, University of Allahabad, Allahabad, India. Chemicals required for nutrient agar and nutrient broth were purchased from HiMedia Laboratories Private Limited.
Synthesis of Silver Nanoparticles
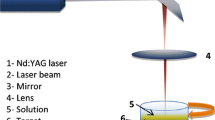
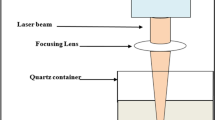
Silver rod of 99.9 % purity was placed at the bottom of a glass vessel containing 20 ml of double-distilled water (Merck, India). The rod was ablated for 3 h by focused beam of 1,064 nm of pulsed Nd:YAG laser (Spectra Physics USA) operating at 40 mJ/pulse energy with 10 Hz repetition rate and 10 ns pulse width. After the ablation, the resultant solution converted in dark yellow color, and sample was collected for further studies.
Characterizations of Silver Nanoparticles
The UV-visible absorption spectrum of synthesized colloidal solution of NPs was acquired with Perkin Elmer Lambda 35, double-beam spectrophotometer. For transmission electron microscopy analysis, a drop has been taken from synthesized colloidal solution of NPs and placed on a carbon-coated copper grid and dried at ambient condition. The transmission electron microscope (TEM) image and selected area electron diffraction (SAED) pattern of the NPs were recorded using a Tecnai G20 microscope.
Zone of Inhibition
The antibacterial susceptibility tests were carried out using agar cup double-diffusion method [31–34]. The microorganisms were cultured overnight at 37 °C in nutrient agar. The final cell concentrations of bacterial inoculants were 106–107 colony-forming unit (CFU)/ml. The colloidal solution of SNPs was delivered into wells, and the plates were incubated at 37 °C for 24 h. The culture plates were observed for the presence of the zone of microbial growth inhibition which was regarded as the presence of antimicrobial action, and this activity was expressed in terms of average diameter of the zone of inhibition measured in millimeters.
Minimum Inhibitory Concentration
Minimum inhibitory concentration (MIC) was determined by the serial dilution method in the nutrient broth [35] using serially diluted NPs. Final concentrations of SNPs from 1 to 10 μg/ml were prepared separately by diluting the stock solution of SNPs. Bacterial inoculums were adjusted to contain ~105 CFU/ml and were incubated at 35 °C for 24 h. The MIC was recorded as the lowest concentration of the NPs showing no visible growth of the test bacterial strains after 24 h of incubation, by measuring optical density [36].
Bacterial Growth Curves
The UV–vis spectrophotometer (Perkin Elmer Lambda 35) was used to determine growth curves. Nutrient broths containing 102 CFU of each microorganism/ml in the growth phase were used. The data were recorded at every 30 min for the first 6 h, and last data were recorded after completing 24 h of experiment for both the cases with and without mixing of SNPs.
Results and Discussion
UV-Visible Absorption of SNPs
The dispersions of the metal SNPs usually display a very intense color due to surface plasmon resonance (SPR) absorption, which can be attributed to the collective oscillation of conduction electrons that is induced by an electromagnetic field. In the UV-visible spectrum, a stable SPR band with symmetric shape was observed at 407 nm for silver colloids (Fig. 1), and the broad band in the UV region originated from an interband transition (245 nm) of the SNPs. The symmetric shape of the SPR band indicates the formation of spherically shaped NPs, and the long tail in the red region indicates the formation of polydisperse size of NPs.
Transmission Electron Microscopy
The TEM image of synthesized SNPs shows nearly spherical and elliptical shape (Fig. 2a) in nature, which is in good agreement with UV-visible absorption a result. The particles size are calculated and found to be ranging from 9 to 27 nm. The distribution of particles size is shown in inset of Fig. 2a. SAED pattern of the SNPs is shown in Fig. 2b that confirms the formation of polycrystalline SNPs.
Zone of Inhibition
Zone of inhibition of SNPs against various gram-positive and gram-negative bacterial strains is shown in Figs. 3, 4, 5, and 6 and summarized in Table 1. For obtaining a more conspicuous view of the zone of inhibition, we used a much higher concentration of SNPs (up to 600 μg/ml) against two gram-positive (B. subtilis and S. aureus) and two gram-negative (K. pneumonia and P. aeruginosa) bacterial strains, and the zone of inhibitions is clearly shown in Fig. 3. Among the used strains, E. aerogenes and E. coli show a maximum susceptibility against SNPs. In these two strains, zone of inhibition appears at 40 μg/ml concentration of SNPs (Fig. 4). Bacterial strains B. subtilis, S. aureus, K. pneumonia, and P. aeruginosa start showing zone of inhibition at 50 μg/ml concentration of SNPs (Fig. 5). Among the studied strains, S. typhi and S. typhimurium shows zone of inhibition at a comparatively higher concentration of 80 μg/ml SNPs (Fig. 6).
Bacterial Growth Curves
The growth curves of the different bacterial strains in the absence and presence of SNPs are shown in Fig. 7. The results clearly show the antibacterial properties of SNPs. The characteristic log phase of bacterial growth was present in the case of bacterial control set (i.e., without the treatment of SNPs), whereas, in case of the bacterial set with the treatments of SNPs, bacterial growth was very much reduced. There was a significant delay in the log phase, and at the log phase of the bacterial growth, the bacterial growth was much reduced over the control. In the case of P. aeruginosa, the log phase was completely absent, whereas in the case of E. aerogenes and E. coli, the bacterial growth suddenly diminished after ca. 5.0–6.0 h. This showed the diverse effect of SNPs on human pathogenic gram-positive and gram-negative bacterial strains.
Minimum Inhibitory Concentration
The MIC, in the present investigation, is ranging from 2 to 6 μg/ml (Table 1). MICs ranging from 13.5 to 1.69 μg/ml were reported for different bacterial strains [29] earlier; thus, the results showed the similarity with the previous investigations. MIC is minimum for E. coli (ca. 2 μg/ml), followed by E. aerogenes, and MIC is maximum for S. typhimurium (6 μg/ml).
Our result clearly showed that the bactericidal property of SNPs synthesized by PLA, against various human enteropathogenic gram-positive and gram-negative bacterial strains, depends upon the concentration of SNPs. Zone of inhibition area appeared at minimum SNP concentration for E. aerogenes and E. coli. MIC was also minimum for E. coli, followed by E. aerogenes, and even growth curve depicted an abrupt decrease in E. coli and E. aerogenes concentration at nearly 5.30 and 6.00 h, respectively. The greater susceptibility of E. aerogenes and E. coli could probably be explained on the basis of their cell wall structure. Both E. aerogenes and E. coli are gram-negative bacteria. The cell wall of gram-negative bacteria is thinner than that of gram-positive bacteria. Thus, gram-negative bacteria are more susceptible to SNPs than gram-positive bacteria; this may be one of the reasons for a higher susceptibility of E. aerogenes and E. coli.
Mechanisms of the bactericidal effects of SNPs are not very clear. Figure 8 shows the various possible mechanisms of antibacterial activity of silver nanoparticles. SNPs may interact with bacterial cell wall. Song et al. [37] reported that SNPs inhibit cell wall synthesis in S. aureus. When SNPs attached to the surface of bacterial cell membrane, its power functions, such as permeability and respiration, are disturbed [25, 38]. Song et al. [37] reported plasmolysis in P. aeruginosa due to the SNPs. Transmission electron microscopy data showed that SNPs adhere to and penetrate into E. coli cells and also able to induce the formation of pits in the cell membrane [28, 39, 40]. SNPs may interact with thio groups of respiratory chain proteins and transport proteins (sulfur-containing proteins), ultimately interfering with their proper functioning [26]. SNPs are able to penetrate the bacteria and cause further damage, possibly by interacting with sulfur- and phosphorus-containing compounds such as DNA [41]. Experimental evidence suggests that DNA loses its replication ability once the bacteria have been treated with silver ions [25]. SNPs release Ag+ ions which interact with cytoplasmic components and nucleic acids to inhibit respiratory chain enzymes and to interfere with membrane permeability [42]. A low concentration of Ag+ ion induces a massive proton leakage through the membrane of Vibrio cholerae, resulting in a collapse of proton-motive force [43]. Ag+ ion interacts with the respiratory chain enzymes of E. coli and inhibits the respiratory chain at a low potential point, possibly the NADH dehydrogenase of complex I [44]. Inside the cell, SNPs would interfere with the bacterial growth-signaling pathway by modulating tyrosine phosphorylation of putative peptide substrates critical for cell viability and division [45]. When SNPs enter the bacterial cell, they form a low molecular weight region inside the bacteria. Thus, the bacteria conglomerate to protect the DNA from the SNPs. Consequently, the SNPs preferably attack the respiratory chain, leading to cell division and, finally, to cell death [46]. SNPs can act as catalysts and generate ROS such as hydrogen peroxide, superoxide radical, and hydroxyl radical in the presence of dissolved oxygen, leading to excess free radical production. Studies done in eukaryotic cells suggest that SNPs inhibit the antioxidant defense by interacting directly with glutathione (GSH), binding GSH reductase or other GSH maintenance enzymes [47], and excess ROS production may produce oxidative stress [48]. The additional generation of free radicals can attack membrane lipids and lead to a breakdown of membrane and mitochondrial function or cause DNA damage, causing bacterial cell death [49].
Conclusions
Pure silver nanoparticles were synthesized by pulsed laser ablation. These SNPs are very potent antibacterial agent against gram-positive and gram-negative human enteropathogenic bacterial strains. The MIC values are found to be very low for both gram-positive and gram-negative bacteria. In comparison to gram-positive bacteria, gram-negative bacteria show a wider range of inhibition, and growth of the gram-negative bacteria is more sensitive with the SNPs.
References
Jones, S. A., Bowler, P. G., Walker, M., & Parsons, D. (2004). Wound Repair and Regeneration, 12, 288–294.
R. Holladay, W. Moeller, D. Mehta, J. Brooks, R. Roy M. Mortenson, Application Number WO2005US47699 20051230 European Patent office 2006.
Silver, S., & Phung, L. T. (1996). Annual Review of Microbiology, 50, 753–789.
Catauro, M., Raucci, M. G., De Gaetano, F. D., & Marotta, A. (2004). Journal of Materials Science Materials in Medicine, 15, 831–837.
Crabtree, J. H., Burchette, R. J., Siddiqi, R. A., Huen, I. T., Handott, L. L., & Fishman, A. (2003). Peritoneal Dialysis International, 23, 368–374.
Cristóbal, L. F. E., Castañón, G. A. M., Martínez, R. E. M., Rodríguez, J. P. L., Marín, N. P., Macías, J. F. R., & Ruiz, F. (2009). Materials Letters, 63, 2603–2606.
Sotiriou, G. A., & Pratsinis, S. E. (2010). Environmental Science and Technology, 44, 5649–5654.
Cao, X. L., Cheng, C., Ma, Y. L., & Zhao, C. S. (2010). Journal of Materials Science Materials in Medicine, 21, 2861–2868.
Shirley, A. D., Sreedhar, B., & Dastager, S. G. (2010). Digest Journal of Nanomaterials and Biostructures, 5, 44–51.
Kaviya, S., Santhanalakshmi, J., Viswanathan, B., Muthumary, J., & Srinivasan, K. (2011). Spectrochimica Acta Part A, 79, 594–598.
Pal, S., Tak, Y. K., & Song, J. M. D. (2007). Applied and Environmental Microbiology, 73, 1712–1720.
Guzmán, M. G., Dille, J., & Godet, S. (2009). International Journal of Chemical and Biomolecular Engineering, 2, 104–111.
Kawashita, M., Tsuneyama, S., Miyaji, F., Kokubo, T., Kozuka, H., & Yamamoto, K. (2000). Biomaterials, 21, 393–398.
Xu, X., Yang, Q., Wang, Y., Yu, H., Chen, X., & Jing, X. (2006). European Polymer Journal, 42, 2081–2087.
Jaiswal, S., Duffy, B., Jaiswal, A. K., Stobie, N., & McHale, P. (2010). International Journal of Antimicrobial Agents, 36, 280–283.
Kora, A. J., Manjusha, R., & Arunachalam, J. (2009). Materials Science and Engineering C, 29, 2104–2109.
Gupta, P., Bajpai, M., & Bajpai, S. K. (2008). The Journal of Cotton Science, 12, 280–286.
Travan, A., Pelillo, C., Donati, I., Marsich, E., Benincasa, M., Scarpa, T., Semeraro, S., Turco, G., Gennaro, R., & Paoletti, S. (2009). Biomacromolecules, 10, 1429–1435.
Swarnkar, R. K., Singh, S. C., & Gopal, R. (2011). Bulletin of Materials Science, 34, 1363–1369.
Singh, M. K., Agarwal, A., Gopal, R., Swarnkar, R. K., & Kotnala, R. K. (2011). Journal of Materials Chemistry, 21, 11074–11079.
Nath, A., Das, A., Rangan, L., & Khare, A. (2012). Science of Advanced Materials, 4, 106–109.
Williams, R. L., Doherty, P. J., Vince, D. G., Grashoff, G. J., & Williams, D. F. (1989). Criticism Review Biocompa, 5, 221–243.
Berger, T. J., Spadaro, J. A., Chapin, S. E., & Becker, R. O. (1976). Antimicrobial Agents and Chemotherapy, 9, 357–358.
Zeng, H., Du, X. W., Singh, S. C., Kulinich, S. A., Yang, S., He, J., & Cai, W. (2012). Advanced Functional Materials, 22, 1333–1353.
Feng, Q. L., Wu, J., Chen, G. Q., Cui, F. Z., Kim, T. N., & Kim, J. O. (2000). Journal of Biomedical Materials Research, 52, 662–668.
Morones, J. R., Elechiguerra, J. L., Camacho, A., Holt, K., Kouri, J. B., Ramírez, J. T., & Yacaman, M. J. (2005). Nanotechnology, 16, 2346–2353.
Hamouda, T., & Baker, J. R., Jr. (2000). Journal of Applied Microbiology, 89, 397–403.
Sondi, I., & Salopek-Sondi, B. (2004). Journal of Colloid and Interface Science, 275, 177–182.
Jones, C. M., & Hoek, E. M. V. (2010). Journal of Nanoparticle Research, 12, 1531–1551.
Guzmán, M. G., Dille, J., & Godet, S. (2012). Nanomedicine, 8, 37–45.
Perez, C., Paul, M., & Bazerque, P. (1990). Acta Biology Medicine Experiment, 15, 113–115.
Deore, S. L., & Khadabadi, S. S. (2008). Rasayan Journal of Chemistry, 1, 887–892.
Zhang, Y. J., Nagao, T., Tanaka, T., Yang, C. R., Okabe, H., & Kouno, I. (2004). Pharmaceutical Bulletin, 27, 251–255.
Bonjar, G. H. S., & Nik, A. K. (2004). Asian Journal of Plant Sciences, 3, 61–64.
Irith, W., Kai, H., & Robert, E. W. (2008). Nature Protocols, 3, 163–175.
Bassam, A. S., Ghaleb, A., Dahood, A. S., Naser, J., & Kamel, A. (2006). Turkish Journal of Biology, 30, 195–198.
Song, H. Y., Ko, K. K., Oh, I. H., & Lee, B. T. (2006). European Cells Materials, 11, 58–59.
Tenover, F. C. (2006). American Journal of Medicine, 119, 3–10.
Choi, O., Deng, K., Kim, N., Ross, L., Surampalli, R., & Hu, Z. (2008). Water Research, 42, 3066–3074.
Raffi, M., Hussain, F., Bhatti, T., Akhter, J., Hameed, A., & Hasan, M. (2008). Journal of Materials Science and Technology, 24, 192–196.
Gibbins, B., & Warner, L. (2005). Medicine Device Diagnostic Industry Magazine, 1, 1–2.
Russell, A. D., & Hugo, W. B. (1994). Progress in Medicinal Chemistry, 31, 351–370.
Dibrov, P., Dzioba, J., Gosink, K. K., & Hase, C. C. (2002). Antimicrobial Agents and Chemotherapy, 46, 2668–2670.
Holt, K. B., & Bard, A. J. (2005). Biochemistry, 44, 13214–13223.
Shrivastava, S., Bera, T., Roy, A., Singh, G., Ramachandrarao, P., & Dash, D. (2007). Nanotechnology, 18(225103), 9.
Rai, M., Yadav, A., & Gade, A. (2009). Biotechnology Advances, 27, 76–83.
Carlson, C., Hussain, S. M., Schrand, A. M., Braydich-Stolle, L. K., Hess, K. L., Jones, R. L., & Schlager, J. J. (2008). Journal of Physical Chemistry B, 112, 13608–13619.
Nel, A., Xia, T., Madler, L., & Li, N. (2006). Science, 311, 622–627.
Mendis, E., Rajapakse, N., Byun, H., & Kim, S. (2005). Life Sciences, 77, 2166–2178.
Acknowledgments
The authors would like to acknowledge Dr. N. P. Lalla of UGC-DAE CSR, Indore, for TEM facility. Authors R. K. Swarnkar and R. Gopal are thankful to the Defense Research and Development Organization (DRDO), New Delhi, for financial assistance.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Pandey, J.K., Swarnkar, R.K., Soumya, K.K. et al. Silver Nanoparticles Synthesized by Pulsed Laser Ablation: as a Potent Antibacterial Agent for Human Enteropathogenic Gram-Positive and Gram-Negative Bacterial Strains. Appl Biochem Biotechnol 174, 1021–1031 (2014). https://doi.org/10.1007/s12010-014-0934-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-0934-y