Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules of 22 nucleotides in length that have been characterized as regulators of messenger RNA (mRNA) regulating a number of developmental processes in plants and animals by silencing genes using multiple mechanisms. miRNAs have been extensively studied in various plant species; however, few information are available about miRNAs in perennial ryegrass, animal feed, and industrial raw materials. In this study, the 12 potential perennial ryegrass miRNAs were identified for the first time by computational approach. Using the newly identified miRNA sequences, the perennial ryegrass mRNA database was further used for BLAST search and detected 33 potential targets of miRNAs. Prediction of potential miRNA target genes revealed their functions involved in various important plant biological processes. Our result should be useful for further investigation into the biological functions of miRNAs in perennial ryegrass. The selected miRNAs representing four families were verified by RT-PCR experiment, indicating that the prediction method that we used to identify the miRNAs was effective.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a small single-stranded endogenous non-coding RNAs with a length of about 22 nt, which cause transcriptional cleavage or translational repression through binding their targets, depending on the extent of complementarity between miRNA and mRNA [1–3]. So far, an increasing number of plant miRNAs have been discovered, and the functions of a small number of them have been elucidated [4, 5]. Increasing evidence showed that plant miRNAs target a large number of genes with functions that are in a range of development processes, including meristem cell identity, leaf organ morphogenesis and polarity, floral differentiation, development, and responses to biotic and environmental stresses [6–10]. Most miRNAs are conserved in animals and plants and from animals to plants [11, 12]. The sequence conservation of miRNAs has laid a sound foundation for predicting and studying miRNAs in non-model plants whose whole genome sequences are unknown, such as expressed sequence tags (ESTs), genome survey sequences (GSS), and nucleotide database [13, 14]. The computational screening of potential miRNAs in plant species has been proved to be successful and more effective for the discovery of new miRNAs that usually cannot be detected by the direct cloning, particularly of those miRNAs at low expression level and/or spatiotemporal expression. As of now, computational approaches have been successfully developed and have accurately predicted miRNA genes in Arabidopsis, rice, and other plant species [15–20]. Moreover, many miRNAs in miRBase have been contributed through computational approach only [21].
Perennial ryegrass is one of the high quality grasses, which was grown and utilized widely in China, especially in southern areas. It can be utilized as fresh, hay, and silage [22]. The wide utilization of perennial ryegrass played an important role in the adjustment of agriculture production system. Meanwhile, perennial ryegrass is grown as a forage crop in many parts of the world [23]. To date, a large number of miRNAs have been reported in many species, but none for perennial ryegrass. In this study, we used the characteristic features of previously known other plants miRNAs to efficaciously prediction novel perennial ryegrass miRNA in the publicly available GSS, EST, and nucleotide database. A total of 12 novel miRNAs were firstly identified belonging to 11 miRNA families, to help us understand the biological processes in which they might be involved, and the miRNA potential targeted genes were also predicted. These findings will be useful for further functional analysis of perennial ryegrass miRNA during development. Novel predicted miRNAs were further verified by RT-PCR experiment, an effective and widely-used method for detecting miRNAs.
Materials and Methods
Referenced Sequence Data
To search for potential perennial ryegrass miRNAs, the known plant miRNAs (http://www.mirbase.org/; Version 20, released June 2013) were used as nucleotide reference set of miRNA sequences. To avoid the redundant or overlapping miRNAs, the repeat mature miRNA sequences were removed, and the remaining sequences were used as query sequences for BLAST search. The 29,293 ESTs, 308 GSSs, and 1,768 nucleotide sequences of perennial ryegrass were obtained from NCBI database (http://www.ncbi.nlm.nih.gov/).
Computational Prediction of miRNAs
A stable version of the BLAST tool was downloaded from NCBI database. BLASTN parameters were the same as that described in the previous papers [24, 25]. The procedure of search for potential miRNAs in the perennial ryegrass was showed in Fig. 1. Four criteria were used to distinguish miRNAs and pre-miRNAs from other kinds of RNAs were as follows: (a) predicted mature miRNAs were allowed to have only 0–4 nucleotide mismatches in sequence with all previously known plant mature miRNAs; (b) pre-miRNAs sequence can fold into an appropriate hairpin secondary structure that contains the ∼22 nt mature miRNA sequence within one arm of the hairpin; (c) miRNA precursors with secondary structures had higher negative minimal free energies (MFEs) and minimal free energy index (MFEIs) than other different types of RNAs by RNA-fold prediction; and (d) pre-miRNA had 15–70 % contents of A+U by SVM (support vector machine) since the unstable structures of pre-miRNAs are needed to produce mature single-stranded miRNAs [26].
Conservation and Phylogenetic Analyses of miRNAs
The candidate miRNAs were analyzed for conservation with their orthologues. The newly identified miRNA156k and the well-known other plant miRNAs orthologues were selected for conservation and were done with the help of publically available web logo: a sequence logo generator [27]. The phylogenetic analyses were investigated for their evolutionary relationships (www.clustal.org/). Evolutionary distances were calculated neighbor-joining (NJ) method following 1,000 bootstrapped replicates. All the analyses were performed using the MEGA 4.0 software [28].
Experimental Verification of Predicted miRNAs
The efficiency of the computational strategy was tested by biological experiments to validate the predicted miRNA genes. The four predicted miRNAs genes were randomly selected from perennial ryegrass for RT-PCR validation. The small RNA samples from perennial ryegrass young leave were isolated using mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer’s instruction. The cDNAs were synthesized from small RNAs by using miRNA specific stem–loop primers according to criteria mentioned as described previously [29, 30]. The stem–loop RT primers and gene specific primers were listed in Table S1. The DNA fragments were directly subcloned into pMD18-T vector (Takara) and sequenced.
The analysis of mature miRNA expression by RT-PCR was carried out. The cDNAs were diluted ten times to perform PCR for expression confirmation and expression pattern analysis. PCRs were performed, respectively, in 20 μl mixture containing 1 μl cDNA, 0.5 μM forward and reverse primers, 10 × PCR buffer, 0.25 μM each of dNTPs (Takara), and 2∪Taq polymerase (Takara), under the following parameters: 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The PCR products were detected by electrophoresis with 3 % agarose gel containing ethidium and photographed under UV light.
Prediction of Potential miRNA Target Genes
In plant, it has been reported that most miRNAs bind to the protein coding region of mRNA targets with perfect or nearly perfect sequence complementarities [31, 32]. The mRNA database of the perennial ryegrass downloaded from NCBI database. The targets were predicted with a plant miRNA potential target by the RNAhybrid program (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) [33]. The parameters employed were described as follows: P value cutoff of 0.05, target duplex free energy△G ≤ −24 kcal/mol. The following criteria were set for predicting the potential perennial ryegrass miRNA target genes: (a) not more than four mismatches between identified miRNA; (b) one mismatch was allowed between position 2nd and 12th; and (c) not more than two consecutive mismatches.
Results and Discussion
Identification of Potential miRNAs
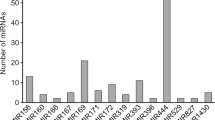
In the plant kingdom, a substantial number of miRNAs are conserved in different plant species. After screening, a total of 12 potential perennial ryegrass miRNAs were predicated for the first time (Table 1). All of the precursors for those mature miRNAs fold into the typical secondary structure of miRNAs, and they are postulated to be important validation parameters for the miRNA genes predicted (Fig. 2). The lengths of the precursors vary in a larger range from 61 to 148 nt with an average of 112 nt. The diversity of the identified miRNAs could be also found in the location of mature miRNA sequences. The sequences of miRNA396h, miRNA156i, miRNA845a, miRNA5021, miRNA6245, and miRNA2937 were located at the 3′ end of the miRNA precursors; whereas, the rest of the sequences of miRNAs were all found at the 5′ end. Predicated miRNAs belong to 11 miRNA families and every miRNA family only has one member, but miRNA156 family has two members. These perennial ryegrass miRNAs were also evaluated for their A + U content, and the results showed that the A + U contents ranged from 18.6 % to 66.8 % in the perennial ryegrass miRNA precursors, which was consistent with previous studies on other plants [24, 34–36]. The identified potential perennial ryegrass miRNAs also have highly negative minimal fold energies (MFEs) between −64.2 and −18.8 kcal/mol. These results showed that the predicted miRNAs were taken with strict screening criteria.
Conservation and Phylogenetic Studies of miRNAs
The perennial ryegrass miRNA156k (Lolium perenne miRNA156K, lpe-MIR156K) conservation and phylogenetic studies were conducted with their orthologues in Oryza sativa (osa), Zea mays (zma), Populus trichocarpa (ptc), Glycine max (gma), Malus domestica (mdm), Manihot esculenta (mes), and Solanum tuberosum (stu). These findings suggest conservation of these miRNAs among dicotyledon and monocotyledon as shown in (Fig. 3). The phylogenetic analysis of the same miRNA (miRNA156k) sequences suggested that L. perenne is more closed to Solanum tuberosum (Fig. 4).
Experimental Verification of Predicted miRNAs
To further validate the expression of the identified miRNAs in perennial ryegrass, RT-PCR analyses were carried out using total RNA isolated from young leave, an effective and widely-used method for detecting miRNAs [37]. The randomly selected four miRNAs, miRNA5021, miRNA396h, miRNA156k, and miRNA5075, were subjected to experimental studies. Results show that the transcripts of the four miRNA genes were successfully detected, demonstrating a high accuracy rate for the computational miRNAs identification (Fig. 5).
Prediction of Potential Targets of miRNAs
The miRNAs target identification is an interesting and demanding step for the new identified miRNAs [33]. The knowledge on target function of the identified perennial ryegrass miRNA will help us gain insight into the important function and regulation of miRNAs in this plant. In our study, a total of 33 targets were identified for the 12 newly identified perennial ryegrass miRNAs (Table 2). Our prediction of target genes for the perennial ryegrass miRNAs revealed that more than one gene was regulated by individual miRNA. This result was similar to the findings in other plant species which suggested that miRNA research should be focused on networks rather than individual connections between miRNA and strongly predicted targets [38, 39]. These targets of perennial ryegrass miRNAs can be separated into several groups. The first group contains targets that are predicted to encode transcription factors. Another group contains miRNA targets encoding a range of different proteins which may play important roles in the aspects of metabolisms, stress response, and signal transduction. The transcription factors are the famous and well-known class of proteins targeted by miRNAs in almost all plant and animal species [40, 41]. The novel identified perennial ryegrass miRNAs also target this class of proteins. For example, the predicted perennial ryegrass targets for miRNAs, 2937 and 3980b-3p, are CONSTANS-like protein, MYB transcriptional regulator. Overall, these findings made us clear that perennial ryegrass miRNAs targeted both transcription factors as well as others specific genes.
Conclusion
With the availability of sequence resources in public databases, computer-based miRNA identification methods have been focused more and more in the recent years due to its advantages of low cost and high efficiency. In the present study, with a computational approach, 12 miRNAs were identified from the EST and GSS databases of perennial ryegrass, which belong to 11 families where miRNA156 family has 2 members and the rest has a single member in each. A total of 33 potential targets were identified, and we found that most of the genes are involved in transcriptional regulation and metabolism, suggesting their essential role in biological processes of perennial ryegrass. The 4 miRNAs out of the 12 that were randomly selected were verified by RT-PCR. Taken together, the knowledge gained from this research will provide an understanding of the essential roles of miRNAs in perennial ryegrass growth and development, stress response, and other biological processes. Our study findings also considerably broaden the scope of understanding the function of miRNA in near future in perennial ryegrass.
References
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell, 136, 669–687.
Tang, G. (2010). Plant microRNAs: an insight into their gene structures and evolution. Seminars in Cell and Developmental Biology, 21, 782–789.
Shukla, L. I., Chinnusamy, V., & Sunkar, R. (2008). The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochimica et Biophysica Acta, 1779, 743–748.
Wu, G. (2013). Plant microRNAs and development. Journal of Genetics and Genomics, 40, 217–230.
Naqvi, A. R., Sarwat, M., Hasan, S., & Roychodhury, N. (2012). Biogenesis, functions and fate of plant microRNAs. Journal of Cellular Physiology, 227, 3163–3168.
Sunkar, R. (2010). MicroRNAs with macro-effects on plant stress responses. Seminars in Cell and Developmental Biology, 21, 805–811.
Valoczi, A., Varallyay, E., Kauppinen, S., Burgyan, J., & Havelda, Z. (2006). Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant Journal, 47, 140–151.
Min Yang, Z., & Chen, J. (2013). A potential role of microRNAs in plant response to metal toxicity. Metallomics, 5, 1184–1190.
Sunkar, R., Li, Y. F., & Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends in Plant Science, 17, 196–203.
de Lima, J. C., Loss-Morais, G., & Margis, R. (2012). MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genetics and Molecular Biology, 35, 1069–1077.
Arteaga-Vazquez, M., Caballero-Perez, J., & Vielle-Calzada, J. P. (2006). A family of microRNAs present in plants and animals. Plant Cell, 18, 3355–3369.
Zeng, C., et al. (2010). Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Research, 38, 981–995.
Zhang, B. H., Pan, X. P., Wang, Q. L., Cobb, G. P., & Anderson, T. A. (2005). Identification and characterization of new plant microRNAs using EST analysis. Cell Research, 15, 336–360.
Sunkar, R., & Jagadeeswaran, G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology, 8, 37.
Lindow, M., & Krogh, A. (2005). Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics, 6, 119.
Kim, H. J., Baek, K. H., Lee, B. W., Choi, D., & Hur, C. G. (2011). In silico identification and characterization of microRNAs and their putative target genes in Solanaceae plants. Genome, 54, 91–98.
Patanun, O., Lertpanyasampatha, M., Sojikul, P., Viboonjun, U., & Narangajavana, J. (2013). Computational identification of microRNAs and their targets in cassava (Manihot esculenta Crantz.). Molecular Biotechnology, 53, 257–269.
Dong, Q. H., et al. (2012). Computational identification of MicroRNAs in strawberry expressed sequence tags and validation of their precise sequences by miR-RACE. Journal of Heredity, 103, 268–277.
Wang, X. J., Reyes, J. L., Chua, N. H., & Gaasterland, T. (2004). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biology, 5, R65.
Archak, S., & Nagaraju, J. (2007). Computational prediction of rice (Oryza sativa) miRNA targets. Genomics, Proteomics & Bioinformatics, 5, 196–206.
Griffiths-Jones, S., Saini, H. K., van Dongen, S., & Enright, A. J. (2008). miRBase: tools for microRNA genomics. Nucleic Acids Research, 36, D154–158.
Wims, C. M., McEvoy, M., Delaby, L., Boland, T. M., & O'Donovan, M. (2013). Effect of perennial ryegrass (Lolium perenne L.) cultivars on the milk yield of grazing dairy cows. Animal, 7, 410–421.
McEvoy, M., O'Donovan, M., & Shalloo, L. (2011). Development and application of an economic ranking index for perennial ryegrass cultivars. Journal of Dairy Science, 94, 1627–1639.
Dezulian, T., Remmert, M., Palatnik, J. F., Weigel, D., & Huson, D. H. (2006). Identification of plant microRNA homologs. Bioinformatics, 22, 359–360.
Han, Y., et al. (2009). Computational identification of microRNAs and their targets in wheat (Triticum aestivum L.). Science in China. Series C, Life Sciences, 52, 1091–1100.
Xu, J. H., Li, F., & Sun, Q. F. (2008). Identification of microRNA precursors with support vector machine and string kernel. Genomics, Proteomics & Bioinformatics, 6, 121–128.
Crooks, G. E., Hon, G., Chandonia, J. M., & Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Research, 14, 1188–1190.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599.
Kou, S. J., et al. (2012). Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Reports, 31, 2151–2163.
Mohammadi-Yeganeh, S., et al. (2013). Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Molecular Biology Reports, 40, 3665–3674.
Numnark, S., Mhuantong, W., Ingsriswang, S., & Wichadakul, D. (2012). C-mii: a tool for plant miRNA and target identification. BMC Genomics, 13(Suppl 7), S16.
Debernardi, J. M., Rodriguez, R. E., Mecchia, M. A., & Palatnik, J. F. (2012). Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genetics, 8, e1002419.
Rehmsmeier, M., Steffen, P., Hochsmann, M., & Giegerich, R. (2004). Fast and effective prediction of microRNA/target duplexes. RNA, 10, 1507–1517.
Zhang, W., Luo, Y., Gong, X., Zeng, W., & Li, S. (2009). Computational identification of 48 potato microRNAs and their targets. Computational Biology and Chemistry, 33, 84–93.
Meyers, B. C., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell, 20, 3186–3190.
Qiu, C. X., et al. (2007). Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene, 395, 49–61.
Mou, G., Wang, K., Xu, D., & Zhou, G. (2013). Evaluation of three RT-qPCR-based miRNA detection methods using seven rice miRNAs. Bioscience, Biotechnology, and Biochemistry, 77, 1349–1353.
Milev, I., Yahubyan, G., Minkov, I., & Baev, V. (2011). miRTour: Plant miRNA and target prediction tool. Bioinformation, 6, 248–249.
Jasinski, S., Vialette-Guiraud, A. C., & Scutt, C. P. (2010). The evolutionary-developmental analysis of plant microRNAs. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 365, 469–476.
Mallory, A. C., & Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nature Genetics, 38(Suppl), S31–36.
Sun, G. (2012). MicroRNAs and their diverse functions in plants. Plant Molecular Biology, 80, 17–36.
Acknowledgments
This research was supported by Natural Science Foundation of China (31302013) and Doctoral Science Foundation (09001578) and Natural Science Foundation (13000904) of Henan University of Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 36 kb)
Rights and permissions
About this article
Cite this article
Huang, Y., Zou, Q., Sun, X.H. et al. Computational Identification of MicroRNAs and Their Targets in Perennial Ryegrass (Lolium perenne). Appl Biochem Biotechnol 173, 1011–1022 (2014). https://doi.org/10.1007/s12010-014-0891-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-0891-5