Abstract
The genes of CS-2 lipase and its cognate foldase were cloned from Pseudomonas aeruginosa CS-2. A stop codon was not found in the lipase gene. The amino acid sequence deduced from the lipase gene from P. aeruginosa CS-2 showed 97.8%, 71.3%, and 71.2% identity with lipases from P. aeruginosa LST-03, P seudomonas mendocina ymp, and Pseudomonas stutzeri A1501, respectively. The co-expression of CS-2 lipase and its cognate foldase of P. aeruginosa CS-2 in E scherichia coli BL21 (DE3) resulted in the formation of a soluble lipase. The recombinant lipase and foldase were purified to homogeneity using nickel affinity chromatography and about 10.2-fold with 40.9% recovery was achieved for the purification of the recombinant lipase. The molecular masses of the lipase and the foldase were estimated to be 35.7 and 38.3 kDa in SDS-PAGE, respectively. The recombinant lipase showed stability in the presence of some organic solvents. The recombinant CS-2 lipase was immobilized and subsequently used for the synthesis of butyl acetate in heptane. The conversion of substrate decreased from 98.2% to 87.4% after 5 cycles in reuse of the immobilized lipase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases (E.C.3.1.1.3) are versatile catalysts and they are being increasing exploited for all kinds of fields including detergents, dairy, diagnostics, oil and lipid processing, and biotransformation [1]. In the last decade, the use of lipases in the presence of organic solvents has dramatically increased in pharmaceuticals, fine chemicals, and agrochemicals [2–4]. Therefore, searching for organic solvent-tolerant lipase is becoming an attractive research field.
By laborious effort, some strains producing organic solvent-tolerant lipases were successfully obtained [5–10]. Furthermore, owing to the pathogenicity of strains and/or the requirement of higher lipase productivity, some lipase genes of these strains were expressed in heterogeneous host [11–15]. However, most of these lipases were overexpressed in the bacterial cytoplasm as inactive inclusion bodies. A refolding procedure was necessary to get active enzymes. One strategy to decrease the production of inclusion bodies is co-expression of a lipase gene and its chaperonin gene [16]. It is believed that some lipases require lipase-specific foldase. These foldases are usually encoded in one operon with their cognate lipases and are able to catalyze the folding of lipases into enzymatically active forms [17]. To date, few reports have been published regarding the expression of lipase genes in Escherichia coli as enzymatically active forms in vivo [18, 19].
Many flavor esters are widely used in the food industry and as solvents and intermediates in chemical and pharmaceutical section. For example, butyl acetate not only contributes strong fruity flavors but is useful in cold lacquers for wood furniture and the topcoats of vehicles and in cellulose paints [20]. Dandavate et al. reported the synthesis of ethyl butyrate ester using the BMV2 lipase immobilized in AOT-based organogel [10]. The production of butyl acetate was investigated by Salah et al. with R hizopus oryzae lipase immobilized in Celite 545 [21].
In the present paper, the genes of lipase and foldase in Psedomonas aeruginosa CS-2, a novel strain isolated from soil, were cloned and co-expressed in E. coli BL21 (DE3). The recombinant lipase and foldase were purified using affinity chromatography, and the purified lipase was immobilized and reuse of the immobilized lipase to synthesize butyl acetate in heptane was attempted.
Materials and Methods
Strains, Plasmids, and Materials
P. aeruginosa CS-2 was isolated from soil. PTG-19T vector (Generay, China) and E. coli XL10-gold (Agilent, USA) were used for gene cloning. E. coli BL21 (DE3) competent cells were purchased from Tiangen and pET28a vector was obtained from Novagen. Ex-Taq DNA polymerase, T4 DNA ligase, restriction endonucleases, and DNA and protein markers were purchased from TaKaRa Biotechnology (Dalian, China). HisTrap FF chromatography columns were from GE Healthcare. Kieselguhr (Celite-545) was purchased from Celite. All the other chemicals were of analytical grade or higher grade. Lipase AYS and Lipase AK were from Amano Enzyme Incorporation.
Preparation of Genomic DNA from P. aeruginosa CS-2
P. aeruginosa CS-2 was grown in a medium (0.5% peptone, 0.3% yeast extract, 0.5% glucose, 0.25% NaCl, 0.05% MgSO4·7H2O, pH 7.5) at 30 °C for 24 h with reciprocal shaking (180 r/min). The cells were harvested by centrifugation at 12,000 r/min for 15 min. Genomic DNA was extracted by using a TIANamp Bacteria DNA Kit (Tiangen Biotech). The purity of genomic DNA was confirmed by agarose gel electrophoresis and the ratio of OD260 and OD280 of DNA.
Cloning and Sequencing of the Genes of Lipase and Foldase in P. aeruginosa CS-2
The genes of lipase and foldase were amplified by polymerase chain reaction using genomic DNA from P. aeruginosa CS-2 as the template. The primers used were 5′-CCTTGGATCCATGAAGAAGAAGTCTCTGCTCC-3′ (forward primer) and 5′-CCTTAAGCTTGCGCTGCTCGGCCTGGCGCAT-3′ (reverse primer). The underlined sequences indicated restriction sites for BamHI and HindIII, respectively. The polymerase chain reaction (PCR) was performed with a cycle for initial denaturation (95 °C, 2 min), 30 cycles at 95 °C (20 s), 62 °C (20 s), and 72 °C (2 min) for amplification and a final elongation cycle (72 °C, 5 min). The PCR products were purified from agarose gels with a quick gel extraction kit (Biodevtech). The purified DNA fragments were ligated with PTG-19T vector (Generay) and the ligated products were then transformed into E. coli XL10-gold. The plasmids of positive clones were extracted and then sequenced.
Co-expression of the Genes of Lipase and Foldase in E.coli BL21 (DE3)
The purified DNA fragments and expression vectors pET28a were digested with BamHI and HindIII. The resulting segments were purified with a quick gel extraction kit (Biodevtech) followed by ligation with T4 DNA ligase to construct recombinant plasmids. The recombinant plasmids were then transformed into E. coli XL10-gold. The recombinant plasmids of positive clones were extracted, which were subsequently transformed into E. coli BL21 (DE3). The transformed E. coli was precultured in a modified LB medium (adding 1% glucose) containing kanamycin (50 μg/ml) at 37 °C with reciprocal shaking (190 r/min) overnight. The resulting cultures were transferred (1%, v/v) into TM medium containing kanamycin (50 μg/ml) and grown at 37 °C with shaking at 190 r/min until OD600 reached 1.0. The cells were induced with 0.05 mM isopropyl-β-d-thio-galactoside (IPTG) and were grown at 20 °C for 8 h after induction. The cells were harvested by centrifugation at 6,000 r/min for 10 min at 4 °C. The total proteins of E. coli BL21 (DE3) and recombinant E. coli BL21 were examined by denaturing polyacrylamide gel electrophoresis.
Expression of Lipase Gene in E.coli BL21 (DE3)
The gene of lipase was amplified by PCR using the genomic DNA of the isolated strain as templates. The primers used were as follows: forward, 5′-CCTTGGATCCATGAAGAAGAAGTCTCTGC-3′ and reverse, 5′-CCTTAAGCTTTTCCGGGAAAGGGCCGGG-3′ .The underlined sequences indicated restriction sites for BamHI (GGATCC) and HindIII(AAGCTT). The PCR began with an initial denaturation for 2 min at 94 °C. Twenty cycles were run, each cycle consisting of 20 s of denaturation at 94 °C, 20 s of annealing at 60 °C, and 50 s of extension at 72 °C followed by a final extension of 3 min at 72 °C. Subsequently, the gene of lipase was individually expressed according to the method described above.
Purification of Soluble Recombinant Protein
About 12 g of wet cells were resuspended in 150 ml of 50 mM Tris–HCl buffer. The cells were then disrupted by sonication. Cell debris was removed by centrifugation at 10,000 r/min for 10 min at 4 °C. The supernatant (300 μl) was loaded onto a HisTrap FF column equilibrated with 20 mM Tris–HCl buffer (pH 7.9) containing 500 mM NaCl and 10 mM imidazole. The column was then washed extensively with 15 ml of the abovementioned buffer to remove unspecific and unbound proteins. The enzymes were eluted with 5 ml of 20 mM Tris–HCl buffer (pH 7.9) containing 500 mM NaCl and 500 mM imidazole at a flow rate of 0.2 ml/min, and the eluted fractions were collected and dialyzed to remove imidazole. The purified enzymes were examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
SDS Polyacrylamide Gel Electrophoresis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed according to the method of Laemmli in a 12% (w/v) polyacrylamide slab gel. The reference proteins were phosphorylase b 97.2 kDa, bovine serum albumin 66.4 kDa, albumin 44.3 kDa, carbonic anhydrase 29.0 kDa, trypsin inhibitor 20.1 kDa, and lysozyme 14.3 kDa [22].
The Tolerance of the Recombinant Lipase and Commercial Lipases Against Various Organic Solvents
The tolerance of the recombinant lipase and commercial lipases (lipases AYS and AK) against various organic solvents was investigated on the following: 1 ml of various organic solvents was added to 1 ml of lipase solution in a sealed glass vial, respectively. The mixture was incubated for 9 and 24 h at 30 °C with shaking at 150 rpm, and then the remaining lipase activity was checked. The relative activity was calculated by regarding the control (no organic solvents in the mixture) as 100%. All assays were performed in triplicates.
Protein Assay
The protein concentration was determined by Bradford dye method using bovine serum albumin as a standard [23].
Assay of Lipase Activity
Lipase activity was assayed using p-NPP as a substrate. Briefly, the substrate was dissolved in 2-propanol (20 mM), mixed with 3 ml Tris–HCl buffer (50 mM, pH 8), followed by incubating them at 50 °C for 5 min. The reaction was initiated by adding 50 μl of a suitably diluted lipase solution or immobilized lipase to the mixture and allowed to proceed at 50 °C for 10 min (the initial velocity was almost constant for up to 20 min). Tris–HCl buffer in place of enzyme was used as the experimental blank for lipase activity assay. The reaction was then terminated by adding 1 ml SDS (0.05%, w/v). The absorbance of liberated p-nitrophenol was recorded at 410 nm. One unit of lipase activity was defined as the amount of enzyme liberating 1 μmol p-nitrophenol per minute under standard assay conditions.
Lipase Immobilization
Kieselguhr (Celite-545) was washed three times with 0.15 M NaOH and 0.15 M HCl, respectively. Celite-545 was then rinsed with deionized water and air-dried at room temperature. Five grams of Celite-545 was added to 50 ml lipase solution, and the mixture was stirred for 4 h at 25 °C under shaking condition (150 rpm). The immobilized lipase was filtered, rinsed three times with deionized water, and dried in a vacuum desiccator. The activity of the immobilized lipase was assayed to be 150.7 U/g.
Reuse of the Immobilized Lipase to Synthesize Butyl Acetate in Heptane
Acetic acid, n-butanol, and heptane were dehydrated by 4 Å molecule sieves. The reaction mixture (10 ml) in a 50-ml sealed round bottom flask contained 0.1 mol/L acetic acid and 0.2 mol/L n-butanol and heptane. The synthesis of butyl acetate was performed at 55 °C under shaking condition (200 rpm) after the addition of 500 mg of immobilized lipase to the reaction mixture. Five hundred milligrams of 4 Å molecular sieves was added to remove water after 8 h. The samples from the reaction mixture were withdrawn and analyzed after 10 h. After each use, the immobilized lipase was separated from the reaction mixture, washed three times with heptane, and used for next cycle.
Assay of Acetic Acid Conversion
The residual acetic acid concentrations were analyzed by volumetric titration [24] using the following formula: acetic acid conversion (in percent) = (initial acetic acid concentrations − residual acetic acid concentrations)/initial acetic acid concentrations × 100.
Results and Discussion
C-Terminal Sequence of CS-2 Lipase from P. aeruginosa CS-2
P. aeruginosa CS-2 was used to produce an organic solvent-tolerant lipase [25]. The accession number of 16S rDNA of P. aeruginosa CS-2 in GenBank was GQ254065. After ultra-filtration, acetone precipitation, and DEAE-Sephadex A50 chromatography, CS-2 lipase was purified. Homogeneity of purified lipase was confirmed by SDS-PAGE. The lipase on the gel was analyzed to determine its C-terminal sequence. The C-terminal sequence of CS-2 lipase was KNASLWDPGRG. Generally, 74–97% of total proteins can be determined by the five residues of C-terminal sequence. No protein was identical with the C-terminal sequence of CS-2 lipase using blastp. The C-terminal region to the conserved α/β hydrolase (i.e., lipase) fold is quite divergent in structure and may confer specific properties to each of the individual enzymes. The C-terminal of Fusarium heterosporum lipase tightened the lipase structure and enhanced lipase production in the post-transcriptional step [26]. Amino acid substitution in the C-terminal region improved the optimum temperature for Rhizopus niveus lipase [27].
Cloning and Analysis of the Genes of CS-2 Lipase and Foldase
Based on the known gene sequences of lipases and foldases from other P. aeruginosa strains, a pair of primers was designed. Restriction sites for BamHI (GGATCC) and HindIII (AAGCTT) were also included in the primers for the following expression of the targeted genes. Using the primers, a DNA fragment of about 2,000 bp in length was amplified by PCR using the chromosomal DNA from P. aeruginosa as a template. The purified DNA fragment was ligated into PTG-19T vector followed by the transformation of the recombinant plasmid into E. coli XL10-gold. Plasmids of positive clones were extracted and the cloned fragment was confirmed by sequencing. The sequence of the cloned fragment was submitted to GenBank with the accession number GU220567. For the cloned lipase gene, a stop codon was not found upstream of a GC-rich palindrome sequence and multiple contiguous thymines, which were indicators of transcription termination. To our knowledge, there were few genes without a terminator codon. Yomo et al. reported that there were no stop codons in the antisense strands of the genes for nylon oligomer degradation [28]. Monnerjahna et al. also reported that a long non-stop reading frame existed on the antisense strand of the grp78 gene of Neurospora crassa, but was not transcribed [29]. To determine the C-terminal sequence of CS-2 lipase, the purified lipase was analyzed by TOF. The sequence corresponding to the C-terminus of CS-2 lipase (KNASLWDPGRG) was located on one side of the hairpin loop within the mRNA transcribed from CS-2 lipase gene. Generally, if the message of an mRNA has no stop codon, the ribosomes stalled on mRNA are rescued by tmRNA, and tmRNA encodes a peptide tag that is incorporated at the end of the aberrant polypeptide and targets it for proteolysis [30]. The stable existence of CS-2 lipase in cells suggested that it was not doomed for proteolysis and another mechanism could work.
The complete amino acid sequence of CS-2 lipase was shown in Fig. 1. The amino acid sequence deduced from the lipase gene from P. aeruginosa CS-2 showed 97.8%, 71.3%, and 71.2% identity with the lipases from P. aeruginosa LST-03, Pseudomonas mendocina ymp, and Pseudomonas stutzeri A1501, respectively. There are a catalytic triad, a conserved G-X-S-X-G sequence and two Asp residues involved in calcium binding among all of the four kinds of lipases. Furthermore, CS-2 lipase has two cysteine residues forming a disulfide bridge which was conserved in subfamily 1.1 and 1.2 lipases [18].
For the CS-02 foldase gene, GTG was used as an initiation codon. CS-02 foldase contains three amino acid residue differences (E126G, A184T, and A210T) as compared to LST-03 foldase.
Co-Expression of Lipase and Foldase of P. aeruginosa CS-2 and Expression of Lipase Individually in E. coli BL21 (DE3)
After the digestion of the amplified fragment by BamHI and HindIII, the genes of CS-2 lipase and foldase were inserted into pET28a vector. To prevent the formation of inclusion bodies, the recombinant cells were cultured at a low temperature (20 °C) after the addition of IPTG, for a higher temperature (e.g., 37 °C) gave rise to easy formation of inclusion bodies [13]. Upon induction with IPTG, both lipase and foldase were expressed in the recombinant E. coli BL21 (DE3) as shown in SDS-PAGE gel (Fig. 2). The co-expression of lipase and foldase in E. coli BL21 (DE3) resulted in the formation of a functional lipase which was confirmed by the activity assay (Table 1). However, no lipase activity was detected in the supernatant of the recombinant E. coli BL21 (DE3) culture. This phenomenon was due to lack of the Xcp-secretion machinery in E. coli BL21 (DE3) [31]. As a result, CS-2 lipase did not secrete into the growth medium. Rosenau et al. reported the secretion of subfamily 1.1 and 1.2 lipases did not proceed properly in heterogonous hosts [17]. Ogino et al. also reported that when the transformant-harboring pLC9, a recombinant plasmid containing LST-03 lipase gene and its foldase gene, was cultured in the LB broth, the lipolytic activity was not detected in the culture supernatant [13]. To our knowledge, there have been only limited researches on the expression of P. aeruginosa 1ipase in E. coli as enzymatically active forms in vivo, although the active expression of various Pseudomonas lipases has been achieved by in vitro refolding with different approaches [13, 32, 33]. In one paper about the functional expression of P. aeruginosa 1ipase in vivo by Madan et al., both the gene of lipase (936 bp) and the gene of foldase (867 bp) were inserted into pET32Xa/LIC and pET29a vectors under the influence of strong T7 promoter, respectively. Upon induction with IPTG, an active lipase was obtained in the E. coli BL21 (DE3) containing pETlipA and pETlipB [19].
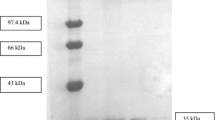
SDS-PAGE pattern of the total proteins of E. coli BL21 (DE3) and the recombinant E. coli BL21 (DE3). Standard proteins of different molecular masses (lane 1), total proteins of E. coli BL21 (DE3) (lanes 2 and 3) and total proteins of recombinant E. coli BL21 (DE3) (lanes 4 and 5) were run in SDS-PAGE (12%) and stained in Coomassie Brilliant blue
When lipase gene was individually expressed in E. coli BL21 (DE3), no activity was found in the supernatant of the disrupted recombinant E. coli BL21 (DE3). These results suggested that foldase was necessary for the formation of an active soluble lipase.
Purification of the Recombinant Lipase and Foldase
The purification of the recombinant lipase and foldase was performed using affinity chromatography (His-tag chromatography). The recombinant lipase was specifically bound to His-tag column due to the presence of His residues at the N-terminus, while the foldase had His residues at the C-terminus. About 10.2-fold with 40.9% recovery was achieved using His-tag chromatography (Table 1). The purified lipase and foldase were confirmed in SDS-PAGE (Fig. 3). The molecular masses of the purified lipase and foldase were determined to be 35.7 and 38.3 kDa, which coincided approximately with the theoretical values calculated from amino acid sequence.
The purification of other organic solvent-tolerant lipases from recombinant E. coli was also reported. Zhang et al. purified a lipase from recombinant E. coli using saturation ammonium sulfate precipitation, Ni-NTA agarose affinity chromatography, and Sephacryl S-200 gel filtration chromatography with 13.5-fold purification and 53.5% yield [18]. Madan et al. succeeded in the purification of an organic solvent-tolerant lipase from engineering E. coli and about 16-fold with 35% recovery was achieved by a single-step purification method using His-tag chromatography [19].
The Tolerance of the Recombinant Lipase and Commercial Lipases Against Various Organic Solvents
Lipases have been increasingly exploited for organic synthesis. The activity and stability of lipase in organic solvents are important for synthetic transformations.
The tolerance of the recombinant lipase against various organic solvents was investigated. The results indicated the recombinant lipase responded differently to different organic solvents (Table 2), which were similar to the effects of various organic solvents on CS-2 lipase. The activity of the recombinant lipase was increased by 22.9% in the presence of acetonitrile after 9 h of incubation, while it was improved by 10.4% in the presence of acetonitrile after 24 h of incubation. Moreover, 62.1% and 78.1% of original activity were retained after 9 h of incubation in the presence of dimethyl sulfoxide (DMSO) and petroleum ether, respectively. The tolerance of the commercial lipase including lipases AYS and AK (un-immobilized lipases) against various organic solvents was also investigated in order to compare the recombinant lipase with commercial lipases. Table 2 showed that the recombinant lipase had an improved tolerance over lipases AYS and AK in the presence of benzene, chloroform, petroleum ether, isooctane, and DMSO after 9 h and 24 h of incubation.
Lipase tolerance to organic solvents differed from one another. The solvent-tolerant lipase from Bacillus sphaericus 205y lost approximately 80% of activity in DMSO and was completely inactivated in acetonitrile [6]. The solvent-tolerant lipase from Burkholderia multivorans V2 was stable in n-hexane up to 24 h retaining about 97.8% of original activity which is further reduced to 53.2% after 48 h of incubation. However, the relative activity of the lipase lowered to about 77.7% after 24 h of incubation and further to about 48.5% after 48 h of incubation in isooctane [10].
Reuse of the Immobilized Lipase to Synthesize Butyl Acetate in Heptane
Repeated use of immobilized enzyme might help to drive down the product cost and make the enzymatic process economically viable. In this study, a method of adsorption was used to immobilize lipase. Celite-545 showed the maximum lipase-binding efficiency (18.5%) with 150.7 U/g of immobilized lipase activity (Fig. 4). The immobilized enzymes were repeatedly used as the biocatalyst for synthesis reaction and subsequently recovered and reused. The reaction variables such as reaction time, molar ratio, temperature, and water activity have been optimized (data not shown). Figure 5 showed that no significant decrease in the conversion yield was observed after 5 cycles of use of the immobilized lipase. Acetic acid conversion only decreased from 98.2% to 87.4%. The loss of activity over the reaction cycles was due to the inactivation of lipase over time. Desorption was almost not taken place for the immobilized enzyme in the net organic solvents. The results indicated the feasibility and suitability of the recombinant lipase immobilized on Celite-545 for the production of butyl acetate. It was reported that the partially purified BMV2 lipase exhibited esterification efficiency in the range of 10–50% for the synthesis of ethyl butyrate, while the BMV2 lipase immobilized in AOT-based organogel showed 1.82-fold higher esterification efficiency than free lipase did [10]. Salah et al. reported that butyl acetate in hexane was produced by using immobilized R. oryzae lipase with conversion yields of 80%. No significant decrease in the synthesis activity was observed for the first 3 cycles of reuse of the immobilized lipase [21]. Verma et al. employed celite-immobilized lipase of Bacillus cereus MTCC 8372 to synthesize ethyl acetate. Batch operational stability tests indicated that immobilized lipase had retained 50% of its original catalytic activity after four consecutive batches of 15 h each [34]. The difference of ester synthesis between the immobilized lipases from B. cereus MTCC 8372 and the immobilized recombinant maybe resulted from the difference of organic-solvent tolerance of two immobilized lipases.
Reuse of the immobilized lipase to synthesize butyl acetate in heptane. The synthesis was carried out in n-heptane containing 0.1 mol/L acetic acid and 0.2 mol/L n-butanol in the presence of immobilized lipase (50 g/L) at 200 rpm and 55 °C for 10 h. During the synthesis process, 4 Å molecular sieve (0.5 g) was added as water absorbent in the reaction system at the eighth hour
References
Hasan, F., Shah, A. A., & Hameed, A. (2006). Enzyme and Microbial Technology, 39, 235–251.
Ghanem, A., & Aboul-Enein, H. Y. (2005). Chirality, 17, 1–15.
Houde, A., Kademi, A., & Leblanc, D. (2004). Applied Biochemistry and Biotechnology, 118, 155–170.
Theil, F. (1995). Chemical Reviews, 95, 2203–2227.
Ogino, H., Miyamoto, K., & Ishikawa, H. (1994). Applied and Environmental Microbiology, 60, 3884–3886.
Hun, C. J., Rahaman, R. N. Z. R. A., & Salleh, A. B. (2003). Biochemical Engineering Journal, 35, 147–157.
Rahaman, R. N. Z. R. A., Baharum, S. N., & Salleh, A. B. (2006). The Journal of Microbiology, 44, 583–590.
Zhao, L. L., Xu, J. H., Zhao, J., Pan, J., & Wang, Z. L. (2008). Process Biochemistry, 43, 626–633.
Ruchi, G., Anshu, G., & Khare, S. K. (2008). Process Biochemistry, 43, 1040–1046.
Dandavate, V., Jinjala, J., Keharia, H., & Madamwar, D. (2009). Bioresource Technology, 100, 3374–3381.
Ogino, H., Hiroshima, S., Hirose, S., Yasuda, M., Ishimi, K., & Ishikawa, H. (2004). Molecular Genetics and Genomics, 271, 189–196.
Ogino, H., Mimitsuka, T., Muto, T., Matsumura, M., Yasuda, M., Ishimi, K., et al. (2004). Journal of Molecular Microbiology and Biotechnology, 7, 212–223.
Ogino, H., Katou, Y., Akagi, R., Mimitsuka, T., Hiroshima, S., Gemba, Y., et al. (2007). Extremophiles, 11, 809–817.
Rahaman, R. N. Z. R. A., Chin, J. H., & Salleh, A. B. (2003). Molecular Genetics and Genomics, 269, 252–260.
Long, Z. D., Xu, J. H., Zhao, L. L., Pan, J., Yang, S., & Hua, L. (2007). Journal of Molecular Catalysis B Enzymatic, 47, 105–110.
Quyen, D. T., Schmidt-Dannert, C., & Schmid, R. D. (1999). Applied and Environmental Microbiology, 65, 787–794.
Rosenau, F., & Jaeger, K. E. (2000). Biochimie, 82, 1023–1032.
Zhang, A. J., Gao, R. J., Diao, N. B., Xie, G. Q., Gao, G., & Cao, S. G. (2009). Journal of Molecular Catalysis B Enzymatic, 56, 78–84.
Madan, B., & Mishra, P. (2010). Applied Microbiology and Biotechnology, 85, 597–604.
Park, Y. C., Shaffer, C. E., & Bennett, G. N. (2009). Applied Microbiology and Biotechnology, 85, 13–25.
Salah, R. B., Ghamghui, H., Miled, N., Mejdoub, H., & Youssef, G. (2007). Journal of Bioscience and Bioengineering, 103, 368–372.
Laemmli, U. K. (1970). Nature, 228, 680–685.
Bradford, M. M. (1976). Analytical Biochemistry, 72, 248–254.
Leitgeb, M., & Knez, Ž. (1990). Journal of the American Oil Chemists’ Society, 67, 775–778.
Peng, R., Lin, J., & Wei, D. (2010). Applied Biochemistry and Biotechnology, 162, 733–743.
Nagao, T., Shimada, Y., Sugihara, A., & Tominaga, Y. (1998). Journal of Biochemistry, 124, 1124–1129.
Kohno, M., Enatsu, M., Funatsu, J., Yoshiizumi, M., & Kugimiya, W. (2001). Journal of Biotechnology, 87, 203–210.
Yomo, T., Urabe, I., & Okada, H. (1992). Proceedings of the National Academy of Sciences of the United States of America, 89, 3780–3784.
Monnerjahn, C., Techel, D., Mohamed, S. A., & Rensing, L. A. (2000). FEMS Microbiology Letters, 183, 307–312.
Keiler, K. C., Shapiro, L., & Williams, K. P. (2002). Proceedings of the National Academy of Sciences of the United States of America, 97, 7778–7783.
Akrim, M., Bally, M., Ball, G., Tommassen, J., Teerink, H., Filloux, A., et al. (1993). Molecular Microbiology, 10, 431–443.
Luo, Y., Zheng, Y., Jiang, Z., Ma, Y., & Wei, D. (2006). Applied Microbiology and Biotechnology, 73, 349–355.
Traub, P. C., Schmidt-Dannert, C., Schmitt, J., & Schmid, R. D. (2001). Applied Microbiology and Biotechnology, 55, 198–204.
Verma, M. L., Azmi, W., & Kanwar, S. S. (2009). Acta Microbiologica et Immunologica Hungarica, 56, 229–242.
Acknowledgments
The financial support from the Ministry of Science and Technology of the People’s Republic of China is gratefully acknowledged (project IDs: 2006AA020203 and 2009CB724703).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Peng, R., Lin, J. & Wei, D. Co-Expression of an Organic Solvent-Tolerant Lipase and its Cognate Foldase of Pseudomonas aeruginosa CS-2 and the Application of the Immobilized Recombinant Lipase. Appl Biochem Biotechnol 165, 926–937 (2011). https://doi.org/10.1007/s12010-011-9309-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-011-9309-9