Abstract
The purpose of this work was to study the combined effect of high hydrostatic pressure (HHP) and enzymatic hydrolysis treatment on the hydrolysis and allergenicity of ginkgo seed proteins (GSPs). Four food-grade proteases (papain, alcalase, pepsin, and neutrase) were used, and HHP (200, 300, and 400 MPa separately) was applied prior to hydrolysis. The extent of hydrolysis was measured with the o-phthaldialdehyde method, SDS-PAGE, and MALDI-TOF-MS, and the allergenicity was assessed with a Western blot and enzyme-linked immunosorbent assay (ELISA). The results showed that HHP could significantly improve the extent of proteolysis by papain, alcalase, or pepsin and reduce the antigenicity of GSP, whereas neutrase showed poor effects at any pressure. Papain and alcalase showed the highest proteolysis at 300 MPa, followed by pepsin at 400 MPa, and all of the obtained hydrolysates showed molecular weights lower than 10 kDa; furthermore, papain or alcalase at 300 MPa as well as pepsin at 400 MPa reduced antigenicity by more than 95 %, and all of the immunoreactive bands disappeared in the obtained hydrolysates. These results suggest that HHP can enhance the hydrolysis of GSP by certain enzymes and reduce the residual antigenicity of the hydrolysates. The obtained hypoallergenic hydrolysates could be used as a source of peptides for food ingredients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ginkgo biloba, one of the oldest species of tree, has existed on the earth for two hundred million years, and 70 % of G. biloba is from China (Deng et al. 2011). As a traditional food and medicine source, the seeds of G. biloba have been used in China for several thousand years (Huang et al. 2010). Ginkgo seeds may be added to desserts, glazed fruit, and alcoholic and non-alcoholic beverages. Furthermore, ginkgo seeds are a traditional Chinese medicinal material and have been recorded in the Compendium of Materia Medica (Tredici 1991). Research shows that ginkgo seeds contain relatively high levels (10–15 %) of proteins, which notably contain much higher levels of essential amino acids than the recommended FAO/WHO standard, making them a source of high-quality protein (Zhou et al. 2012). Ginkgo seed proteins (GSPs) exhibit many biological activities, including anti-oxidation, anti-aging, anti-tumor, and anti-bacterial activities; thus, GSPs exhibit high nutritional and medicinal value (Huang et al. 2004). However, eating GSPs could result in allergic reactions in humans, mainly in infants or children, who are more sensitive to GSPs. The clinical symptoms of these reactions include nausea, vomiting, abdominal pain, diarrhea, dysphoria, exanimation, convulsions, dyspnea, mydriasis, and sometimes death (Yang and Wu 2009). However, methods to reduce and eliminate the allergenicity of GSPs have not yet been studied, which has limited the development of hypoallergenic GSPs and the suitable use of this precious protein source in the food industry.
Food processing can alter the allergenic and functional properties of proteins by hiding, destroying, or disclosing allergenic epitopes via changes in the conformation or digestibility of proteins (Takagi et al. 2003). Enzymatic hydrolysis is the most commonly used technological process to reduce the allergenicity of food proteins (Bonomi et al. 2003). During hydrolysis, some of the peptide or disulfide bonds are broken, resulting in the collapse of conformational and sequential epitopes, which consequently reduces or eliminates the allergenicity of food protein (Calvo and Gomez 2002). In addition to their lower antigenicity, protein hydrolysates have many advantages compared with intact proteins, such as improved solubility and facilitated intestinal absorption (Ziegler et al. 1998). A number of studies have evaluated the effects of enzymatic hydrolysis on the allergenicity and functional properties of food proteins, such as whey, soybean, peanut, rice, egg, and chickpea proteins, which were hydrolyzed with trypsin, chymotrypsin, pepsin, alcalase, neutrase, or papain. The results showed that enzymatic proteolysis reduced the allergenicity of these proteins by more than 90 % while improving their functional properties, such as the thermostability, emulsifying activity, solubility, and digestibility (Kananen et al. 2000; Lee et al. 2007; Maleki et al. 2000; Watanabe et al. 1990; Kovacs-Nolan et al. 2000; Cabanillas et al. 2010).
High hydrostatic pressure (HHP), an emerging technology for food processing also known as “cold treatment,” is a valuable alternative to thermal treatments to reduce the immunoreactive properties of food proteins (Peñas et al. 2006c). HHP can modify the tertiary and quaternary structures of proteins and reduce the immunoreactive properties of food proteins (Pereda et al. 2008). Recently, HHP treatment has been widely used in combination with proteolysis to produce hydrolysates with lower residual antigenicity and improved functional properties. Food proteins reportedly exhibit enhanced proteolytic digestibility after HHP. HHP may cause protein unfolding, which can enhance the susceptibility of proteins to enzymatic action by exposing new cleavage sites that allow the proteases to reach otherwise buried hydrolysis sites, thereby generating hydrolysates with lower immunoreactivity (Eisenmenger and Reyes-De-Corcuera 2009). Many studies have reported that HHP enhances enzymatic proteolysis for various proteins, such as whey proteins, soybean proteins, egg proteins, bovine whey proteins, peanut proteins, and β-lactoglobulin (Chicón et al. 2009; Li et al. 2012; Quirós et al. 2007; Peñas et al. 2006a, b; Dong et al. 2011; Stapelfeldt et al. 1996). After enzymatic hydrolysis is assisted by HHP processing, these protein hydrolysates exhibited non-antigenic properties that were superior to those of proteins only treated with enzymatic hydrolysis.
To date, the influence of enzymatic hydrolysis assisted by HHP treatment on the allergenicity and hydrolysis properties of hydrolysates of GSP has not yet been reported. Therefore, this study examined the effects of the enzymatic hydrolysis of GSP by different food-grade enzymes in combination with prior HHP processing on the extent of hydrolysis and the antigenicity of the hydrolysis products to produce hypoallergenic hydrolysates with suitable properties for the food industry.
Materials and Methods
Plant Materials
The seeds of G. biloba were purchased from the Taixin Market of Jiangsu province, and GSPs were obtained according to the method described by Huang et al. (2004). Briefly, the Ginkgo seeds were freeze-dried, crushed, and then defatted with petroleum ether at 4 °C for 10 h. The defatted meal was then air-dried and homogenized in Tris–HCl buffer (Tris-buffered saline (TBS), 0.2 mol/L, pH 7.4). The homogenate was centrifuged at 5000 rpm and 4 °C for 15 min, and the resultant supernatant was then added to ammonium sulfate at a final concentration of 80 % to precipitate proteins. After stirring overnight, the proteins were collected by centrifugation at 5000 rpm for 30 min and then dialyzed with a 3.5 kDa molecular weight cutoff dialysis bag. The dialysate was then freeze-dried to obtain the purified GSP. The GSP powder was dissolved in MilliQ water at a concentration of 50 mg/mL and used as substrate for the enzymatic hydrolysis.
High Hydrostatic Pressure Treatment
A high-pressure apparatus (model UHPF-750 MPa-3 L; KEFA Hitech Food Machine Co. Ltd., Baotou, China) featuring a hydraulic cell with an inner capacity of 3 L (100 mm in diameter and 300 mm in height) and a water jacket for temperature control was used in this study. The water was used as the pressure-transmitting medium in the machine, and its temperature was maintained at 25 °C. During the pressure processing, the GSP solutions were subjected to HHP treatment at 200, 300, or 400 MPa for 20 min. The target pressure was achieved within 1–2 min, held for 20 min, and released to atmospheric pressure within 1–2 min. Each sample was subjected to three treatments.
Enzymatic Hydrolysis Experiments
Hydrolysis was performed at atmospheric pressure (0.1 MPa) for 120 min using untreated substrate and substrate treated with HHP. The GSP samples were subjected to hydrolysis using one of four food-grade enzymes: pepsin (E.C.3.4.23.1, Sigma, USA), neutrase (E.C.3.4.24.28, Sigma, USA), alcalase (E.C.3.4.21.62, Novozymes, Denmark), and papain (E.C. 3.4.22.2, Sigma, USA). Before starting the hydrolysis, the pH of the substrate solution was adjusted to 1.5 for pepsin, 7 for neutrase, 8 for alcalase, and 6.5 for papain. The reactions were performed at the optimal temperatures for each enzyme: 40 °C for pepsin, 45 °C for alcalase and neutrase, and 55 °C for papain. Fifteen milliliters of GSP solution (5 %) and a 1:20 (w/v) enzyme to substrate (E:S) ratio was used for the proteolysis. Blank samples were subjected to the same conditions in the absence of enzyme. Hydrolysis was performed in triplicate, and the reaction was stopped by heating the hydrolysates for 5 min at 90 °C. The hydrolysates were stored at −20 °C until analysis.
Determination of the Degree of Hydrolysis
The degree of hydrolysis was determined by quantifying the α-aminogroups released in the hydrolysis reaction with an o-phthaldialdehyde (OPA) spectrophotometric assay, as described by Church et al. (1983). The OPA solution was prepared by combining 25 mL of 100 mM sodium tetraborate, 2.5 mL of 20 % SDS, 40 mg of OPA in 1 mL of methanol, and 100 μL of β-mercaptoethanol to a total volume of 50 mL. A 50 μL aliquot of the hydrolysates was added to 1 mL of OPA solution. The solution was mixed by inversion and incubated for 2 min at room temperature, and the absorbance was measured at 340 nm in a spectrophotometer (UV-1601, Shimadzu, Japan).
SDS-PAGE Analyses
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to Huang et al. (2010) using a PhastSystem apparatus (Amersham Bioscience, Piscataway, NJ) with ready-made slab gels (PhastGel Gradient 4–20 %) and ready-made buffer strips (PhastGel SDS Buffer Strips). Prior to electrophoresis, each sample was diluted to a concentration of 1 mg/mL with 0.01 M Tris–HCl buffer (pH 8.0) containing 2.5 % SDS, 20 % glycerin, 5 % mercaptoethanol, and 0.5 % bromophenol blue and heated in a boiling water bath for 10 min. After loading 20 μL of sample, the gels were run at 70 V for 30 min followed by a constant voltage of 120 V for 45–60 min. A low molecular weight marker ranging from 5 to 170 kDa (Sigma-Aldrich, Madrid, Spain) was used. After running, the gels were stained with 0.1 % PhastGel Blue R solution (Merck, Darmstadt, Germany) containing 20 % methanol and 10 % acetic acid and destained with this solvent.
MALDI-TOF-MS Analysis
The molecular weights of GSP and its hydrolysates were analyzed using MALDI-TOF-MS, as described previously (Wang and Yang 2001). The apparatus was set up as follows: nitrogen laser, wavelength 337 nm; linear-flight distance, 1.2 m; acceleration voltage, 20 kV; detector voltage, 25 kV. α-Cyano (α-cyano-4-hydroxyeinnamic acid) was used as the matrix as follows: α-Cyano was placed into a tube and mixed with 40–100 μL of 0.1 % TFA/50 % acetonitrile solution by vortexing. The tube was then centrifuged, leaving a small amount of insoluble α-Cyano at the bottom of tube under ideal conditions. The resultant supernatant, a saturated solution, was used for the experiments. Internal and external standards were prepared using distilled water. The samples were dissolved in deionized water to a concentration of 40 pmol /μL. One microliter of sample was mixed with a matrix solution of the same volume. The matrix-embedded sample was dried on a stainless steel target for analysis. Ten picomoles of CytoC were used for the internal calibration.
Western Blotting Assay
Western blotting was carried out on a Trans-blot Electrophoretic Transfer Cell according to the method described by Shimakura et al. (2005). The proteins and hydrolysates separated by SDS-PAGE were electrotransferred from the gel to the PVDF membrane (Millipore). The membrane was washed with Tween-PBS (0.01 M phosphate buffer, pH 7.0, containing 0.15 M NaCl and 0.05 % Tween 20) and blocked with 2 % skim milk in Tween-PBS at 37 °C overnight. After washing with Tween-PBS, the membrane was incubated for 2 h at 37 °C with mouse anti-GSP serum (obtained from Professor Yin Lu, Nanjing University of Traditional Chinese Medicine; diluted 1:100 with Tween-PBS) as a primary antibody, washed in TBS containing 0.5 mL/L Tween20 (TBST), and then incubated with horseradish peroxidase-conjugated goat anti-mouse IgE (Sigma Aldrich, St Louis, MO; diluted 1:500 with Tween-PBS) as a secondary antibody for 2 h at 37 °C. Finally, the immunoblots were detected by immersing the membrane in 20 mL of TBS containing 12 mg of 4-chloro-1-naphthol, 4 mL of ethanol, and 12 μL of 300 mL/L H2O2 for 5 min at 37 °C. The membrane images were then analyzed on a gel scanner (Sharp JX-330, Pharmacia Biothec).
Enzyme-Linked Immunosorbent Assay
An indirect competitive enzyme-linked immunosorbent assay (ELISA) was applied to determine the antigenicity according to the method described by Julià et al. (2007). The individual wells of a 96-well microtiter plate were coated with 100 μL of sample (50 μg/mL) in coating buffer (0.015 mol/L Na2CO3, 0.035 mol/L NaHCO3, pH 9.6) and incubated overnight at 4 °C. The plates were subsequently blocked with 50 mL/L skim milk in phosphate-buffered saline (PBS) for 1 h at 37 °C. Mouse antiserum (diluted to 1:400 in PBS) was then added to each well. After incubation at 37 °C for 1 h, 100 μL of goat anti-mouse IgE conjugated with horseradish peroxidase (diluted to 1:1000 in PBS) was added to each well, and the plate was then incubated for 1 h at 37 °C to detect the bound immunogen. Between each step, the wells were washed three times with PBS containing 0.5 mL/L Tween 20. The color was developed by adding 100 μL of substrate solution (0.5 mg/mL o-phenylene diamine, 0.3 mL/L H2O2 in 0.1 mol/L citrate buffer, pH 5.5) to each well and incubating the plate for 15 min at 37 °C. The reaction was terminated by adding 50 μL of 2 mol/L H2SO4. The absorbance at 490 nm was read using a Bio-Rad 1860 microplate reader (Bio-Rad, CA, USA).
Statistical Analysis
The SPSS program (SPSS 11.5 for windows, SPSS Inc. Chicago, IL, USA) was used to statistically analyze the results. A one-way linear analysis of variance (ANOVA) was performed, and the significance level set to p ≤ 0.05. Means were compared with Duncan’s test.
Results and Discussion
Effects of Enzymatic Hydrolysis Assisted by HHP Treatment on the Hydrolysis of GSP
The degree of hydrolysis of GSP after HHP treatment is shown in Fig. 1. Compared with the control, which was hydrolyzed at atmospheric pressure (0.1 MPa), the extent of proteolysis by papain, alcalase, pepsin, or neutrase significantly increased (p ≤ 0.05) after the HHP treatment. Papain, alcalase, or pepsin exhibited significantly higher (p ≤ 0.05) degrees of GSP hydrolysis after HHP treatment at 300 and 400 MPa than at 200 MPa. Moreover, the highest levels of hydrolysis were obtained at 300 MPa for papain or alcalase and at 400 MPa for pepsin. However, the extent of hydrolysis considerably decreased for papain or alcalase after HHP treatment at 400 MPa compared with 300 MPa, but this decrease was not significant. In addition, the extent of GSP hydrolysis by neutrase was much lower than those by papain, alcalase, or pepsin and minimally differed from that of the control, which was hydrolyzed at atmospheric pressure (0.1 MPa). These results suggest that neutrase is not suitable for the hydrolysis of GSP.
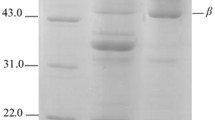
The SDS-PAGE profiles of GSP and its hydrolysates for HHP-treated and untreated samples are shown in Fig. 2. The electrophoretogram of GSP showed bands with apparent molecular weights (MW) ranging from 5 to 100 kDa, including distinct protein bands of high intensity with MWs of approximately 5, 10, 22, 30, 39, 46, and 90 kDa; the same MWs of protein have been reported for ginkgo seeds in the literature (Yang et al. 2011). As expected, the electrophoretograms of hydrolysates confirmed that the hydrolysis at atmospheric pressure was less effective than HHP. In the case of papain or alcalase (Fig. 2a, b), compared to native GSP, when hydrolysis is at atmospheric pressure (0.1 MPa), only some protein bands of low content with MW of 10, 22, and 90 kDa disappeared. Similarly, when hydrolysis was conducted after 200 MPa treatment, the 10, 22, 39, and 90 kDa protein bands disappeared. However, when hydrolysis was conducted after HHP at 300 MPa, the resultant hydrolysates showed bands with MWs lower than 10 kDa, and the SDS-PAGE pattern did not show traces of any intact protein for samples treated with papain or alcalase at 300 MPa. In contrast, in hydrolyzed samples treated with HHP at 400 MPa, additional high-intensity bands were observed at 78 kDa for papain and 57 kDa for alcalase. These bands were absent in the other samples, suggesting that they were formed at 400 MPa. In samples hydrolyzed by pepsin (Fig. 2c) at atmospheric pressure (0.1 MPa), only the protein bands at 90 kDa disappeared. For samples hydrolyzed after 200 and 300 MPa treatment, the 46- and 90-kDa protein bands disappeared and some of the remaining bands showed decreased intensity. However, most samples hydrolyzed after 400 MPa HHP treatment obtained bands with MWs lower than 15 kDa. These results confirm that the hydrolysis of papain and alcalase are maximized by 300 MPa HHP treatment, whereas the hydrolysis of pepsin is maximized by 400 MPa HHP treatment, as evidenced by the production of smaller peptides. These findings agree with the highest degree of hydrolysis observed with the OPA method. Most native protein bands remained in samples hydrolyzed by neutrase (Fig. 2d) at atmospheric pressure (0.1 MPa), and these samples did not markedly differ from native GSP. After HHP treatment, neutrase-hydrolyzed samples showed most of the bands of the intact protein; only the 39-kDa band disappeared and the 30-kDa band exhibited decreased intensity. This result agreed with the degree of hydrolysis observed by the OPA method.
SDS-PAGE of native GSP and its hydrolysates obtained by hydrolysis with papain (a), alcalase (b), pepsin (c), and neutrase (d). M molecular weight standard solution, GSP ginkgo seed proteins, HA hydrolysis at atmospheric pressure (0.1 MPa), H200 hydrolysis at 200 MPa, H300 hydrolysis at 300 MPa, H400 hydrolysis at 400 MPa
To determine the accurate MW of GSP and its hydrolysates, the MALDI-TOF-MS profiles of native protein and hydrolyzed peptides were investigated before and after HHP treatment, and the results are shown in Table 1. The MS spectra of GSP showed a mass range of 5 to 100 kDa and major protein peaks with masses of 5.3, 9.3, 22.5, 30.6, 39.3, 45.1, and 90.3 kDa. Moreover, for samples hydrolyzed by papain or alcalase at atmospheric pressure (0.1 MPa), the protein peaks with masses of 5.3, 30.8, 39.1, and 44.5 kDa remained, and hydrolysates obtained at 200 MPa showed peaks with masses of 5.1, 27.8, 29.1, and 30.7 kDa. However, most samples hydrolyzed after HHP treatment at 300 MPa showed peaks with masses lower than 10 kDa, whereas those subjected to 400 MPa HHP obtained showed additional peaks with masses of 78.5 kDa for papain and 57.3 kDa for alcalase. For samples hydrolyzed by pepsin, the main protein peaks with masses of 5.3, 9.3, 30.6, 38.7, and 45.1 kDa remained in samples treated at atmospheric pressure (0.1 MPa), and samples treated at 200 and 300 MPa showed peaks with masses of 5.3, 9.3, 29.1, and 39.3 kDa. However, most samples treated with HHP at 400 MPa showed peaks with masses lower than 15 kDa. For samples hydrolyzed by neutrase at atmospheric pressure (0.1 MPa), most protein peaks remained, and these samples did not significantly differ from the intact protein. Most of these peaks also remained after HHP treatment, and only some peaks exhibited reduced intensity. These results agreed with the SDS-PAGE profiles.
In food proteins, enzymatic hydrolysis has been shown to break peptide bonds, and HHP treatment has been shown to break disulfide bonds, which will transform the intact proteins into smaller fragments of peptides (Toldrà et al. 2011). In this study, four enzymes, papain, alcalase, pepsin, and neutrase, were used to hydrolyze GSP. The hydrolysis of each protease is specific and their target sites differ; consequently, the efficiency of hydrolysis varies. Alcalase, an endopeptidase, is least specific and can hydrolyze aromatic and basic amino acids, such as Asp, Glu, His, Gly, Lys, Phe, Tyr, Trp, and Leu. Papain is a sulfhydryl endopeptidase that hydrolyzes both aromatic and basic amino acids, such as Phe, Tyr, Lys, Leu, Asp, Ser, and Gly. Pepsin is an endopeptidase that cleaves peptide bonds near aromatic amino acids, including Glu, His, Leu, Tyr, Trp, and Phe, to produce peptides. Neutrase is a metalloendopeptidase that preferentially cleaves bonds near the carboxyl terminal between hydrophobic amino acids, including Leu and Tyr (Sabadin et al. 2012). In the present study, the degree of hydrolysis indicated by SDS-PAGE and MALDI-TOF-MS analyses indicated that papain, alcalase, and pepsin exhibited good proteolytic activity and were able to hydrolyze all intact proteins into peptides with MWs lower than 10 kDa. However, the efficiency of neutrase hydrolysis was poor, and most of the proteins in ginkgo seed remained intact after neutrase hydrolysis. These results are likely due to the composition and content of amino acids in GSP as well as differences in the target sites and enzymatic activity of each protease. Moreover, the enzymatic hydrolysis of GSP was significantly improved after HHP treatment compared with the hydrolysis at atmospheric pressure. HHP treatment can reportedly increase the susceptibility of food proteins to proteolytic enzymes. This increase may be related to the denaturation, unfolding, or dissociation of the proteins into monomers in the presence of protease, which allowed the enzyme to access the binding sites (Knudsen et al. 2002). Furthermore, when GSP was hydrolyzed with papain and alcalase assisted by HHP treatment, the efficiency of hydrolysis was highest at 300 MPa, and increasing the pressure to 400 MPa did not improve proteolysis. On the contrary, peptides at new molecular weights that were not evident in other samples were observed. A similar phenomenon has been reported by Peñas et al. (2006a, b), who found decreases in the degree of dairy whey protein hydrolysis by alcalase when the pressure was increased from 300 to 400 MPa, as evidenced by new protein bands in the SDS-PAGE analysis. This phenomenon can be explained as follows: HHP treatments can induce the unfolding of protein and create new free thiol residues, which appear to be stable at certain pressures. However, the pressure is exceeds this range, some of these thiol residues will aggregate and recombine to result in sulfhydryl-disulfide exchange reactions or new sulfhydryl-disulfide binding by oxidation and aggregation (Wang et al. 2008).
Effects of Enzymatic Hydrolysis Assisted by HHP Treatment on Allergenicity of GSP
Specific antibodies can recognize certain site(s) on allergenic food proteins called epitopes to provoke an immune response. The epitopes are either linear or conformational: they may consist of a few amino acids in the primary structure or a unique three-dimensional motif of the protein structure, respectively (Sathe et al. 2005). The food-processing conditions affect the allergenicity by altering the immunoreactive epitopes of allergenic proteins. Enzymatic hydrolysis has been shown to destroy the structure of linear epitopes, and HHP processing can destroy the structure of conformational epitopes. Thus, enzymatic hydrolysis assisted by HHP can effectively reduce the allergenicity of food proteins by simultaneously destroying linear and conformational epitopes (Mills et al. 2009). In our study, the allergenicity of GSP after enzymatic hydrolysis assisted by HHP treatment was assessed by Western blotting and enzyme-linked immunosorbent assays.
Western blotting was performed using a serum pool from mice that were allergic to GSP, and the results are shown in Fig. 3. The raw GSP contained allergenic proteins with MWs of 22, 30, 46, and 90 kDa. Hydrolysis combined with HHP reduced the total IgE-reactive bands in GSP. Hydrolysis by papain or alcalase (Fig. 3a, b) at atmospheric pressure (0.1 MPa) removed the immunoreactive bands at 22 and 90 kDa, whereas the bands at 30 and 46 kDa remained; at 200 MPa, the 30 and 46 kDa remained in papain-hydrolyzed samples, whereas only the 30 kDa was visible in alcalase-hydrolyzed samples. However, when HHP was conducted at 300 and 400 MPa, all immunoreactive bands disappeared: the Western blotting pattern did not show traces of any intact allergenic proteins in samples treated with papain or alcalase at 300 and 400 MPa. In samples hydrolyzed by pepsin (Fig. 3c) at atmospheric pressure (0.1 MPa), the main immunoreactive bands at 22, 30, and 46 kDa remained, and only the 90-kDa band disappeared. In samples hydrolyzed by this enzyme after 200 MPa treatment, the 46-kDa band disappeared, and only the 22- and 30-kDa bands remained. After 300 MPa HHP treatment, the 22- and 30-kDa bands were less visible in hydrolysates, and all immunoreactive bands disappeared in hydrolyzed samples after HHP at 400 MPa. In samples hydrolyzed by neutrase (Fig. 3d) at atmospheric pressure (0.1 MPa), most immunoreactive bands remained, and these samples did not markedly differ from the raw GSP. After HHP treatment, the hydrolysates retained most of the immunoreactive bands (at 22, 30, 46, and 90 kDa) and only some immunoreactive bands were less visible.
Western blotting of native GSP and its hydrolysates obtained by hydrolysis with papain (a), alcalase (b), pepsin (c), and neutrase (d). M molecular weight standard solution, GSP ginkgo seed proteins, HA hydrolysis at atmospheric pressure (0.1 MPa), H200 hydrolysis at 200 MPa, H300 hydrolysis at 300 MPa, H400 hydrolysis at 400 MPa
Clinical studies have confirmed that most abnormal immune responses caused by food proteins are due to IgE (Mekore 1996). Therefore, the investigations of allergies to food proteins have primarily focused on food allergies regulated by IgE. To further assess the residual immunochemical reactivity in hydrolysates of GSP in our study, the IgE-binding activities of hydrolysates were tested with ELISA assays, and the results are shown in Fig. 4. HHP reduced the immunoreactivity of most hydrolysates compared with the raw protein. HHP also significantly reduced the immunoreactivity of hydrolysates compared with hydrolysis at atmospheric pressure (0.1 MPa). Specifically, the binding capacity to IgE in the papain and alcalase hydrolysates was minimized at 300 MPa, corresponding to a maximum antigenicity reduction of 95.5 % for papain and 96.8 % for alcalase. However, HHP treatment at 400 MPa resulted in a small increase in the immunochemical reactivity of hydrolysates. Increasing pressure gradually decreased the immunoreactivity of pepsin hydrolysates. Specifically, the binding capacity to IgE was minimized in the hydrolysates obtained at 400 MPa, corresponding to a maximum antigenicity reduction of 95.2 % and an almost complete loss of allergenicity. Moreover, neutrase hydrolysates showed a small decrease in the immunochemical reactivity after HHP treatment compared with the control, which was hydrolyzed at atmospheric pressure (0.1 MPa). These decreases were not significant at 200, 300, and 400 MPa, indicating that neutrase does not effectively reduce the allergenicity of GSP. These results are consistent with the Western blotting profiles.
The IgE-binding activities determined by ELISA assay, of GSP untreated (control) and treated by HHP (200, 300, or 400 MPa, for 20 min at 25 °C) prior to hydrolysis with papain, alcalase, pepsin, and neutrase. Different letters above the bars for each enzyme indicate significant differences (p ≤ 0.05)
The results of the present study suggest that enzymatic hydrolysis by papain, alcalase, or pepsin, but not neutrase, significantly reduced the allergenicity of GSP. The differences in the effects of enzymes may be related to their specificity, which is related to the ability of the enzyme to cleave the antigenic determinants in allergenic proteins (Heinzmann et al. 1999). Notably, the application of HHP to GSP prior to enzymatic hydrolysis may result in larger decreases in allergenicity than hydrolysis at atmospheric pressure. HHP reportedly induces structural unfolding and protein denaturation and may even dissociate some proteins into subunits, which might favor the exposure of conformational antigenic epitopes to proteases activated during enzymatic hydrolysis (Silva et al. 2006). Consequently, these epitopes may be more accessible to enzymes, allowing these antigenic epitopes to be destroyed. Moreover, proteolysis by papain or alcalase at 400 MPa increased the allergenicity of proteins compared with proteolysis at 300 MPa. Similar results have been found in soybean proteins, which showed an antigenicity increase of 15 % after alcalase proteolysis at 400 MPa compared with 300 MPa. This increase in antigenicity may result from the unfolding of the structure, which could expose hydrophobic groups, intermolecular-disulfide bonds and antigenic epitopes buried inside the native molecule, leading to the increased allergenicity of proteins (Sathe et al. 2005).
Conclusions
From the results of our study, we conclude that HHP treatment can enhance the enzymatic hydrolysis of GSP depending on the type of enzyme used. Papain, alcalase, or pepsin can effectively hydrolyze GSP into small peptides. Specifically, papain and alcalase were most efficient at 300 MPa, followed by pepsin at 400 MPa. However, GSP was resistant to proteolysis by neutrase, irrespective of the pressure applied. Furthermore, HHP treatment significantly reduced the antigenicity of GSP hydrolysates, and this reduction also depends on the type of enzyme used. Specifically, papain or alcalase hydrolysis at 300 MPa and pepsin hydrolysis at 400 MPa resulted in an almost complete loss of allergenicity, whereas neutrase did not markedly reduce the allergenicity of GSP at any pressure. Finally, the combination of HHP treatment and enzymatic hydrolysis could be an important tool for reducing or removing the immunoreactivity of GSP. Our results aid the design of hypoallergenic hydrolysates from GSP, which may be used as base ingredients of food. However, this application requires further optimization.
References
Bonomi, F., Fiocchi, A., Frøkiær, H., Gaiaschi, A., Iametti, S., Poiesi, C., & Rovere, P. (2003). Reduction of immunoreactivity of bovine β-lactoglobulin upon combined physical and proteolytic treatment. Journal of Dairy Research, 70, 51–59.
Cabanillas, B., Pedrosa, M. M., Rodríguez, J., Gonzalez, A., Muzquiz, M., Cuadrado, C., Crespo, J. F., & Burbano, C. (2010). Effects of enzymatic hydrolysis on lentil allergenicity. Molecular Nutrition and Food Research, 54, 1266–1272.
Calvo, M. M., & Gomez, R. (2002). Peptidic profile, molecular mass distribution and immunological properties of comercial hypoallergenic infant formulas. Milchwissenschaft, 57, 187–190.
Chicón, R., Belloque, J., Alonso, E., & López-Fandiño, R. (2009). Antibody binding and functional properties of whey protein hydrolysates obtained under high pressure. Food Hydrocolloids, 23, 593–599.
Church, F. C., Swaisgood, H. E., Porter, D. H., & Catignani, G. L. (1983). Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. Journal Dairy Science, 66, 1219–1221.
Deng, Q. C., Wang, L., Wei, F., Xie, B. J., Huang, F. H., Shi, J., Huang, Q. D., Tian, B. Q., & Xue, S. (2011). Functional properties of protein isolates, globulin and albumin extracted from Ginkgo biloba seeds. Food Chemistry, 124, 1458–1465.
Dong, X., Zhao, M., Shi, J., Yang, B., Li, J., Luo, D., & Jiang, Y. (2011). Effects of combined high-pressure homogenization and enzymatic treatment on extraction yield, hydrolysis and function properties of peanut proteins. Innovative Food Science and Emerging Technologies, 12, 478–483.
Eisenmenger, M. J., & Reyes-De-Corcuera, J. I. (2009). High pressure enhancement of enzymes: a review. Enzyme and Microbial Technology, 45, 331–347.
Heinzmann, A., Blattmann, S., Spuergin, P., Forster, J., & Deichman, K. A. (1999). The recognition pattern of sequential B cell epitopes of beta–lactoglobulin does not vary with the clinical manifestations of cow’s milk allergy. International Archives of Allergy and Immunology, 120, 280–286.
Huang, W., Xie, B., Wang, Y., Yang, E. N., & Luo, R. (2004). Study on separation and purification of protein from ginkgo seed and its antioxidant activity. Scientia Agdcultura Sinica, 37, 1537–1543.
Huang, W., Deng, Q. C., Xie, B. J., Shi, J., Huang, F. H., Tian, B. Q., Huang, Q. D., & Xue, S. (2010). Purification and characterization of an antioxidant protein from Ginkgo biloba seeds. Food Research International, 43, 86–94.
Julià, S., Sánchez, L., Pérez, M. D., Lavilla, M., Conesa, C., & Calvo, M. (2007). Effect of heat treatment on hen’s egg ovomucoid: an immunochemical and calorimetric study. Food Research International, 40, 603–612.
Kananen, A., Savolainen, J., MaKkinen, J., Perttila, U., Myllykoski, L., & Pihlanto-Leppala, A. (2000). Influence of chemical modification of whey protein conformation on hydrolysis with pepsin and trypsin. International Dairy Journal, 10, 691–697.
Knudsen, J. C., Otte, J., Olsen, K., & Skibsted, L. H. (2002). Effect of high hydrostatic pressure on the conformation of β-lactoglobulin A as assessed by proteolytic peptide profiling. International Dairy Journal, 12, 791–803.
Kovacs-Nolan, J., Zhang, J. W., Hayakawa, S., & Mine, Y. (2000). Immunochemical and structural analysis of pepsin-digested egg white ovomucoid. Journal of Agricultural and Food Chemistry, 48, 6261–6266.
Lee, H. W., Keum, E. H., Lee, S. J., Sung, D. E., Chung, D. H., Lee, S. I., & Oh, S. (2007). Allergenicity of proteolytic hydrolysates of the soybean 11S globulin. Journal of Food Science, 72, 168–172.
Li, H., Zhu, K., Zhou, H., & Peng, W. (2012). Effects of high hydrostatic pressure treatment on allergenicity and structural properties of soybean protein isolate for infant formula. Food Chemistry, 132, 808–814.
Maleki, S. J., Chung, S. Y., Champagne, E. T., & Raufman, J. P. (2000). The effects of roasting on the allergenic properties of peanut proteins. Journal of Allergy and Clinical Immunology, 106, 763–768.
Mekore, Y. A. (1996). Introduction to allergic diseases. Critical Reviews in Food Science and Nutrition, 36, 1–18.
Mills, E. N. C., Sancho, A. I., Rigby, N. M., Jenkins, J. A., & Mackie, A. R. (2009). Impact of food processing on the structural and allergenic properties of food allergens. Molecular Nutrition and Food Research, 53, 963–969.
Peñas, E., Snel, H., Floris, R., Préstamo, G., & Gomez, R. (2006a). High pressure can reduce the antigenicity of bovine whey protein hydrolysates. International Dairy Journal, 16, 969–975.
Peñas, E., Préstamoa, G., Baezac, M. L., Martínez-Moleroc, M. I., & Gomez, R. (2006b). Effects of combined high pressure and enzymatic treatments on the hydrolysis and immunoreactivity of dairy whey proteins. International Dairy Journal, 16, 831–839.
Peñas, E., Préstamo, G., Polo, F., & Gomez, R. (2006c). Enzymatic proteolysis, under high pressure of soybean whey: analysis of peptides and the allergen Gly m 1 in the hydrolysates. Food Chemistry, 99, 569–573.
Pereda, J., Ferragut, V., Buffa, M., Guamis, B., & Trujillo, A. J. (2008). Proteolysis of ultra-high pressure homogenised treated milk during refrigerated storage. Food Chemistry, 111, 696–702.
Quirós, A., Chichón, R., Recio, I., & López-Fandiño, R. (2007). The use of high hydrostatic pressure to promote the proteolysis and release of bioactive peptides from ovalbumin. Food Chemistry, 104, 1734–1739.
Sabadin, I. S., Villas-Boas, M. B., de Lima Zollner, R., & Netto, F. M. (2012). Effect of combined treatment of hydrolysis and polymerization with transglutaminase on β-lactoglobulin antigenicity. European Food Research and Technology, 235, 801–809.
Sathe, S. K., Teuber, S. S., & Roux, K. H. (2005). Effects of food processing on the stability of food allergens. Biotechnology Advances, 23, 423–429.
Shimakura, K., Tonomura, Y., Hamada, Y., Nagashima, Y., & Shiomi, K. (2005). Allergenicity of crustacean extractives and its reduction by protease digestion. Food Chemistry, 91, 247–253.
Silva, J. L., Cordeiro, Y., & Foguel, D. (2006). Protein folding and aggregation: two sides of the same coin in the condensation of proteins revealed by pressure studies. Biochemica et Biophysica Acta, 1764, 443–451.
Stapelfeldt, H., Petersen, P. H., Kristiansen, K. R., Qvist, K. B., & Skibsted, L. H. (1996). Effect of high hydrostatic pressure on the enzymatic hydrolysis of β-lactoglobulin B by trypsin, thermolysin and pepsin. The Journal of Dairy Research, 63, 111–118.
Takagi, K., Teshima, R., Okunuki, H., & Sawada, J. (2003). Comparative study of in vitro digestibility of food proteins and effect of preheating on the digestion. Biological and Pharmaceutical Bulletin, 26, 969–973.
Toldrà, M., Parés, D., Saguer, E., & Carretero, C. (2011). Hemoglobin hydrolysates from porcine blood obtained through enzymatic hydrolysis assisted by high hydrostatic pressure processing. Innovative Food Science and Emerging Technologies, 12, 435–442.
Tredici, P. D. (1991). Ginkgos and people—a thousand years of interaction. Arnoldia, 51, 2–15.
Wang, H. X., & Yang, S. C. (2001). The development of electrospray ionization mass spectrometry (ESI-MS) in studies of non-covalent protein complexes. Acta Pharmaceutica Sinica, 36, 315–320.
Wang, X. S., Tang, C. H., Li, B. S., Yang, X. Q., Li, L., & Ma, C. Y. (2008). Effects of high pressure treatment on some physicochemical and functional properties of soy protein isolates. Food Hydrocolloids, 22, 560–567.
Watanabe, M., Miyakawa, J., Ikezawa, Z., Suzuki, Y., Hirao, T., Yoshizawa, T., & Arai, S. (1990). Production of hypoallergenic rice by enzymatic decomposition of constituent proteins. Journal of Food Science, 55, 781–783.
Yang, J. T., & Wu, C. E. (2009). Advance in allergic composition and mechanism of allergy of Ginkgo biloba seeds. Food Science and Technology, 6, 282–286.
Yang, J. T., Wu, C. E., Li, Y. Y., Jia, S. Q., Fan, G. J., & Peng, F. R. (2011). Identification and purification of an allergic glycoprotein from Ginkgo biloba kernel. Agricultural Sciences in China, 10, 631–641.
Zhou, H., Chen, X. J., Wang, C. Z., Ye, J. Z., & Chen, H. X. (2012). Purification and characterization of a novel ∼ 18 kDa antioxidant protein from Ginkgo biloba seeds. Molecules, 17, 14778–14794.
Ziegler, F., Nitenberg, G., Coudray-Lucas, C., Lasser, P., Giboudeau, J., & Cynober, L. (1998). Pharmacokinetic assessment of an oligopeptide-based enteral formula in abdominal surgery patients. The American Journal of Clinical Nutrition, 67, 124–128.
Acknowledgments
The authors gratefully acknowledge the Foundation Research Project of Jiangsu Province (The Natural Science Fund, BK20141073), the financial support by the Sanxin forestry project of Jiangsu Province (LYSX[2015]25) and the China Basic Research Foundation of National Commonwealth Research Institute, CAF(CAFYBB2014QA021).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, H., Wang, C., Ye, J. et al. Effects of Enzymatic Hydrolysis Assisted by High Hydrostatic Pressure Processing on the Hydrolysis and Allergenicity of Proteins from Ginkgo Seeds. Food Bioprocess Technol 9, 839–848 (2016). https://doi.org/10.1007/s11947-016-1676-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11947-016-1676-3