Abstract
Background
Thiopurines, commonly used to treat autoimmune conditions and cancer, can be limited by life-threatening leucopenia. However, whether NUDT15 (nucleoside diphosphate-linked moiety X-type motif 15) is associated with thiopurine-induced leucopenia in Asians is controversial.
Methods
Relevant studies in English that were published until July 10, 2016 were identified through PubMed, EMbase, and other web knowledge databases. Study quality was assessed according to the Newcastle-Ottawa Scale (NOS) criteria. Summary risk ratio (RR) and 95% confidence intervals (CI) were estimated based on a fixed-effects model or a random-effects model, depending on the absence or presence of significant heterogeneity.
Results
Seven studies of 1138 patients met our inclusion criteria. Random-effects model meta-analysis provided evidence that T carriers of NUDT15 c.415C>T were significantly correlated with high incidences of thiopurine-induced leukocytopenia [CT + TT vs. CC: RR = 3.79, 95%CI (2.64 ~ 5.44), P < 0.00001]. This correlation was especially strong in TT patients, where it was found to be significantly increased by 6.54-fold compared with CC patients [TT vs. CC: RR = 6.54, 95%CI (3.34 ~ 12.82), P < 0.00001]. We also found that the NUDT15 c.415C>T variant was common in Asians and Hispanics, but rare in Europeans and Africans; the frequency of the NUDT15 c.415C>T distribution varied substantially by race/ethnicity.
Conclusion
The results of this meta-analysis confirm that NUDT15 c.415C>T may be an important predictor of thiopurine-induced leukocytopenia in Asians. Genotype targeting of NUDT15 c.415C>T before initiating thiopurine treatment may be useful to limit leukocytopenia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thiopurines (mercaptopurine, thioguanine, and azathioprine) remain one of the most important and extensively prescribed drugs for cancer and autoimmune disease. Myelosuppression is the most common adverse reaction of thiopurines, and includes macrocytosis, leucopenia megaloblastic anemia, thrombocytopenia, and pancytopenia [1]. Azathioprine (AZA) is rapidly metabolized following oral administration to form 6-mercaptopurine, (6-MP) which in turn crosses cell membranes and is converted intracellularly into a number of thiopurine analogues, including 6-thioguanine nucleotides (6-TGNs). There are three major competing routes for 6-MP. One important route is catalyzed by the enzyme thiopurine methyltransferase (TPMT). Most researchers have speculated that the cytotoxicity caused by thiopurines is due to DNA damage produced by the concentrations of 6-TGNs, and the wide interpatient variation in 6-TGN concentration is largely accounted for by variation in TPMT activity [2]. Thiopurine methyltransferase (TPMT) catabolizes an important thiopurine metabolic pathway and its genetic polymorphism affects thiopurine-induced toxicity [2–5]. A total of 31 TPMT allele genetic polymorphisms have been identified which are, or may be associated with, decreased or absent TPMT enzyme activity [6]. TPMT*2, *3A, *3B, *3C, and *8 take up approximately 95% of all TPMT variants known to result in TPMT deficiency [7]. The predominant TPMT variant in Asian populations is *3C, with an allele frequency of 2.3% [8, 9]. However, thiopurine-induced leukocytopenia is common in Asian populations [10–12]. Therefore, we believe that thiopurine-induced leucopenia can be affected by multiple factors. Some factors, such as TPMT and other genes might play a role in the development of the leucopenia caused by thiopurines.
Recently, a number of studies suggested that NUDT15 c.415C>T was a novel predictor of thiopurine-induced leukocytopenia in Asians [13–15]. However, these studies have two major limitations: the sample size of the observational studies was relatively small and the statistical power was very insufficient. Furthermore, no meta-analysis has estimated the relationship between NUDT15 c.415C>T and thiopurine-induced leukocytopenia in Asians. Therefore, it is necessary to perform a quantitative synthesis of the existing genetic association studies, despite their inconsistent results and inadequate power. The purpose of this paper is to systematically accumulate and quantitatively analyze the data regarding the genetic association between NUDT15 c.415C>T and thiopurine-induced leukocytopenia.
Methods
Data sources and search strategy
We performed a computerized literature search of PubMed, EMbase, and Web of Knowledge databases (we search for articles published up until 10 July 2016) by using the Boolean combinations of the key terms “NUDT15 OR MTH2” AND “polymorphisms OR polymorphism OR SNP” AND ‘leukocytopenia OR myelosuppression” AND “Azathioprine OR 6-MP OR thiopurine OR thionucleosides.” The Medical Subject Headings (MeSH) or keywords were used when the searching database had this option available. The language of the published papers was restricted in English, and only human studies were included. The bibliographies of the included articles were examined to identify additional studies.
Study selection
Two authors (Ailing Zhang and Jing Yang) independently selected the studies and retrieved data by using a standardized form. All disagreements were resolved by group consensus. For inclusion, the criteria were as follows: (1) patients were treated with thiopurines and then the association between NUDT15 c.415C>T and thiopurine-induced leukocytopenia was investigated; (2) patients were genotyped for NUDT15 c.415C>T; (3) leukopenia was graded by common toxicity criteria as follows: grade 2, 2000 ~ 3000 mm−3; grade 3, 1000 ~ 2000 mm−3; and grade 4, <1000 mm−3 [16]. Studies were excluded from the primary analysis if any of the following were met: (1) the patient populations were selected on the basis of their reported leukocytopenia or NUDT15 c.415C>T genotype status; (2) letters, case reports, review articles, studies with insufficient information and duplicate studies were excluded.
For the systematic review of the frequencies of the genotype and allele of NUDT15 c.415C>T, all articles had to meet our inclusion and exclusion criteria. Inclusion criteria: (1) included a cohort study or case-control study; (2) the article explicitly listed the genotype and allele frequencies of NUDT15 c.415C>T. Exclusion criteria: (1) reviews, meta-analysis, letters or case reports; (2) literature without information on NUDT15 c.415C>T frequencies; (3) the retrieval returned duplicate publications.
Data extraction and quality assessment
The following information was extracted from each included study: the first author’s last name, year of publication, geographic origin, study design, age and gender of patients, thiopurine regimens, disease type, leukocyte classification criteria, patient demographics, the method of SNP detection, and the main results of NUDT15 c.415C>T among patients. The methodological quality of the included studies were evaluated separately by two authors (Jingli Lu and Hua Wang) using the Newcastle-Ottawa quality assessment scale (NOS) criteria [17]. When the results were different, consensus was reached. The NOS criteria were based on three aspects: (1) subject selection: 0 ~ 4; (2) comparability of study groups: 0 ~ 2; (3) clinical outcome: 0 ~ 3. Total NOS scores ranged from 0 to 9 with a score ≥6 indicating good quality.
Statistical analysis
Rigorous statistical analysis was performed using the Review Manager (Revman) program version 5.2 (Cochrane Collaboration, Oxford, UK) in the meta-analysis and the STATA statistical software (Version 12.0, Stata Corporation, College Station, TX, USA) for sensitivity analyses and Egger’s linear regression test. The risk ratio (RR) with a corresponding 95% confidence interval (95%CI) was calculated to evaluate the association between the NUDT15 c.415C>T polymorphism and thiopurine-induced leukocytopenia. The Z test was used to estimate the statistical significance of pooled RRs. Between-study heterogeneity was assessed by Cochran’s Q statistic and I 2 test [18]. If the Q test exhibited a P > 0.05 or the I 2 test was <50%, indicating that these studies were homogeneous, the fixed-effects model was used; otherwise, the random-effect model was used. We also made use of sensitivity analyses to explore sources of heterogeneity. P < 0.05 represented statistically significant heterogeneity. Funnel plots and Egger’s linear regression test was used to investigate the potential publication bias [19]. Pooled data were performed to analyze the genotype and allele frequencies of NUDT15 c.415C>T.
Results
Study characteristics and selection
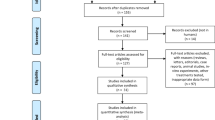
Our primary searches by search terms independently yielded 32 articles. Among these articles, two publications were excluded as they were duplicates and 12 articles were deemed not relevant after reading the abstract. Then, we reviewed the full texts of the remaining 18 articles. Letters or case reports (n = 2), not reporting an association between NUDT15 and leukopenia (n = 3), insufficient information (n = 4), Japanese language (n = 1), and overlapping with another included study (n = 1) were excluded after reading the full text. Finally, seven remaining studies were included in the meta-analysis (Fig. 1) [13, 20–25]. Our study included a total of 1138 patients with Crohn’s disease, ALL or IBD. Among them, 311 patients carried the NUDT15 415T allele. The sample size of each trial ranged from 69 to 346 (Table 1). One study was a case-control study and six articles were cohort studies. Their scores ranged from 6 to 9.
Association between NUDT15 c.415C>T and thiopurine-induced leukocytopenia
For the NUDT15 c.415C>T polymorphism, quantitative synthesis from seven studies showed significant differences in the comparisons of CT + TT vs. CC [RR = 3.79, 95%CI (2.64 ~ 5.44), P < 0.00001, Fig. 2a], CT vs. CC [RR = 3.41, 95%CI (2.44 ~ 4.77), P < 0.00001, Fig. 2b], TT vs. CC [RR = 6.54, 95%CI (3.34 ~ 12.82), P < 0.00001, Fig. 2c]. However, no significant difference was found when comparing TT vs. CT [RR = 1.80, 95%CI (0.93 ~ 3.51), P = 0.08, Fig. 2d]. The funnel plots and Egger’s test demonstrated no evidence of publication bias for CC vs. CT + TT (P = 0.118) (Fig. 3). The result of sensitivity analysis, which omitted individual studies to assess their influence on the pooled estimates, indicated that no single study could influence the pooled RRs (Fig. 4).
The genotype and allele frequencies of NUDT15 c.415C>T in Asians
We assessed the literature to determine frequencies of NUDT15 c.415C>T and identified 11 studies [13–15, 20, 22–27]. The following information was extracted, including the first author’s last name, year of publication, geographic origin, disease type, the number of patients, genotype and allele data. Details on demographic and other data are summarized in Table 2. All the studies clearly report the ethnic origin of patients, and most of patients were East Asians. Only one study included Hispanics and Europeans. Pooling the same ethnic populations of the NUDT15 c.415C>T genotype, the genotype frequencies of NUDT15 c.415C>T (CC, CT and TT) in Asians were 75.84%, 21.76%, and 2.39%, respectively. The genotype frequencies were 92.34%, 7.21%, and 0.45% in Hispanics; 99.51%, 0.49%, and 0% in Europeans; and 100%, 0, 0% in Africans. The respective allele frequencies of NUDT15 c.415C>T (C and T) in Asians were 86.72% and 13.28%, respectively; 95.95% and 4.05% in Hispanics; 99.76% and 0.24% in Europeans; and 100% and 0% in Africans.
Discussion
The current meta-analysis demonstrated that the NUDT15 c.415C>T allele is strongly associated with an increased risk of developing leucopenia in patients who are given thiopurines. It also revealed that the risk of developing leucopenia among thiopurine users with NUDT15 c.415C>T is significantly increased by 3.79-fold compared to patients without the gene. The analysis of genotype and allele frequencies of NUDT15 c.415C>T showed that the NUDT15 c.415C>T variant was most common in Asians and Hispanics, and rare in Europeans and Africans. The genotype and allele distribution of NUDT15 c.415C>T varied substantially by race/ethnicity. The higher prevalence of the NUDT15 variant in East Asians may contribute to over-representation of thiopurine intolerance in this population. However, more studies, including those with a greater number of Europeans and Africans, are needed to evaluate the frequencies of NUDT15 c.415C>T.
As prodrugs, thiopurines are enzymatically converted into the active end-metabolite of thiopurines, 6-thioguanine nucleotides (6-TGNs), consisting of 6-thioguanine-monophosphate (6-TGMP), 6-thioguanine-diphosphate (6-TGDP) and 6-thioguanine-diphosphate-triphosphate (6-TGTP) [28]. 6-TGTP is further reduced to 6-deoxythioguanosine-triphosphate (6-TdGTP). Then, 6-TGTP and 6-TdGTP are incorporated into RNA or DNA to trigger futile mismatch repair and, eventually, apoptosis [29–31]. Thiopurines have a narrow therapeutic index owing to frequent toxicity. NUDT15 (nucleoside diphosphate-linked moiety X-type motif 15), also known as MTH2, is a 164-amino-acid protein which can convert 8-oxo-dGTP and 8-oxo-dGDP to 8-oxo-dGMP, thereby removing the oxidatively damaged guanine nucleotides from cells to minimize DNA damage [32]. Studies from comprehensive in vitro and in vivo studies strongly indicate that NUDT15 can convert 6-TGTP to 6-TGMP and TdGTP to TdGMP and then prevent the incorporation of these thiopurine metabolites into DNA (DNA-TG) and negatively regulate thiopurine activation and, consequently, cytotoxicity [27].
NUDT15 c.415C>T is a missense variant in the NUDT15 gene (rs116855232, encoding p.Arg139Cys) [27]. Yang et al. found that c.415C > T was significantly associated with AZA-induced leukopenia (OR = 35.6, p = 4.88^10−94) among 978 individuals [23]. Meanwhile, our meta-analysis also found that patients carrying CT or TT for the NUDT15 c.415C>T risk alleles experience especially excessive thiopurine toxicity compared to those with the CC genotype. However, Yang et al. reported that individuals homozygous for the risk allele encoding p.Arg139Cys were exquisitely sensitive to mercaptopurine and tolerated only 8% of the standard dose, while this NUDT15 variant alone explained 22% of variance in thiopurine tolerance [27]. Based on these facts, NUDT15 c.415C>T variants may be another important predictor for thiopurine-induced leucopenia.
The mechanism of NUDT15 c.415C>T gene mutation related thiopurine-induced early severe hair loss and leukopenia is controversial. So far, several researchers had proposed some ideas. For instance, a gene-dose effect for the risk allele. Jurkat cells transfected with the variant (CT or TT) showed increased AZA-induced toxicity and signs of apoptosis compared to cells transfected with wild type NUDT15 (CC). These findings suggested that NUDT15 is a pharmacogenetic determinant for thiopurine-induced leukopenia in diverse populations [23]. However, Asada et al. did not agree with this view. They suggested that if NUDT15 c.415C>T induces loss-of-function, accumulation of oxidative stress and increased apoptosis in bone marrow progenitor cells might be a potential mechanism of NUDT15 c.415C>T-related thiopurine-induced leucopenia [13]. Moriyama et al. found that NUDT15 c.415C>T variants resulted in partial loss of nucleotide diphosphatase activity and excessive levels of toxic thiopurine active metabolites [33]. Further studies are ongoing in our group to elucidate how this gene is involved in the pathogenesis of thiopurine-induced leucopenia.
Our meta-analysis also has a number of potential limitations. For instance, due to the small number of studies, except for Asians, our study did not include all the data required to assess the relationship between NUDT15 c.415C>T genetic polymorphisms and thiopurine-induced leucopenia. Although our search strategy is relatively comprehensive, it still cannot cover all relevant studies. Hence, it is possible that larger and better designed studies might have identified an impact on thiopurine-induced leucopenia. The monitoring of signs and symptoms in these patients is still needed.
This meta-analysis suggests that patients carrying the NUDT15 T risk allele are at greater risk of thiopurine-induced leucopenia compared with the C homozygotes. Suzuki et al. [26] also found that NUDT15 can predict the dose reduction of 6-MP for children with acute lymphoblastic leukemia. However, a larger study is required to clarify this observed dosing effect in variant carriers.
Conclusion
In conclusion, based on our meta-analysis, NUDT15 c.415C>T gene polymorphisms may be an important predictor of thiopurine-induced leukocytopenia in Asians. Furthermore, the frequency of TPMT variants was rare, while thiopurine-induced leukocytopenia is common in Asians. Therefore, genotyping for NUDT15 c.415C>T before initiating thiopurine treatment may be useful to avoid leukocytopenia.
References
Anstey A, Lear JT (1998) Azathioprine: clinical pharmacology and current indications in autoimmune disorders. BioDrugs 9:33–47
Lennard L, Van Loon JA, Weinshilboum RM (1989) Pharmacogenetics of acute azathioprine toxicity: relationship to thiopurine methyltransferase genetic polymorphism. Clin Pharmacol Ther 46:149–154
Evans WE, Horner M, Chu YQ, Kalwinsky D, Roberts WM (1991) Altered mercaptopurine metabolism, toxic effects, and dosage requirement in a thiopurine methyltransferase-deficient child with acute lymphocytic leukemia. J Pediatr 119:985–989
Lennard L, Lilleyman JS, Van Loon J, Weinshilboum RM (1990) Genetic variation in response to 6-mercaptopurine for childhood acute lymphoblastic leukaemia. Lancet 336:225–229
Lennard L, Van Loon JA, Lilleyman JS, Weinshilboum RM (1987) Thiopurine pharmacogenetics in leukemia: correlation of erythrocyte thiopurine methyltransferase activity and 6-thioguanine nucleotide concentrations. Clin Pharmacol Ther 41:18–25
Relling MV, Gardner EE, Sandborn WJ et al (2013) Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther 93:324–325
Roberts RL, Barclay ML (2015) Update on thiopurine pharmacogenetics in inflammatory bowel disease. Pharmacogenomics 16:891–903
McLeod HL, Pritchard SC, Githang'a J et al (1999) Ethnic differences in thiopurine methyltransferase pharmacogenetics: evidence for allele specificity in Caucasian and Kenyan individuals. Pharmacogenetics 9:773–776
Collie-Duguid ES, Pritchard SC, Powrie RH et al (1999) The frequency and distribution of thiopurine methyltransferase alleles in Caucasian and Asian populations. Pharmacogenetics 9:37–42
Kim JH, Cheon JH, Hong SS et al (2010) Influences of thiopurine methyltransferase genotype and activity on thiopurine-induced leukopenia in Korean patients with inflammatory bowel disease: a retrospective cohort study. J Clin Gastroenterol 44:e242–e248
Tajiri H, Tomomasa T, Yoden A et al (2008) Efficacy and safety of azathioprine and 6-mercaptopurine in Japanese pediatric patients with ulcerative colitis: a survey of the Japanese Society for Pediatric Inflammatory Bowel Disease. Digestion 77:150–154
Takatsu N, Matsui T, Murakami Y et al (2009) Adverse reactions to azathioprine cannot be predicted by thiopurine S-methyltransferase genotype in Japanese patients with inflammatory bowel disease. J Gastroenterol Hepatol 24:1258–1264
Asada A, Nishida A, Shioya M et al (2016) NUDT15 R139C-related thiopurine leukocytopenia is mediated by 6-thioguanine nucleotide-independent mechanism in Japanese patients with inflammatory bowel disease. J Gastroenterol 51:22–29
Lee YJ, Hwang EH, Park JH et al (2016) NUDT15 variant is the most common variant associated with thiopurine-induced early leukopenia and alopecia in Korean pediatric patients with Crohn’s disease. Eur J Gastroenterol Hepatol. doi:10.1097/MEG.0000000000000564
Liang DC, Yang CP, Liu HC et al (2015) NUDT15 gene polymorphism related to mercaptopurine intolerance in Taiwan Chinese children with acute lymphoblastic leukemia. Pharmacogenomics J 16:536–539
N. NCI (2009) National Cancer Institute, common terminology criteria for adverse events v4. NIH publication, Bethesda 09-7473
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25:603–605
Zintzaras E, Ioannidis JP (2005) HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics 21:3672–3673
Peters JL, Sutton AJ, Jones DR et al (2006) Comparison of two methods to detect publication bias in meta-analysis. JAMA 295:676–680
Chiengthong K, Ittiwut C, Muensri S et al (2016) NUDT15 c.415C>T increases risk of 6-mercaptopurine induced myelosuppression during maintenance therapy in children with acute lymphoblastic leukemia. Haematologica 101:e24–e26
Kakuta Y, Naito T, Onodera M et al (2015) NUDT15 R139C causes thiopurine-induced early severe hair loss and leukopenia in Japanese patients with IBD. Pharmacogenomics J 16:280–285
Tanaka Y, Kato M, Hasegawa D et al (2015) Susceptibility to 6-MP toxicity conferred by a NUDT15 variant in Japanese children with acute lymphoblastic leukaemia. Br J Haematol 171:109–115
Yang SK, Hong M, Baek J et al (2014) A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet 46:1017–1020
Zhu X, Chao K, Wang X et al (2016) NUDT15 R139C genotype is a determinant of thiopurines-induced leukopenia in Chinese patients with Crohn’s disease. Gastroenterology 150:S318
Shah SA, Paradkar M, Desai D et al (2016) Nudt15 C415t variant as a predictor for thiopurine induced toxicity in Indian patients. J Gastroenterol Hepatol. doi:10.1111/jgh.13494
Suzuki H, Fukushima H, Suzuki R et al (2016) Genotyping NUDT15 can predict the dose reduction of 6-MP for children with acute lymphoblastic leukemia especially at a preschool age. J Hum Genet 61:797–801
Yang JJ, Landier W, Yang W et al (2015) Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol 33:1235–1242
Meijer B, Mulder CJ, de Boer NK (2016) NUDT15: a novel player in thiopurine metabolism. J Gastrointestin Liver Dis 25:261–262
Ebbesen MS, Nersting J, Jacobsen JH et al (2013) Incorporation of 6-thioguanine nucleotides into DNA during maintenance therapy of childhood acute lymphoblastic leukemiaThe influence of thiopurine methyltransferase genotypes. J Clin Pharmacol 53:670–674
Fotoohi AK, Coulthard SA, Albertioni F (2010) Thiopurines: factors influencing toxicity and response. Biochem Pharmacol 79:1211–1220
Hedeland RL, Hvidt K, Nersting J et al (2010) DNA incorporation of 6-thioguanine nucleotides during maintenance therapy of childhood acute lymphoblastic leukaemia and non-Hodgkin lymphoma. Cancer Chemother Pharmacol 66:485–491
Takagi Y, Setoyama D, Ito R et al (2012) Human MTH3 (NUDT18) protein hydrolyzes oxidized forms of guanosine and deoxyguanosine diphosphates: comparison with MTH1 and MTH2. J Biol Chem 287:21541–21549
Moriyama T, Nishii R, Perez-Andreu V et al (2016) NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 48:367–373
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Source of funding
This study was funded by the Science and Technology Planning Project of Henan Province, China (grant number 201601001).
Rights and permissions
About this article
Cite this article
Zhang, A.L., Yang, J., Wang, H. et al. Association of NUDT15 c.415C>T allele and thiopurine-induced leukocytopenia in Asians: a systematic review and meta-analysis. Ir J Med Sci 187, 145–153 (2018). https://doi.org/10.1007/s11845-017-1608-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-017-1608-x