Abstract
The symptoms of plant diseases are evident in different parts of a plant; however leaves are found to be the most commonly observed part for detecting an infection. Researchers have thus attempted to automate the process of plant disease detection and classification using leaf images. Several works utilized computer vision technologies effectively and contributed a lot in this domain. This manuscript summarizes the pros and cons of all such studies to throw light on various important research aspects. A discussion on commonly studied infections and research scenario in different phases of a disease detection system is presented. The performance of state-of-the-art techniques are analyzed to identify those that seem to work well across several crops or crop categories. Discovering a set of acceptable techniques, the manuscript highlights several points of consideration along with the future research directions. The survey would help researchers to gain understanding of computer vision applications in plant disease detection.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Agriculture in any country depends on the quality and quantity of farming products, especially plants. The detection of plants disease (i.e., unusual growth or dysfunction) thus compelled many researchers to employ image processing techniques to ease this difficult task [1,2,3,4,5,6,7,8,9,10]. Depending upon the cause, a plant may have a specific type of infection out of a range of diseases. This fact further complicates the applicability of computer vision techniques in their proper recognition [11, 12]. Different plants disease detection techniques are proposed and a survey of traditional and innovative techniques is also presented in literature [13, 14]. Popular traditional techniques include molecular, serological, and deoxyribose nucleic acid (DNA). Volatile organic compounds and imaging & spectroscopic techniques are also utilized innovatively to automate the detection process. Such innovative techniques are faster and do not need personnel monitoring. Research by Zhang and Meng [15] reported an accuracy of 87.99% (using an imaging technique) and 86.87% (using human experts on screen) for automatic detection of citrus canker on leaves. Their study further supports the usage of image processing techniques to automatically detect plant diseases at an early stage [15]. Thus, the articles considered in this study are those which have utilized innovative imaging techniques to identify a plant infection using leaf images.
Starting with a discussion on different types of diseases in plants (Sect. 2), a general architecture of a plant disease detection system is presented (Sect. 3). The performance of these systems depends greatly on the classifiers employed. Therefore an attempt to summarize the existing literature based on culture types and classification models employed is done (Sect. 4). Also, the applicability of various classification models to detect infected leaves in different cultures is considered. The manuscript tries to analyze the most studied and the best available system for a specific culture. Several discussions (Sect. 5) and research objectives (Sect. 6) are also highlighted. The reviews presented here would be of great help not only to researchers and experts in this domain but for plant pathologists too, for early detection of plant diseases effecting leaves. In addition, a large population of farmers who wish to utilize automatic systems for quantity as well as quality productions may also be benefitted under proper supervision.
2 Types of Plant Diseases
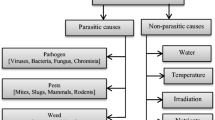
Plant diseases originated from living organisms are biotic [16]. Fungi, bacteria, and viruses are the main causes of different forms of biotic diseases. Abiotic, in contrast, are produced by non-living ecological circumstances such as hail, spring frosts, weather conditions, burning of chemicals, etc. Abiotic diseases are non-infectious, non-transmissible, less dangerous, and are mostly avoidable. The manuscript thus considers the biotic diseases and their categorization with a few common forms is shown in Fig. 1. A range of works exists for various fungal and bacterial diseases, but those under viral category are not focused much in literature [17, 18]. Spots (caused either through fungi or bacteria), mildew, and rust are the top three types which are most commonly considered for identification and classification. In addition, deficiency of nutrients is explored for automation. Section 4 presents further details on these observed facts.
3 Plant Disease Detection System
Figure 2 shows architecture of a simple plant disease detection system with following modules: acquisition, pre-processing, segmentation, feature extraction, and classification (or recognition). It has two phases, the training and the testing. Training phase starts with capturing an image of a specific part like leaves, stems, roots, and branches. Images may or may not be pre-processed to correct various geometric misrepresentation, grey level correction, noise reduction, and blur improvement. Segmentation separates the regions of interest from background and identifies one or more regions from an infected training leaf image. Lastly, feature vectors of the regions of interest are extracted and used to train the classifier. In testing phase, a test leaf image passes through pre-processing, segmentation, and feature extraction modules. The trained classifier identifies the test image as an infected or a healthy sample. The necessity of all the modules is discussed later in this section with a brief summary of different techniques utilized or proposed for each module.
A simple plant disease detection system with imaging techniques. Testing phase images are from [19]
The effectiveness and applicability of these systems are popularly assessed using accuracy as a performance measure. It is also known as precision, classification rate, recognition rate, and success rate [19]. In addition, a few existing works evaluated performance using prediction time [20] and mean square error [21]. Higher accuracy values with smaller prediction time and lower mean square error prove superiority of one system over another.
3.1 Acquisition
Image acquisition is important as accuracy of the system depends greatly on image samples used for training. In this domain, researchers have used a few known datasets, namely IPM Images, PlantVillage Images, and APS Image database [22,23,24,25]. Images with theoretical details are also available at the website of University of Minnesota Extension [26]. Some of the works are privileged to access datasets of research centers IRRI and INIBAP [27, 28]. A few have observed single culture for a period of time instead of a full-fledged dataset [29, 30]. A few have used scanned images [31, 32]. A range of studies have used self-collected image datasets taken either under controlled environmental conditions or in field with complex backgrounds. For effective control of illumination, lighting, and intensity images are also acquired inside laboratories or a sampling box [33,34,35,36]. Infected soybean leaves are placed on white base to remove background complexity [18]. On contrary, some works have captured images with complex background in the field [3, 37, 38].
The quality of image samples depends on camera type and its orientation. Most of the studies used digital cameras with optical axis perpendicular to the leaf plane, but a few have used specialized techniques. CCD color cameras with different specifications are combined with software tools to capture images [37, 39, 40]. Android mobile is also used to capture a leaf at some fixed distance [41]. A multispectral CDD camera with portable spectroradiometer is also employed for soybean leaves [42]. Recently a hyperspectral imaging system is utilized for tomato leaves [43]. It’s obvious that processing images collected under controlled environment is easier. Equipments and techniques used provide different image details. The performance of a plant disease detection system thus varies with background of the acquired image as well as its capturing conditions [11].
3.2 Pre-processing
During pre-processing, distortion removal improves images which ease further processing. Popular pre-processing techniques include color space conversion, cropping, smoothing, and enhancement. Depending upon image quality the functionality of this module varies. As per literature color space conversion is followed by filtering and enhancement. Hue, saturation, and value (HSV) is commonly used color space as it closely resembles human color sensing properties [44,45,46,47,48]. HSI (I for intensity), another similar color space is also used [1, 49,50,51,52]. Works utilizing different color spaces like YCbCr, Hue-Max–Min–Diff, CIE 1976 L*a*b*, RGB, and CIE 1976 uniform chromaticity scale diagram do exists [20, 53, 54]. Automatic segmentation comparable to manual is achieved by converting RGB to a new color space H, I3a and I3b [28].
After color space transformation, filters are applied for desired enhancements, like better contrast and brightness. Noise occurrence is very general, thus such systems popularly use median [27, 37, 55, 56] and rank filters [57]. Laplacian filter is used for sharpening [45]. Apart from these, techniques like histogram equalization and Gabor wavelets are also used for filtering and controlling varying lighting conditions [3]. Concept of anisotropic diffusion is recently presented for enhancement [46]. Other commonly used filters are spatial low pass filter, neighborhood mean, and frequency low-pass filter [58]. Cropping is also important if images are captured in an uncontrolled environment with complex backgrounds. It can be done either manually [45] or automatically using functions [5, 7, 59].
3.3 Segmentation
Segmentation divides image into regions with robust correlation along with the objects of interest. Features of an effectively segmented image help in an easy identification of healthy or infected samples, for instance, number of histogram peaks [19]. Edges, thresholds, locality or color based segmentation techniques are shown to work well with plant disease detection systems. Edge based techniques, like Sobel operator and canny edge detectors are employed in range of studies [2, 33, 34, 57, 60, 61]. A few studies have exploited genetic algorithms [3, 62] and Grab-cut segmentation [63]. Methods based on the concept of entropy and Otsu methods are popular threshold based segmentation techniques [47, 64,65,66]. A manual setting of threshold is explored for an effective segmentation of disease spots in HSI space [67]. An integration of seeded region growing (SRG) concept with local threshold is also utilized for an automatic and efficient segmentation of leaf spot [68].
The infected leaf area shows significant color differences from its original color and this leads to the development of spot color based segmentation [37]. Also, studies discover k-means clustering better than Sobel, prewitt and canny based segmentation approaches [5, 7, 46, 55, 57]. However, k-mediods based segmentation is found to be more robust to noise [69]. A unique Fermi energy based segmentation technique using color and grey level intensity values, is also used [4]. It has shown to work better than Otsu and k-means based methods. A combination of saliency region threshold and k-means algorithm is also utilized to directly extract the diseased leaf area [70]. It is found better than mean shift and unsupervised optimal fuzzy C-means clustering. A study suggests k-means clustering over fuzzy C-means or expectation maximization for accurate leaf disease detection [71].
In conclusion, determination of threshold value is an important step in segmentation. Incorrect threshold determination may infer inaccurate segmentation which leads to an erroneous system [72].
3.4 Feature Extraction
Images are usually interpreted as color, texture, and shape features. Color is commonly defined as moments and histograms. Properties like, contrast, homogeneity, variance, and entropy, can be attached to texture. Similarly, for shape, roundness, area, eccentricity and concavity characteristics are identified. Heterogeneous datasets demand combination of features, but for plants disease detection system texture is identified as the best [73].
Classical gray level co-occurrence matrix (GLCM) and its spatial variants are utilized to compute texture parameters like, energy, entropy, moment of inertia, etc., of an infected area [39, 53]. After color space conversion, a spatial gray level dependence matrix (SGDM) of H image is also employed to extract several parameters [46, 49]. A hybrid feature combing two or more texture features based on discrete cosine transform (DCT), structure, Fourier transform, difference operators, and Wavelet packet decomposition is built for efficient disease detection [36, 74]. Fourier based fractal descriptors from each lesion are observed to give good results [45].
Some studies have combined texture with color (histograms or moments) as well as shape (area, perimeter, length, width, compactness, rectangularity, roundness, and elongation) features to detect type of plant leaf diseases [37, 39]. Combination is found to improve the system performance [33]. A few have eliminated texture and worked only with color and shape. Shape features are computed along with mean, median, standard deviation, Quartile 1, Quartile 3, and average brightness of RGB color space [4, 21]. Another research presented Eigen vector based extraction to detect cotton leaf diseases [75]. Recently, local descriptors such as speeded-up robust features (SURF), histogram of oriented gradients (HOG), scale-invariant feature transform (SIFT), dense SIFT (DSIFT), pyramid histograms of visual words (PHOW) are explored and compared for better detection as well as classification of soybean diseases [31]. The reported results show that usage of PHOW leads to a better system.
3.5 Classification or Recognition
Classification is an important module in plant disease detection systems. This manuscript considers systems that detect plant diseases using an image, thus classification here is defined as a process of categorizing plant leaf images based on identified diseases. Firstly images from a training set are used to train the classifier; the trained classifier then classifies or recognizes test set images. Researchers have explored a range of machine learning methods to identify diseases in several cultures. The classifier should differentiate between a healthy and an unhealthy leaf image [76].
Machine learning methods are categorized as supervised and unsupervised [77]. The training set for supervised methods has inputs and the corresponding response values. In contrast, unsupervised methods build inferences for absent labeled responses in the training set. Semi-supervised, a special class of supervised methods, utilizes a mix of labeled and unlabeled training data. Classification techniques popularly explored in the domain of plant disease identification are shown in Fig. 3. A few works have lead to accurate identification using other techniques based on features, fuzzy logic, etc. as shown in Fig. 3. Section 4 presents a discussion on various classifiers explored for identifying plant diseases in different cultures. On the basis of observations made, an attempt to identify the best known system, the most studied culture as well as the most popular classifier is made.
4 Classification: A Review
Heterogeneity in leaf images greatly affects the performance of classifiers to identify and classify infected leaves. This work thus analyzes classifiers after categorizing crops (or cultures) as is shown in Fig. 4. The following sub-sections discuss performance of different classifiers with respect to various cultures in each category. Many articles explore leaf diseases for a single culture and some focus on diseases irrespective of the culture. Latter works, termed as ‘Assorted Cultures’ in this work, use datasets consisting of heterogeneous cultures. Figure 5 shows the current state of research in different crops. Clearly, food grain crops are studied the most and very few studies have focused on floriculture crops. Several unexplored crops do exist because either they are unknown or their images are unavailable.
4.1 Cash Crops
Cotton has always been a very popular raw material in textile industries and its crisis has to be dealt rightly. Survey shows that 80 to 90% of cotton diseases can be recognized just by observing leaves appearances [78]. The study focusing on attainment of desired accuracy in machine vision based disease recognition systems is conducted [79]. It trains SVM on several combinations of features to identify the best one [79]. The work obtains a set of informative features for a dataset containing spots, stains or strikes infected leaf images. The study declares texture as the best discriminators (83%) and the worst is shape (55%). The most appropriate set of 45 features reports maximum classification accuracy of 93.1%. Another work extracts features using Eigen vectors [75]. The image space is decomposed into sub-spaces then features are regularized and extracted in each of the sub-spaces separately. The system is trained using nearest neighborhood classifier and achieves 90% accuracy in detecting the red spot. The proposed feature improves accuracy and can be utilized to identify other cotton leaf infections too. Another system employs back propagation neural networks (BPNN) with adaptive learning characteristics to detect mildew (powdery, downy) and leafminer diseases [78]. Obtained results show appropriateness of neural networks in accurately identifying cotton leaf diseases. A slow snake segmentation based feed forward BPNN is developed to detect myrothecium, bacterial blight, and alternaria diseases [80]. The system employs Hu moments for training using Levenberg–Marquardt optimization and reports an average accuracy of 85.52%. The performance and robustness of the system can be enhanced by using other features at a cost of increased training and testing timings.
BPNN is utilized to develop an automatic disease detection system for another globally important cash crop, groundnut [81]. Groundnut that bridges vegetable oil deficit in most of the countries usually suffers from cercospora. The system uses color and texture features to detect four phases cercospora, cercospodium personatum, phaeoisariopsis, and altemaris of this disease. Reporting an accuracy of 97.41%, the work proves relevance of neural networks in automation of plant disease detection systems. Another work combines morphology with heuristics designed using specialist knowledge. The system measures early and late leaf spots caused by Cercosporidium personatum and Cercospora arachidicola fungi in peanut [82]. Trained using CMYK color channel, this system is automatic, practical, quick, and computationally effective. Moreover, it uses only two training images (one from each spot) and is tested with 124 early and 114 late leaf spots images. Still the system reports great results. Production of any crop gets affected by deficiencies also; a study thus attempts to detect different stages of deficiencies in groundnut using geometric moments [83]. The system can assist farmers in deficiency detection as well as estimation of its stage, which is difficult to be done with naked eyes. The system reports an accuracy of 93% and can successfully be applied to other cultures.
Table 1 summarizes all the researches covered under cash crops in this study. For cotton, SVM is shown to achieve maximum accuracy of 93.1% in detecting spots, stains and strikes. However, BPNN detects cercospora in groundnut with an accuracy of 97.14%. In summary, neural networks, i.e. BPNN can be considered as the most preferred classifier. Also spots are the commonly explored disease in case of cash crops. Researchers have reported an accuracy of more than 90% in correct identification and classification of spots.
4.2 Horticulture Crops
4.2.1 Fruits
The important commercial group in this category is citrus plants. Some popular citrus crops are the tangerines, limes, oranges, grapefruits, and lemons. These crops are mainly affected by melanose, scab, canker, downy mildew, powdery mildew, greasy spots, and anthracnose. Using normal and infected grapefruit leaves, both front and back, a generalized square distance classifier is designed for four classes: normal, melanose, scab, and greasy spot [34]. Eight statistical classification models based on combinations of texture based HSI color features are compared. Model built using intensity features reduces accuracy for leaf fronts but not with backs. Moreover, hue and saturation based model performs better (95.8%) than intensity based model (81%). Models using HSI texture features or reduced hue and saturation features reported 100% accuracy. The models are computationally efficient, robust to light variations, and are best suited to examine diseased leaves under laboratory conditions. The study suggests cameras that control lighting levels which in turn helps to reduce impact of low lighting conditions on hue and saturation. Another work focuses on automatic identification and classification of infected grapefruit leaf using multiple artificial intelligence techniques [3]. The system uses self organizing feature map (SOFM) and BPNN for pre-processing; modified SOFM, genetic algorithms (GA), and SVM for segmentation; and SVM again for classifying leaf samples as normal, rust, and scab. The system reports 97.8% accuracy. This work presents a complex but effective blend of several techniques. The final resulting features are filtered using Gabor for improved SVM performance. As a result acceptable accuracy (scab—83.5% and rust—82.5%) is observed. In contrast, other work uses a simple feed forward BPNN to detect downy and powdery mildew diseases [46]. The system is robust to lighting effect as it employs only hue component. As a result 100% accuracy is observed on small dataset of 33 images. The developed system can also be utilized to detect other leaf diseases like anthracnose. The study suggests replacement of k-means to improve its aptness in accurate lesion extraction. Another system attempts to detect downy and powdery mildew using a combination of PCA reduced color, texture, and shape features to train several neural networks (BPNN, RBF-NN, GRNN, and PNN) [55]. The developed system reports 100% fitting accuracy in each of the four cases. GRNN and PNN are found to be the best (94.29%) for fungal disease detection followed by BPNN (approximately 91%) and RBF-NN (80%).
Instead of focusing on leaf disease detection, an approach to discover potassium deficiency in six red grapes varieties viz. cabernet sauvignon, cabernet franc, merlot, malbec, shiraz, and tempranillo, is developed [84]. The study mainly compares the performance of histogram and k-nearest neighbor (k-NN) based segmentation techniques. The study reveals inability of histogram based techniques to distinguish colors and hence found them suitable for grayscale images only. But for images with shadows or taken in less controlled environment conditions, k-NN based techniques are preferred. The presented approach can also be used to identify other deficiencies after some minor revisions, like addition of sample color classes to the database and designing of rules to categorize the symptoms. A two phase system combining techniques of image processing, K-means, and fuzzy set theory is developed to identify downy mildew [85]. The first phase utilizes k-means for feature reduction. Fuzzy value is then computed for each cluster feature with respect to the number of infected images. Features with fuzzy values larger than the predefined threshold are used for detection. If average of the retained fuzzy values for an image is greater than or equal to some predefined threshold then the sample is infected. This fuzzy system reports a classification accuracy of 87.09% thus proves the effectiveness of fuzzy set theory in the domain of automatic detection of leaf diseases.
Similarly, performances of k-NN, Naive Bayes (NB), LDA, and Random Forest Tree (RFT) are observed to automatically detect lemon leaf diseases [59]. Forty sample images from each category (greasy spot, scab, and melanose) are collected in addition to normal leaves. Texture features of 50% of images for training classifiers. The study observes following classifier arrangement in increasing order of classification accuracy: k-NN (77.5%), NB (95%), RFT (97.5%), and LDA (98.75%). Moreover, the study also reveals that it’s easier to classify normal and greasy spot leaf samples as compared to scab and melanose (least classification rate) leaf samples.
A novel two-level feature descriptor is presented to detect and classify orange leaves as normal or infected with canker, black spot, scab, and melanose [15]. Primarily, the work is meant for images collected in fields. The descriptor employs enhanced AdaBoost (SceBoost) to separate background and combines color-texture zone-based local features to get the final descriptor. Experimental comparisons with existing descriptors prove effectiveness of the proposed descriptor. Further several classifiers, namely, Adaboost, RBF-NN, k-NN, and SVM are trained with this descriptor. SVM reports a minimum classification rate of 63% and Adaboost achieves a maximum accuracy, i.e., 88%. Results obtained by the proposed approach are found to be closer to those obtained by human experts, which confirms the feasibility of the system.
For performance comparison, summary of all the studies related to fruit crops is given in Table 2. It can easily be observed that mildew in grape fruit is explored the most in past 10 years. Accordingly, outstanding results (100% detection using feed forward BPNN [46] and statistical analysis of HSI [34]) are obtained for small datasets (≤ 40 images). However, the superiority of multi-class SVM is discovered by looking at the number of training and testing images [3]. It reports an accuracy of ≈ 83% on a large dataset of more than 1500 images. The results obtained for other fruits are not as good as those for grapes. Also scab is found to be the most studied disease followed by spots, melanose, and mildew. For this category of crops, although the best performance is reported with feed forward BPNN, but others like SVM, NB, LDA, random forest, and statistical analysis have also displayed the potential of their applicability.
4.2.2 Vegetables
Chili, a high-risk horticultural good, generally gets affected by diseases caused by bacteria, micro-organisms, and pests. An accurate and fast system for early detection of infected chili leaves is designed [16]. Instead of concentrating on some specific set of diseases, the system examines each plant on a scale of healthiness. The criterion to measure healthiness is based on percentages of a few colors in a leaf image. Tested on 107 samples, the system reports acceptable results. Although, the main focus is to reduce the usage of harmful chemicals by early recognition of potential problems in plants.
Next in this category are cucumbers which have valuable nutritional benefits, such as the hydrating properties. Its production is commonly affected by powdery mildew, downy mildew, brown spot, angular leaf spot, blight, and anthracnose leaf infections. A study compares linear, polynomial, radial basis, and sigmoid kernel based SVM with artificial NN to identify powdery and downy mildews [56]. Each of the four kernels is individually trained with color, texture and shape features. The highest recognition performance is reported by linear kernel, utilizing least number of vectors, in all the considered cases and the color features are shown to have the lowest running time. Similarly, best results are achieved with color features followed by a combination of texture and shape. The experimental results show that SVM is more appropriate than ANN for efficient recognition of powdery and downy mildew in cucumber leaves. Another similar work presented a system to detect downy mildew, angular leaf spot and brown spot [86]. Radial basis function based SVM generates higher recognition rates than sigmoid and polynomial kernel based SVMs. Study suggests the usage of each spot image in an infected leaf during training for an improved system efficiency. Applicability of ANN to detect fungal infections (downy mildew and powdery mildew) is proved by means of an autonomous device [87]. It is based on Levenberg–Marquardt back-propagation algorithm; and perceives leaf symptoms using normalized thermal and textural parameters. The developed device can detect an infection and an hour post inoculation using images. In another such attempt PNN is trained using a 38 dimensional vector (24 color, 4 shape, 5 texture, and 5 meteorological features) [88]. The study focuses to enhance the recognition accuracy of downy mildew, blight and anthracnose infected leaf images acquired under different environmental conditions. Achieving a recognition rate of 91.08% using 300 image samples shows the ability of combined features to successfully train PNN.
Tomato is another important commercial crop which frequently gets infected and leads to low production quality. Common tomato leaf diseases are bacterial (canker, spec, spot), anthracnose, fungal blight, viral curl, etc. A system to automatically detect nitrogen and potassium deficiencies in tomato culture is presented by uniquely combining GA for feature selection and fuzzy k-NN for classification [36]. A fine set of color-texture features is used to train fuzzy k-NN. The results present a classification accuracy of 90 and 85% for nitrogen and potassium deficient leaves. Study shows that the chosen feature set using GA gives more accuracy as compared to the whole feature set. Moreover, a binary tree classification framework is presented to identify a nutrient deficient leaf. The developed system is claimed to successfully identify disease 6–10 days before the actual disease symptoms become visible to an expert. Another work uses a unique hyperspectral imaging concept to automate yellow leaf curl detection without visible scars [43]. Efficiency of the system to distinguish nine texture features of healthy and infected leaf samples, computed in different spectrums, is shown using receiver operator characteristic (ROC). The study signifies that leaf edges are more prone to diseases than its midrib area. System accuracy varies with the employed texture feature ranging from 87.2 to 92.3%. Another study uses simple color descriptors to train 1NN classifier [54]. The main focus is to compare color structure, scalable color, and color layout descriptors to detect mycotic infection (early blight). Using nested-leave-one-out cross validation method, the study summarizes that the classification accuracy of color structure is optimum. The work also suggests usage of texture features for improved accuracy. Similarly, another work compares the performance of SVM with different kernel functions: linear, RBF, MLP, and polynomial [76]. The main aim is to differentiate healthy-unhealthy tomato leaves using texture features. The system is efficient as it is trained on 400 images only and tested on 800 images. The highest classification accuracy of 99.83% is achieved for SVM with linear kernel function. Decision trees are also explored to classify healthy and diseased leaves infected from bacterial canker, bacterial leaf spot, fungal late blight, septoria leaf spot, and leaf curl [89]. The system reported an average recognition accuracy of 78%. The authors attempted the same objective using fuzzy and BPNN as well [90]. An improvement of 9.2% in average recognition accuracy is observed with BPNN.
The vegetable category of horticulture crops is well explored for chili, cucumber, and tomato. The same is shown by the summary results in Table 3. SVM-Linear, 1-NN, and BPNN are observed to perform the best by reporting 100% accuracy in detecting the cucumber and tomato leaf diseases. Various versions of SVM are popularly explored in this crop category followed sequentially by neural networks and nearest neighbor classifiers. It is clearly visible that more than 90% accuracy is achieved in nearly 50% of the studies covered in this manuscript. For tomato, all diseases are equally explored; however for cucumber mildew is the most explored as well as the most correctly detected and classified leaf disease.
4.3 Floriculture Crops
Oil palm easily gets infected by several leaf diseases like wilt, rots, streaks, blast, colored spots, and blight, etc. These diseases are mainly caused due to virus, bacteria or nutrient deficiencies. A system suggesting suitable fertilizer to cure several infections caused due to macro- or micronutrients deficiencies is developed [58]. Deficiency caused due to Nitrogen, Phosphorous, Potassium, Boron, Magnesium, Manganese, and Zinc is considered. A fuzzy classifier is developed using color and shape features, and rules are designed after interviewing domain experts. It provides a nondestructive way to identify deficiency, improve productivity, and optimize fertilizers usage. Contrary, another work attempts to identify leaf diseases showing visual symptoms like hawar leaf, anthracnose, and leaf spot [21]. Each pixel identified as a spot is used to extract color and shape features. NN with different number of hidden neurons (3, 6, and 12) are trained with the extracted features. The best classification accuracy of 87.75% is achieved using a NN with 6 hidden neurons.
Not only commercial crops, but a range of decorative crops are important too. Popular decorative crops are maple and hydrangeas characterized by large and heavy flower heads. Leaf diseases like anthracnose, wilt, leaf spot, and powdery mildew are common in this crop category, occasionally leaf scorch is also observed. Combining techniques of NN and fuzzy logic, a two phase automatic system is presented to detect leaf spot and leaf scorch diseases [5]. First phase utilizes NN to identify maple or hydrangeas and the second phase classifies the disease using fuzzy logic. Disease severity is graded using five fuzzy rules. The experimental results show effectiveness of the system over manual recognition. The study may assist agricultural experts to automate decisions like identification of correct pesticide, their quantity, etc. Next important decorative crop is phalaenopsis from an orchid family. It is infected the most from bacterial soft rot, phytophthora black rot, and bacterial brown spot. For detecting these diseases in initial stages of crop, an automatic system that analyzes seedlings is developed [39]. It utilizes two classifiers, Bayes for differentiating leaves from a container, and BPNN for classification. BPNN trained with color-texture features reports a classification accuracy of 89.6% and a good infected leaf detection accuracy of 97%. The system is not able to detect covered blade infections but can be of great help to make automatic observations in greenhouses.
The past 10 years studies included in the manuscript for floriculture crops are reviewed in Table 4. The maximum accuracy of only 90.9% is reported for Phalaenopsis. Leaf disease detection and classification is not explored much for floriculture crops. Spots are cleverly explored in all the studies using either the concepts of neural networks or fuzzy logic. Although, the fuzzy logic implementation is supported by visual examination, but the results presented in the corresponding works are quite acceptable. As far as neural networks are concerned, BPNN outperforms others.
4.4 Food Grains
A large variety of food grains is studied in literature, thus for effective presentation a few sub-groups are formed. They are: Clover and corn; Legumes; and Rice and wheat. The details of works identified in each sub-group are included in following sub-sections.
4.4.1 Clover and Corn
A pixel classification method to detect ozone-induced visible injuries on clover leaves is presented [20]. The study aims to identify an efficient and robust approach for leaf surface injury detection. The system individually uses four classifiers trained on different color and texture features. The classification approaches used are fit to a pattern multivariate image analysis combined with T2 statistics, residual sum of square statistics, k-means clustering, and linear discriminant analysis (LDA). Evaluations made using manually segmented images as the ground-truth prove that LDA is superior to other approaches. Other observations show computational efficiency and robustness of LDA to variable backgrounds as well as degrees of injury. Also, LDA trained on color feature is observed to detect leaf surfaces injury rapidly, thus is suitable for real-time applications.
A few hazardous corn leaf diseases are leaf blight (turcicum, maydis, sheath), banded leaf, mildew (powdery and downy), and bacterial stalk rot. Among these leaf blight and mildew are focused the most. A system to recognize corn leaf spot diseases, i.e., leaf blight, sheath blight, and southern leaf blight is designed using YCbCr color space [53]. After recognizing the infected region, texture features using spatial GLCM are extracted to train BPNN. The developed system achieves classification accuracy 98%. Another work developed a histogram feature based system to identify and grade turcicum leaf blight disease [91]. The proposed methodology is capable to identify different corn diseases. For example, the classification accuracies reported to detect downy mildew and powdery mildew are 83.5 and 95.2%, respectively. Another work uses locality sensitive discriminant analysis (LSDA) to develop a supervised, robust, and orthogonal nonlinear dimensionality reduction algorithm, named orthogonal locally discriminant projection (OLDP), for plant disease detection [6]. The system is based on 1NN classifier and is trained for five corn leaf diseases. Results presented on real (infected) leaf images of maize reports 93.42 classification accuracy using 18 training images. Another system is developed using k-NN classifier trained with color, texture, and shape features of infected leaf regions for five types of corn leaf diseases [48]. The results are compared with three more systems having different classifiers, i.e., color-texture features with neural network, principal component analysis with neural networks, and Bayesian. The reported classification rate of more than 90% with 18 training samples shows the applicability of k-NN classifier over other combinations. A histogram based system combining SRG and curvelet modulus correlation (CMC) algorithms is also developed to detect six maize diseases: leaf blight, rust spots, gray leaf spot, curvularia leaf spot, brown patch, and small spot [92]. A histogram template is used to detect disease category of the target image. Using n-fold cross-validation, the system reports a classification accuracy of 94.45%.
Summary of clover and corn food grains is given in Table 5. Clover leaf diseases are not explored much in literature. Among the results from available classifiers, LDA provides the highest accuracy of 95% followed by K-means and statistical methods. For corn leaf diseases a range of classifiers are studied and BPNN is observed to achieve the highest accuracy of 98% in leaf spots detection using a small dataset of 40 images. Surprisingly, the results reported using feature based classifiers prove their applicability in corn leaf disease detection. A histogram based system reports an average accuracy of 91.8% and another similar feature based (SRG and CMC) system reports the second highest average accuracy, i.e., 94.45% for corn culture. One more clear observation is the larger size of the dataset considered in feature based systems (253–744 images) as compared to other available works. This further clarifies the superiority of these systems. Also spots are found to be the most commonly studied corn leaf disease. Considering only the reported accuracy values, BPNN can be seen as the preferred classifier for group of food grains too.
4.4.2 Legumes
Digital color imaging finds its usage in diagnosing nutrient deficiencies in plants by observing changes in leaf color. This also provides a way for system automation. One such work attempts to identify macronutrient (Nitrogen, Potassium, Phosphorus and Magnesium) deficiencies in three legume species, viz. pea, yellow lupine, and faba bean [52]. Leaf color variations are observed in L*a*b* and HSI color spaces with Euclidean distances. Remarks presented show that crop species direct the phenomenon of changes in leaf color caused due to some deficiency. Study reveals that potassium deficiency is greatly visible in pea and faba bean where as yellow lupine responded to phosphorus deficiency the most.
Another popular crop in this category is soybean. In recent years, various soybean diseases, like fungal (brown spot, frog eye, rust etc.), bacterial (pustule and blight), and viral (bean pod mottle virus) are explored for automatic detection. A few systems able to work on images captured in fields with different conditions are developed [41, 74]. Using images acquired with a mobile phone, a method detects and classifies two soybean diseases, i.e., brown spot and frog eye [41]. For 50 testing samples, the k-NN classifier trained with shape based feature vector is shown to identify brown spot and frog eye with 70 and 80% accuracy. In addition, two more diseases rust and bacterial blight, are also detected in another work [74]. LDA is trained using a combination of structural texture and normalized DCT based features. The system reports an average classification accuracy of 89.9%. The proposed hybrid feature is said to classify other infections like downy mildew and sudden death syndrome well. Further, the system can be used for rice, beans, cotton, fruits, vegetables, etc. A system based on concepts of reference histogram, correlations, and likelihood function is developed for automatic detection of nine diseases, namely, bacterial blight, rust, phytotoxicity, stem canker, corynespora leaf spot, myrothecium leaf blight, downy and powdery mildew, and septoriabrown spot [93]. Results are presented using a confusion matrix. Diseases with least confusion are myrothecium leaf blight and downy mildew. It is also observed that for good classification accuracy in a system dealing with many diseases consideration of external parameters is keenly required. A disease independent and level estimation method is developed based on three parameters, i.e., ratio of infected area, lesion color index, and damage severity index [94]. The system accurately identifies various leaf diseases; they are rust, bacterial blight, brown spot, sudden death syndrome, frog eye, and downy mildew. An effective and fast disease detection method based on local descriptors and bag-of visual words is presented [31]. Five local descriptors (SURF, HOG, DSIFT, SIFT, and PHOW) are compared on a real-world dataset containing 300 healthy and 900 infected leaf samples (mildew, rust tan, or rust RB). The system uses SVM and is evaluated on correct classification rate (CCR) metric. The results prove dominance of PHOW over others and its applicability to color spaces. The study also reveals that local descriptors can classify mildew more accurately than classifying rust RB or rust tan. The method is general enough and can easily be used for other crops. Another study trains SVM with SIFT features to develop an autonomous decision support system [95]. The approach uses leaf shape to identify its species and can also classify the sample as healthy or infected. The system reports an average classification accuracy of 93.79%. The main focus is to effectively assist farmers’ using minimal amount of input information, i.e., only an image.
Besides detection, a system to grade disease severity is developed for reducing the usage of pesticides or other control measures [18, 25, 29]. A neural network based system for classifying downy mildew, frog eye, and bacterial pustule infections reports an accuracy of 93.3% [25]. Another study presented a severity grading system using K-means clustering to automatically detect diseases (bacterial leaf blight, septoria brown spot, and bean pod mottle virus) [18]. Efficacy of the system is evaluated by comparing the results with manual technique. One more system aims to study color distribution and pixel relationship at every stage of disease growth [29]. Observations are made for 25 days using local and global features of rust infected leaf images. Again percentage disease index (PDI) based on severity levels is computed for disease categorization. The minimum PDI of 0.2 and maximum of 95.5 are observed on 6th and 25th day respectively. The study reveals that higher PDI indicates a decrease in spatial relationship among color and gray pixels due to lesser contribution of green color region.
Table 6 summarizes several studies to detect and classify leaf diseases in legumes species such as pea, yellow lupine, faba bean, and soybean. Considering 57 leaf images infected from rust, brown spot, bacterial blight, and frog eye, LDA is observed to be 100% accurate in detecting brown spot and rust. Similar observation is obtained for detecting myrothecium leaf blight using the concepts of reference histogram, correlations, and likelihood function. But nothing can be said much in later case as dataset contains only 2 images. On contrary, the same concept reports 9% accuracy for stem canker detection using a larger dataset of 22 images. In all, this concept needs more supporting observations for stronger recommendation in future studies. However, the results obtained with SVM classifier is the highest, i.e. 98% followed by NN and feature based systems which have reported 93% accuracy. Also automatic detection of various diseases is attempted for soybean culture mainly, blight, rust, brown spots, and frog eye are explored the most.
4.4.3 Rice and Wheat
Majority of the diseases in rice can easily be recognized by observing the appearances of spots around the infected areas. Moreover unbalanced mineral compositions cause several deficiencies which lead to a disease. Both of these causes are explored by the researchers to automate the detection process. A prototype system using BPNN is developed to identify Nitrogen, Iron, Magnesium, Potassium, Boron, and Manganese deficiencies in rice [27]. The system combines the outcomes of two BPNNs trained individually with color and texture features. The segmentation mappings obtained at the output layers categorize 88.56% of pixels accurately. Another variant of NN, self organizing map neural network (SOM-NN) is employed to detect brown spot and rice blast diseases [65]. The network is trained using gray feature values of spots and is tested on RGB as well as Fourier transform features. The system performed better with RGB features. Results generated on images transformed in frequency domain are inferior to those obtained with original images. A faster version of NN, probabilistic neural network (PNN) trained using fractal texture descriptors are also explored [45]. The system reports good classification accuracy for four diseases, namely, tungro (97.96%), leaf blast (83%), bacterial leaf blight (96.25%) and brown spot (92.31%). The observations show that color variability in leaf blast leads to higher misclassification rates. In fact, the method presented fails to differentiate diseases with similar color characteristics. To get efficiency in such cases, other features like shape are also required.
SVM is also explored to identify rice bacterial leaf blight, rice blight, and sheath blight diseases [37]. Radial basis function based SVM models trained individually on various features (texture, shape, and their combination) are presented and compared for efficient identification of bacterial leaf blight, rice blight, and sheath blight. Maximum overall classification accuracy of 97.2% is observed when combination of features is employed. On other hand minimum overall classification accuracy is achieved with shape features, as rice blight and sheath blight spots have instable shape. The study thus recommends usage of shape and texture features for accurate disease detection not only for rice, but for other crops too. Another work developed a two stage system and compares Bayes classifier with SVM [19]. In first stage, system identifies healthy or infectious sample. In infectious case, the second stage classifies the sample as brown spot and blast. The system accurately recognizes 92% of healthy samples and performs better for brown spot than blast. Besides, the classification accuracy of Bayes (79.5%) is superior to SVM (68.1%). The study also reveals time efficiency of Bayes over SVM. Another work compares performance of k-NN with SVM using an automatic disease detection and classification system [96]. The identification phase utilizes Haar features to train AdaBoost classifier and reports a detection accuracy of 83.33%. The second phase compares SIFT trained k-NN with SVM in classifying test leaf samples as brown spot, leaf blast, and bacterial blight. This study reports that k-NN (93.33%) is better than SVM (91.10%). Also, authors claimed that the system helps in early disease identification.
A study designed a set of membership functions to automatically identify rice sheath blight, brown spot, and rice blast diseases [33]. The system utilizes the nearest neighbor classification concept to put a test sample in appropriate disease class. It is fast and provides good results with reasonably high-quality images. The system recognizes brown spot (85%) more successfully as compared to other two, although the reported average classification accuracy is 70%. Another study interviewed agricultural experts and designed production rules with forward chaining method to detect brown spot, narrow brown spot and blast diseases [72]. The main focus is to reveal the importance of threshold in local entropy threshold and Otsu method segmentation techniques. The developed system achieves 94.7% classification accuracy using local entropy threshold as it deals effectively with different intensities and illumination issues. A novel idea of Fermi energy is introduced to segment an image and then a rule generation algorithm using classification rule mining techniques is presented [4]. The developed rule based classifier reports a classification accuracy of 92.29% for identifying brown spot and blast. Comparison with various traditional classifiers, like C4.5, NB, Part, Kstar, SMO and bagging, further proves its efficacy in accurate plant diseases detection. Capability of fuzzy c-mean clustering algorithm is also utilized to detect blast fungal disease and the associated production loss for rice crop [97]. This pixel based approach separates leaf region into three classes: healthy pixels, medium and highly affected by blast pixels. The main focus is to estimate the loss of production due to blast instead of its detection. The system reports 85% accuracy and is meant to assist farmers for precision farming using decision support systems.
Wheat, another most produced food grain, is observed to get affected generally by any form of rust diseases. Common types of wheat rust are powdery mildew, stripe, septoria leaf spot, tan spot, and snow mold. Researchers have proposed several solutions for automatic and accurate classification of these rust categories at early stages. In one such attempt, a combination of color, texture, and shape features are used to train BPNN, radial basis function NN (RBF-NN), generalized regression NN (GRNN), and PNN [55]. All the NN are compared on accurately classifying the stripe rust and leaf rust fungal diseases. The study reports the least accuracy for RBF-NN and the optimum performance is achieved when BPNN is trained on PCA reduced features. The fitting and prediction accuracy of the system is 100%. PCA usage is optional in this system, but mandatory in case disease recognition is performed via Internet. In addition, the study suggests replacing PCA with other dimension reduction method in case non-linear features are employed. Improved rotation kernel transformation based directional feature (IRKT) is developed to classify stripe rust and powdery mildew [47]. Experimental results show that IRKT is noise insensitive and provides better edge related information. Compared to edge orientation histograms (EOH), IRKT classifies stripe rust (97.5%) more correctly. However, both EOH and IRKT report same classification accuracy for powdery mildew. Overall it can be summarized that IRKT can successfully be used to recognize a range of wheat diseases. Two systems are developed to detect and recognize four types of rust, viz., powdery mildew, septoria leaf spot, tan spot and snow mold [51, 98]. The system employing fuzzy c-means is simple, fast, and focuses on identifying a set of best suited features [98]. First phase separates a set of diseased leaf images and second phase classifies a test sample. The system reports low recognition accuracy (56%) and also requires images of all types of infections during training. To resolve this issue along with improved accuracy, another system based on BPNN is presented. It is trained on a combination of color and texture features [51]. The system estimated 290 out of 342 test samples accurately. The improved system also attempted to rate the severity of rust infection in addition to diagnosis. The applicability of the system is proved through manual examination by two experienced doctors.
Table 7 summarizes all the studies considered for rice and wheat food grains. For both grains, neural network based classifiers are shown to achieve the best results. In particular, BPNN reports highest accuracy of 100% in detecting wheat leaf diseases (stripe rust and leaf rust) on 100 images dataset. Similarly, for much lesser images (only 40), PNN is observed to detect tungro rice leaf disease with 97.76% accuracy. Researchers have reported an accuracy > 60% in case of rice and > 50% in case of wheat to correctly identify as well as classify diseases. As far as diseases are considered, blast and brown spots are equally studied for rice leaves; also rust is explored in all the works for wheat leaves. Considering number of images, the best performing classifier is Bayes for rice and BPNN for wheat. Both have reported accuracies ranging from 80 to 85% for larger datasets. Here also BPNN can easily be observed to outperform others.
4.5 Assorted Cultures
There exist a range of works focusing on automatic detection of a common leaf disease affecting a group of cultures. All such works use dataset containing leaf images infected from some specific diseases irrespective of culture. One such work utilizes image processing techniques to detect and classify five leaf diseases, late scorch, early scorch, ashen mold, small whiteness, and cottony mold [50]. The system trains a 10 hidden layer based feed forward BPNN with optimized color-texture feature set obtained from infected leaf region. Five such models considering various color components (HSI, HS, H, S, and I) are compared. The model using HS components reports a maximum classification accuracy of 89.5%. The study also shows that computational complexity improves if intensity (I) is not considered. Another work proposes a 2D Fourier transform based wilting index for early detection of temporary wilting caused due to drought stress [99]. Inspired from leaf morphology, the index depends on curvatures of the points on a 3D laser scanned leaf image surface. The applicability of wilting index is shown using zucchini leaves and is also suitable for Cucurbita pepo leaves, as both the species have same leaf shape. However, generalization of index is questioned due to variability of plants, their leaf shapes, and wilting morphologies. The study motivates all researchers in this domain to explore leaf morphology in 3D space to extract physiologic information using mathematical tools. An intelligent and specialized image sequence capture device is integrated to capture a series of images for automatic spore detection [64]. The obtained set of images are processed and identified as powdery mildew spores using BPNN. The proposed approach counts the number of spores after detection. Using 155 training samples BPNN reports 95.5% accuracy, but the accuracy obtained for 89 testing samples is only 63.6%.
Infections may affect any part of a plant. Based on this fact, various fungal diseases are examined in different crop categories, viz. fruit, vegetable, commercial, and cereal using separate models [63]. The work considers infections in leaf, stem, and fruits of various cultures in four categories; vegetable crops (beans, bengal gram, soybean, sunflower, and tomato); commercial crops (chili, cotton, and sugarcane); cereal crops (jowar, wheat, and maize); and fruits (grape, mango, and pomegranate). The model presented for vegetable crops uses local binary patterns of both sides of a segmented leaf to analyze several infections (anthracnose, blight, rust, and mildew). For this category neuro-k-NN classifier, a combination of BPNN and k-NN is introduced and compared with ANN. Neuro-k-NN (91.54%) reports better average classification accuracy. PCA reduced discrete wavelet transform (DWT) features are utilized to train Mahalanobsis distance based PNN classifiers for commercial crops. PNN reports an average classification accuracy of 86.48% to detect anthracnose, rot, powdery mildew, alternaria leaf spot, smut, gray mildew, and wilt infections. The system developed for cereal crops employs different combinations of color, texture, and shape features to train SVM. In first stage radon transform differentiates between a healthy and diseased plant followed by SVM classification to identify an infection as leaf blight, leaf spot, powdery mildew, leaf rust, and smut. The results show that color-texture feature based training is most appropriate for this category as it reports the maximum average classification accuracy of 85.33%. The study considers infections affecting fruits as well. The corresponding model achieves an average classification accuracy of 94.085% and classifies the test sample as normal or infected (partial, moderate, and severe). The presented architecture can be used in remote monitoring of crops to detect diseases at early stages. Moreover, this work is effective yet complex and challenging due to varying outdoor conditions. Another work compares the performance of ANN with SVM on different features (color, texture, and their combination) [17]. Both the systems are trained using 900 images taken from plant pathology department, Dharwad. They can identify an agriculture crop test sample as fungal, bacterial, nematodes, viral, deficiency and normal. ANN models report an average classification accuracy ranging from 82 to 87%, but those based on SVM performs better for all the features. The maximum and minimum classification accuracies achieved by SVM models are 84 and 92%, respectively. Here as well, SVM performs the best when trained with a combination of color and texture features. The performance of SVM trained using texture features is also studied for other cultures to detect a range of diseases [49]. The approach is designed to detect blight, sun burn, scorches, spots, bacterial/fungal infections, and mold in leaves of different cultures (banana, beans, jackfruit, lemon, guava, mango, potato, sapota, and tomato). Trained on H image based texture features, the minimum distance and SVM classifiers report accuracies of 86.77 and 97.74%, respectively. The study again made the same observation of “SVM is better”. In contrast with the trend, one study analyzes the effect of Salmonella Typhimurium (human pathogenic bacterium) on immune system of Arabidopsis leaves [100]. Although the study uses popular and successful linear SVM trained using color features. The system classifies infected foreground region pixels with 95.8% accuracy and additionally refines the final image using neighborhood-check method. The system is shown to provide accurate results for the considered dataset and can also be extended to detect other diseases. An effective blend of color features (histograms and transformations) with pairwise-correlation based classification is presented for disease recognition [101]. Dataset used to design the system contains 82 biotic and abiotic stresses of 12 plant species. The performance is evaluated by means of confusion matrix prepared for each plant species. The obtained results are not very impressive and can be improved by considering some measures related to capturing of images. All the required points of consideration are also discussed thoroughly by the author [11]. This work is very similar to the one discussed in Sect. 4.4.2 for soybean.
Focusing uneducated farmers, a human-mobile interface (HMI) is designed recently that can assist them in automatic examination of fields at any phase, just on a mobile click [7]. The initial steps (pre-processing and segmentation) are implemented at the client mobile device, and the remaining steps (feature extraction and classification) are performed on the pathology server. Final result is intimated to the user using short messaging service (SMS). In the present state, the interface runs only on Android operating system using concepts of Gabor, GLCM, and k-NN classifier. It is able to identify infections as leaf spots and leaf blotch with a classification accuracy of 93%.
Similarly, a deep convolution NN model for crop disease diagnosis on large scale using a smart-phone is presented [23]. The study focuses on popular AlexNet and GoogLeNet architectures. Applicability of system is proved using a subset of PlantVillage dataset containing 54,306 images of 14 crop species infected from 26 diseases. As per expectations, the approach reported an accuracy of 99.35%. Moreover, for an online collected heterogeneous image dataset the system achieves 31.4% accuracy. The study suggests usage of diverse training set to attain feasible results in case of general purpose large datasets. Working on the same domain of deep convolutional NN, a system to recognize 13 types of leaf diseases is developed using a popular Caffe framework [102]. The system is tested on a dataset of 30,000 images prepared after suitable transformations of more than 3000 original images collected online. For the considered diseases, system reports an overall classification accuracy of 96.3%, more specifically the obtained values range from 91.11 to 98.21%. The study reveals that augmentation process is more important than fine-tuning to obtain desired overall accuracy.
The current research trend is focused much in the development of a system that is capable of detecting and classifying a range of diseases over a variety of cultures. To highlight this observation, Table 8 presents the summary of all the studies covered for assorted cultures. A gradual increase in the number of diseases being identified by a system is clearly visible in Table 8. As far as classifiers are concerned, this category has explored neural networks and SVM the most. SVM is observed to achieve > 90% accuracy in any case, but different types of neural networks have shown varied performances with accuracies ranging from 31 to 100%. Also neural networks further prove their applicability for larger datasets. It is evident that deep convolution neural networks can effectively be used in systems dealing with some thousands of images (as large as 55,000). Although BPNN has reported 100% accuracy, but in the absence of dataset size, 96.3% accuracy reported by deep convolution neural networks for 33,469 images is observed as the best.
5 Discussions and Summary
5.1 Image Acquisition and Database Size
The leaf images database for a particular infection in a specific culture is very difficult to obtain. This fact is clearly visible by limited size of image databases used in the studies. Only a few works (less than 10%) have used large size databases ranging in thousands [3, 19, 23, 31, 94, 100,101,102]. Also, the ratio training is to testing images, is varying a lot from one work to another. A common observation is the use a large proportion of database images during training than that of the testing phase. However, some exceptions are always there [51, 53, 78]. It is observed that an efficient acquisition of a leaf image is the need of an hour. If captured in real-life (i.e., an uncontrolled environment) then its acceptance would automatically be increased. As per current state-of-the-art works, images taken using a mobile are slowly gaining popularity. Another issue of concern is varying stages of some leaf infections, which complicates the process of image acquisition further. Although, some complicated yet effective image acquisition techniques as well as a single click image systems are presented, but much more is still supposed to be done. The concept of using a leaf back can also be considered in sensor based system for proper detection of an infection in early stages. Because of all these reasons, a transition from the consideration of a specific culture disease to a disease common to a set of cultures can easily be observed in the domain of plant leaf disease detection systems. This transition may help in overcoming several issues related to database size.
5.2 Techniques Employed in Pre-processing, Segmentation, and Feature Extraction Modules
The nature of database images plays an important role in selecting appropriate techniques to perform the task of pre-processing and segmentation efficiently. Among several techniques, one that is suitable for a particular form of acquisition usually serves the purpose. A large variability span is observed in algorithms available under different modules. Similar observations are made for a feature extraction module. In other words, standardization of techniques is yet to be achieved. This observation is palpable in an automatic plant leaf disease detection system as it is a type of content based image retrieval system only. Also, it is observed that proposing a universal technique for an individual module is very difficult for these types of systems. As per the current trends, dependency of techniques employed in any module of the system is very high on the database being used by that system.
5.3 Difficulties in Classification Module
Automation of plant leaf disease detection and classification system is been focused by the researchers since a very long time. Highly acceptable results are reported in some of the studies considering a few numbers of images. Also a range of classifiers are explored in this domain. As observed that the classifiers back propagation neural network, support vector machine, and linear discriminant analysis perform better across all the cultures followed by random forest tree, feature based, Naive Bayes, probabilistic neural network, k-nearest neighbor, multi layer perceptron, and rule mining. Talking specifically, then among all the considered systems 41% have used either SVM or a feature based classifier. Both these classifier types are used equally in the past 10 years. The next classifier that is used popularly in 17% of the articles is BPNN and k-NN is utilized in 14% of the studies. The remaining classifiers are used in remaining 28% of the articles and are excluded from this discussion due to their lesser number. Recently, the deep convolution neural networks are used in systems working with assorted cultures. They are yet to be explored in systems pertained to a single culture. Proper utilization of convolution networks may help in improving the effectiveness of a system on large databases.
5.4 Limitations of Available Systems
Although image analysis and hyperspectral imaging techniques are better than methods that rates disease severity visually [103]. But systems designed using imaging techniques are not perfect as well. Efficiency of any system depends greatly on the quality of training data; indirectly it is actually the number of training images and their extracted features. So it can be said that a well trained system is highly efficient. But, all the existing systems have a well-defined set of requirements which are essential to be fulfilled for accurate performance. If one or the other constraint is not fulfilled, then the considered system may produce inaccurate results which lead to inappropriate disease detection. For example, the problem of over-training or over-fitting is commonly observed in the studied systems that improperly employ powerful techniques of NN, SVM, and GA. In such a scenario, researchers must think of hybrid and adaptive systems designed with flexible set of requirements instead of fixed one. In addition, some generalized techniques working on a group of heterogeneous environments must be developed. Also an in-depth knowledge of several techniques as well as proper usage of classy tools cannot be compromised for efficiency. All these issues of concern actually fall in the area of domain adaptation, which in itself is a very popular research problem these days.
6 Future Scope
The manuscript summarizes various studies to automate identification and classification of plant leaf diseases using machine learning and image processing techniques. The survey shows well-acceptance of a huge range of computer vision techniques in this domain and thus makes it a wide area of research in near future. Here are some research points which may help to enhance current state-of-the-art.
6.1 Disease Stage Identification and Quantification
Usually, a disease has certain stages, but as per the survey most of the researches have focused mainly on disease detection and their classification. Thus, the design and implementation of a system that can detect a particular stage of a disease would be of great interest. In addition, these systems should possess capability to suggest a suitable measure depending on the identified disease stage. Detection of a disease in an early stage, also known as disease forecasting, may help agriculturists to take proper precautions and thus reduce damage percentage. Another related area of research is quantification, i.e., detecting the infected proportion of a culture. This research objective is particularly important as it controls the amounts of pesticides or other chemicals to be used for disease prevention. In the present scenario, chemicals are applied periodically without any prior analysis of infection or quantification. This practice may have harmful effects on human health. Effective application of image processing methods would help in determining, if chemicals are required or not. In case of an accurate quantification, the analysis would further be used to control the quantity of chemicals to be applied.
6.2 Development of New Applications
Various solutions do exist in literature, but the corresponding system is not available for a public use. Only a few Web portals and mobile based applications are accessible to provide an online assistance for a specific disease set of a particular culture. To the best of our knowledge, Leaf Doctor and Assess software are available but they work on images with black background only [104]. Thus, development of an online system, for plant disease detection and then classification, may also form another research objective in this domain. The availability of any such software would help farmers to a great extent. In the near future, these systems may replace the requirement of specialist suggestions in initial stages of infections. For helping farmers in the remote area, the system may also provide an option of “analysis report generation” which can further be send to an expert for getting proper suggestions.
6.3 Accurate Classification
Disease identification is bit simpler than its proper classification. Sometimes it becomes difficult for an expert to classify a particular infection with 100% confidence. Development of systems that can categorize various fungal, viral, and bacterial diseases correctly may also be focused. Literature considers minerals or nutrients deficiencies as another form of plant disease. The development of systems that can effectively differentiate between an infection and a deficiency may be another interesting topic of research. This can be considered as a very difficult objective because from expert perspective separating an infected leaf from a deficient leaf is a complex task.
6.4 Real World Application
Most of the works presented in the manuscript considers images that are collected offline by picking leaves. This is somewhat destructive. The research conducted till date identifies diseases under some specified conditions. To the best of our knowledge, none of the studies perform disease identification in real world scenario with acceptable accuracy. The real world implementation of these studies may be attempted to get a practical method. For example, one may think of a real time system which uses the concept of continuous remote monitoring of a field area for disease identification. This research objective is also related directly to several computational complexity and memory requirement issues.
6.5 Reliability of Fully Automatic Systems
Another issue of concern is the semi-automatic nature of existing systems. Attempts have been made for complete automation of these systems to mimic judgmental ability of an expert. But at some stage or other an agriculture engineer (or a plant pathologist) intervention is often required to keep a check on accuracy. One possible solution to this is an intelligent blend of the expert system concept into the computer vision and machine learning techniques. An attempt to develop such a system may also be of great interest to researchers in this domain.
References
Bagde S, Patil S, Patil S, Patil P (2015) Artificial neural network based plant leaf disease detection. Int J Comput Sci Mob Comput 4(4):900–905
Weizheng S, Yachun W, Zhanliang C, Hongda W (2008) Grading method of leaf spot disease based on image processing. In: IEEE international conference on computer science and software engineering, Wuhan, Hubei, December 8, pp 491–494
Meunkaewjinda A, Kumsawat P, Attakitmongcol K, Srikaew A (2008) Grape leaf disease detection from color imagery using hybrid intelligent system. In: IEEE 5th international conference on electrical engineering/electronics, computer, telecommunications and information technology ECTI-CON., Krabi, pp 513–516
Phadikar S, Sil J, Das AK (2013) Rice diseases classification using feature selection and rule generation techniques. Comput Electron Agric 90:76–85
Rastogi A, Arora R, Sharma S (2015) Leaf disease detection and grading using computer vision technology and fuzzy logic. In: IEEE 2nd international conference on signal processing and integrated networks SPIN, pp 500–505
Zhang S, Zhang C (2013) Orthogonal locally discriminant projection for classification of plant leaf diseases. In: IEEE international conference on computational intelligence and security CIS, Leshan, pp 241–245
Prasad S, Peddoju SK, Ghosh D (2016) Multi-resolution mobile vision system for plant leaf disease diagnosis. Signal Image Video Process 10(2):379–388
Patil JK, Kumar R (2011) Advances in image processing for detection of plant diseases. J Adv Bioinform Appl Res 2(2):135–141
Khirade SD, Patil AB (2015) Plant disease detection using image processing. In: IEEE international conference on computing communication control and automation (ICCUBEA), pp 768–771
Barbedo JGA (2013) Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus 2(1):1–12
Barbedo JGA (2016) A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst Eng 144:52–60
Prashar K, Talwar R, Kant C (2015) A review on efficient identification of american cotton leaf diseases through training set. Int J Comput Appl 132(7):32–39
Martinelli F, Scalenghe R, Davino S et al (2015) Advanced methods of plant disease detection. A review. Agron Sustain Dev 35(1):1–25
Sankaran S, Mishra A, Ehsani R, Davis C (2010) A review of advanced techniques for detecting plant diseases. Comput Electron Agric 72(1):1–13
Zhang M, Meng Q (2011) Automatic citrus canker detection from leaf images captured in field. Pattern Recognit Lett 32(15):2036–2046
Husin ZB, Aziz AHBA, Shakaff AYBM, Farook RBSM (2012) Feasibility study on plant chili disease detection using image processing techniques. In: IEEE 3rd international conference on intelligent system modeling and simulation ISMS., Kota Kinabalu, pp 291–296
Pujari JD, Yakkundimath R, Byadgi AS (2016) SVM and ANN based classification of plant diseases using feature reduction technique. Int J Interact Multimed Artif Intell 3:6–14
Jadhav SB, Patil SB (2015) Grading of soybean leaf disease based on segmented image using k-means clustering. Int J Adv Res Electr Commun Eng 4(6):1816–1822
Phadikar S, Sil J, Das AK (2012) Classification of rice leaf diseases based on morphological changes. Int J Inf Electron Eng 2(3):460–463
Kruse OMO, Prats-Montalbán JM, Indahl UG, Kvaal K, Ferrer A, Futsaether CM (2014) Pixel classification methods for identifying and quantifying leaf surface injury from digital images. Comput Electron Agric 108:155–165
Aji AF, Munajat Q, Pratama AP, Kalamullah H, Setiyawan J, Arymurthy AM (2013) Detection of palm oil leaf disease with image processing and neural network classification on mobile device. Int J Comput Theory Eng 5(3):528–532
Ipm images. https://www.ipmimages.org/about/. Accessed 15 May 2017
Mohanty SP, Hughes D, Salathé M (2016) Using deep learning for image-based plant disease detection. Front Plant Sci 7:1–10
APS Image database. https://imagedatabase.apsnet.org/search.aspx. Accessed 16 May 2017
Gharge S, Singh P (2016) Image processing for soybean disease classification and severity estimation. In: Shetty N, Prasad N, Nalini N (eds) Emerging research in computing, information, communication and applications. Springer, New Delhi, pp 493–500
University of Minnesota Extension. https://www.extension.umn.edu/. Accessed 16 May 2017
Sanyal P, Bhattacharya U, Parui SK, Bandyopadhyay SK, Patel S (2007) Color texture analysis of rice leaves diagnosing deficiency in the balance of mineral levels towards improvement of crop productivity In: IEEE 10th international conference on information technology ICIT, Orissa, pp 85–90
Camargo A, Smith JS (2009) An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosyst Eng 102(1):9–21
Pujari JD, Yakkundimath RS, Jahagirdar S, Byadgi AM (2015) Quantitative detection of soybean rust using image processing techniques. J Crop Prot 5(1):75–87
Rumpf T, Mahlein AK, Steiner U, Oerke EC, Dehne HW, Plümer L (2010) Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput Electron Agric 74(1):91–99
Pires RDL, Gonçalves DN, Oruê JPM, Kanashiro WES, Rodrigues JF, Machado BB, Gonçalves WN (2016) Local descriptors for soybean disease recognition. Comput Electron Agric 125:48–55
Barbedo JGA (2014) An automatic method to detect and measure leaf disease symptoms using digital image processing. Plant Dis 98(12):1709–1716
Anthonys G, Wickramarachchi N (2009) An image recognition system for crop disease identification of paddy fields in Sri Lanka. In: IEEE international conference on industrial and information systems ICIIS, Sri Lanka, pp 403–407
Pydipati R, Burks TF, Lee WS (2006) Identification of citrus disease using color texture features and discriminant analysis. Comput Electron Agric 52(1):49–59
Bauer SD, Korč F, Förstner W (2011) The potential of automatic methods of classification to identify leaf diseases from multispectral images. Precis Agric 12(3):361–377
Xu G, Zhang F, Shah SG, Ye Y, Mao H (2011) Use of leaf color images to identify nitrogen and potassium deficient tomatoes. Pattern Recognit Lett 32(11):1584–1590
Yao Q, Guan Z, Zhou Y, Tang J, Hu Y, Yang B (2009) Application of support vector machine for detecting rice diseases using shape and color texture features. In: IEEE international conference on engineering computation ICEC, Hong Kong, pp 79–83
Mirik M, Michels GJ, Kassymzhanova-Mirik S, Elliott NC, Catana V, Jones DB, Bowling R (2006) Using digital image analysis and spectral reflectance data to quantify damage by greenbug (Hemitera: Aphididae) in winter wheat. Comput Electron Agric 51(1):86–98
Huang KY (2007) Application of artificial neural network for detecting Phalaenopsis seedling diseases using color and texture features. Comput Electron Agric 57(1):3–11
Oberti R, Marchi M, Tirelli P, Calcante A, Iriti M, Borghese AN (2014) Automatic detection of powdery mildew on grapevine leaves by image analysis: optimal view-angle range to increase the sensitivity. Comput Electron Agric 104:1–8
Shrivastava S, Hooda DS (2014) Automatic brown spot and frog eye detection from the image aptured in the field. Am J Intell Syst 4(4):131–134
Cui D, Zhang Q, Li M, Zhao Y, Hartman GL (2009) Detection of soybean rust using a multispectral image sensor. Sens Instrum Food Qual Saf 3(1):49–56
Lu J, Cui D, Jiang H (2013) Discrimination of tomato yellow leaf curl disease using hyperspectral imaging. American Society of Agricultural and Biological Engineers, Kansas City, Missouri, July 21-July 24, p 1
Kumbhar NP, Dhaygude SB (2013) Crop disease detection using CBIR. Int J Emerg Technol Comput Appl Sci 3(2):113–118
Asfarian A, Herdiyeni Y, Rauf A, Mutaqin KM (2013) Paddy diseases identification with texture analysis using fractal descriptors based on Fourier spectrum. In: IEEE international conference on computer, control, informatics and its applications IC3INA, Jakarta, pp 77–81
Sannakki SS, Rajpurohit VS, Nargund VB, Kulkarni P (2013) Diagnosis and classification of grape leaf diseases using neural networks. In: IEEE 4th international conference on computing, communications and networking technologies ICCCNT, Tiruchengode, pp 1–5
Wang L, Dong F, Guo Q, Nie C, Sun S (2014) Improved rotational kernel transformation directional feature for recognition of wheat stripe rust and powdery mildew. In: IEEE 7th international conference on image and signal processing CISP, Dalian, pp 286–291
Zhang SW, Shang YJ, Wang L (2015) Plant disease recognition based on plant leaf image. J Anim Plant Sci 25(Suppl. 1):42–45
Arivazhagan S, Shebiah RN, Ananthi S, Varthini SV (2013) Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric Eng Int CIGR J 15(1):211–217
Bashish AD, Braik M, Bani-Ahmed S (2010) A framework for detection and classification of plant leaf and stem diseases. In: IEEE international conference on signal and image processing ICSIP, Chennai, pp 113–118
Majumdar D, Kole DK, Chakraborty A, Majumdar DD (2015) An integrated digital image analysis system for detection, recognition and diagnosis of disease in wheat leaves. In: 3rd international ACM internat. Symposium on women in computing and informatics, New York, pp 400–405
Wiwart M, Fordoński G, Żuk-Gołaszewska K, Suchowilska E (2009) Early diagnostics of macronutrient deficiencies in three legume species by color image analysis. Comput Electron Agric 65(1):125–132
Kai S, Zhikun L, Hang S, Chunhong G (2011) A research of maize disease image recognition of corn based on BP networks. In: proceedings of third international confeence on measuring technology and mechatronics automation ICMTMA. IEEE, Shanghai, pp 246–249
Molina JF, Gil R, Bojacá C, Gómez F, Franco H (2014) Automatic detection of early blight infection on tomato crops using a color based classification strategy. In: IEEE XIX symposium on image, signal processing and artificial vision, September, pp 1–5
Wang H, Li G, Ma Z, Li X (2012) Application of neural networks to image recognition of plant diseases. In: IEEE international conference on systems and informatics ICSAI, Yantai, pp 2159–2164
Youwen T, Tianlai L, Yan N (2008) The recognition of cucumber disease based on image processing and support vector machine. In: IEEE congress on image and signal processing CISP’08, May, vol 2, pp 262–267
Shinde RC, Mathew CJ, Patil CY (2015) Segmentation technique for soybean leaves disease detection. Int J Adv Res 3(5):522–528
Hairuddin MA, Tahir NM, Baki SRS (2011) Overview of image processing approach for nutrient deficiencies detection in elaeis guineensis. In: IEEE international conference on system engineering and technology ICSET, Shah Alam, pp 116–120
Bandi SR, Varadharajan A, Chinnasamy A (2013) Performance evaluation of various statistical classifiers in detecting the diseased citrus leaves. Int J Eng Sci Technol 5(2):298–307
Xia C, Lee JM, Li Y, Song YH, Chung BK, Chon TS (2013) Plant leaf detection using modified active shape models. Biosyst Eng 116(1):23–35
Bankar S, Dube A, Kadam P, Deokule S (2014) Plant disease detection techniques using canny edge detection & color histogram in image processing. Int J Comput Sci Inf Technol 52(2):1165–1168
Singh V, Misra AK (2015) Detection of unhealthy region of plant leaves using image processing and genetic algorithm. In: IEEE international conference on advances in computer engineering and applications ICACEA, pp 1028–1032
Pujari JD, Yakkundimath R, Byadgi AS (2015) Image processing based detection of fungal diseases in plants. Proc Comput Sci 46:1802–1808
Danping W, Botao W, Yue Y (2013) The identification of powdery mildew spores image based on the integration of intelligent spore image sequence capture device. In: IEEE 9th international conference on intelligent information hiding and multimedia signal processing IIH-MSP, Beijing, pp 170–180
Phadikar S, Sil J (2008) Rice disease identification using pattern recognition techniques. In: IEEE 11th international conference on computer and information technology ICCIT, Khulna, pp 420–423
Masood R, Khan SA, Khan MNA (2016) Plants disease segmentation using image processing. Int J Mod Educ Comput Sci 8(1):24–32
Cui D, Zhang Q, Li M, Hartman GL, Zhao Y (2010) Image processing methods for quantitatively detecting soybean rust from multispectral images. Biosyst Eng 107(3):186–193
Pang J, Bai ZY, Lai JC, Li SK (2011) Automatic segmentation of crop leaf spot disease images by integrating local threshold and seeded region growing. In: IEEE international conference on image analysis and signal processing, October, pp 590–594
Shire A, Jawarkar U, Manmode MM (2015) A review paper on: agricultural plant leaf disease detection using image processing. Int J Innov Sci Eng Technol 2(1):282–285
Gui J, Hao L, Zhang Q, Bao X (2015) A new method for soybean leaf disease detection based on modified salient regions. Int J Multim Ubiquitous Eng 10:45–52
Krishnan M, Sumithra MG (2013) A novel algorithm for detecting bacterial leaf scorch of shade trees using image processing. In: IEEE Malaysia international conference on communications MICC, Kuala Lumpur, pp 474–478
Kurniawati NN, Abdullah SNHS, Abdullah S, Abdullah S (2009) Investigation on image processing techniques for diagnosing paddy diseases. In: IEEE international conference on soft computing and pattern recognition SOCPAR., Malacca, pp 272–277
Gavhale KR, Gawande U (2014) An overview of the research on plant leaves disease detection using image processing techniques. IOSR J Comput Eng 16(1):10–16
Shrivastava S, Singh SK, Hooda DS (2014) Statistical texture and normalized discrete cosine transform-based automatic soya plant foliar infection cataloguing. Br J Math Comput Sci 4(20):2901–2916
Gurjar AA, Gulhane VA (2012) Disease detection on cotton leaves by eigenfeature regularization and extraction technique. Int J Electr Commun Soft Comput Sci Eng 1(1):1–4
Mokhtar U, El Bendary N, Hassenian AE, Emary E, Mahmoud MA, Hefny H, Tolba MF (2015) SVM-based detection of tomato leaves diseases. In intelligent systems. Springer International Publishing, Berlin, pp 641–652
Machine Learning. https://en.wikipedia.org/wiki/Machine_learning. Accessed 17 May 2017
Ghaiwat SN, Arora P (2015) Cotton leaf disease detection by feature extraction. In: Research advances in the integration of big data and smart computing, Oct 13:89
Camargo A, Smith JS (2009) Image pattern classification for the identification of disease causing agents in plants. Comput Electron Agric 66(2):121–125
Rothe PR, Kshirsagar RV (2015) Cotton leaf disease identification using pattern recognition techniques. In: IEEE international conference on pervasive computing (ICPC), January, pp 1–6
Ramakrishnan M (2015) Groundnut leaf disease detection and classification by using back probagation algorithm. In: IEEE international conference on communications and signal processing (ICCSP), April, pp 0964–0968
Barbedo JGA (2013) Automatically measuring early and late leaf spot lesions in peanut plants using digital image processing. In: Congresso brasileiro de agroinformática vol. 9
Nisale SS, Bharambe CJ, More VN (2011) Detection and analysis of deficiencies in groundnut plant using geometric moments. World Acad Sci Eng Technol 5:608–612
Rangel BM, Fernández MA, Murillo JC, Ortega JC, Arreguín JM (2016) KNN-based image segmentation for grapevine potassium deficiency diagnosis. In: IEEE international conference on electronics, communications and computers (CONIELECOMP) Feb 24, pp 48–53
Kole DK, Ghosh A, Mitra S (2014) Detection of downy mildew disease present in the grape leaves based on fuzzy set theory. In: Advanced computing, networking and informatics, Volume 1, pp 377–384, Springer International Publishing
Jian Z, Wei Z (2010) Support vector machine for recognition of cucumber leaf diseases. In: IEEE international conference on advanced computer control, ICACC, March, vol 5, pp 264–266
Asefpour Vakilian K, Massah J (2013) An artificial neural network approach to identify fungal diseases of cucumber (Cucumis sativus L.) plants using digital image processing. Arch Phytopathol Plant Prot 46(13):1580–1588
Yun S, Xianfeng W, Shanwen Z, Chuanlei Z (2015) PNN based crop disease recognition with leaf image features and meteorological data. Int J Agric Biol Eng 8(4):60–68
Sabrol H, Kumar S (2016) Intensity based feature extraction for tomato plant disease recognition by classification using decision tree. Int J Comput Sci Inf Secur 14(9):622–626
Sabrol H, Kumar S (2016) Fuzzy and neural network based tomato plant disease classification using natural outdoor images. Indian J Sci Technol 9(44):1–8
Samanta D, Ghosh A (2012) Histogram approach for detection of maize leaf damage. Int J Comput Sci Telecommun 3(2):26–28
Luo J, Geng S, Xiu C, Song D, Dong T (2015) A curvelet-SC recognition method for maize disease. J Electr Comput Eng 2015:25
Barbedo JGA, Godoy CV (2015) Automatic classification of soybean diseases based on digital images of leaf symptoms. SBI AGRO
Shrivastava S, Singh SK, Hooda DS (2015) Color sensing and image processing-based automatic soybean plant foliar disease severity detection and estimation. Multimed Tools Appl 74(24):11467–11484
Dandawate Y, Kokare R (2015) An automated approach for classification of plant diseases towards development of futuristic Decision Support System in Indian perspective. In: IEEE international conference on advances in computing, communications and informatics (ICACCI), August, pp 794–799
Mohan KJ, Balasubramanian M, Palanivel S (2016) Detection and recognition of diseases from paddy plant leaf images. Int J Comput Appl 144(12):34–41
Verma T, Satpathy SK, Sharma LK (2011) A step towards precision farming of rice crop by estimating loss caused by leaf blast disease using digital image processing and fuzzy clustering. Int J Comput Trends Technol pp 1–6
Majumdar D, Ghosh A, Kole DK, Chakraborty A, Majumder DD (2014) Application of fuzzy c-means clustering method to classify wheat leaf images based on the presence of rust disease. In: International conference on frontiers of intelligent computing: theory and applications (FICTA), Springer International Publishing, pp 277–284
Cai X, Sun Y, Zhao Y, Damerow L, Lammers PS, Sun W, Tang Y (2013) Smart detection of leaf wilting by 3D image processing and 2D Fourier transform. Comput Electron Agric 90:68–75
Schikora M, Neupane B, Madhogaria S, Koch W, Cremers D, Hirt H, Kogel KH, Schikora A (2012) An image classification approach to analyze the suppression of plant immunity by the human pathogen Salmonella Typhimurium. BMC Bioinform 13(1):171
Barbedo JGA, Koenigkan LV, Santos TT (2016) Identifying multiple plant diseases using digital image processing. Biosyst Eng 147:104–116
Sladojevic S, Arsenovic M, Anderla A, Culibrk D, Stefanovic D (2016) Deep neural networks based recognition of plant diseases by leaf image classification. Comput Intell Neurosci pp 1–11
Bock CH, Poole GH, Parker PE, Gottwald TR (2010) Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. CRC Crit Rev Plant Sci 29(2):59–107
Pethybridge SJ, Nelson SC (2015) Leaf Doctor: a new portable application for quantifying plant disease severity. Plant Dis 99(10):1310–1316
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors of this paper have no potential conflict of interest.
Rights and permissions
About this article
Cite this article
Kaur, S., Pandey, S. & Goel, S. Plants Disease Identification and Classification Through Leaf Images: A Survey. Arch Computat Methods Eng 26, 507–530 (2019). https://doi.org/10.1007/s11831-018-9255-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11831-018-9255-6