Abstract
In this paper, we address a comprehensive study on disease recognition and classification of plant leafs using image processing methods. The traditional manual visual quality inspection cannot be defined systematically as this method is unpredictable and inconsistent. Moreover, it involves a remarkable amount of expertise in the field of plant disease diagnostics (phytopathology) in addition to the disproportionate processing times. Hence, image processing has been applied for the recognition of plant diseases. The paper has been divided into two main categories viz. detection and classification of leafs. A comprehensive discussion on the diseases detection and classification performance is presented based on analysis of previously proposed state of art techniques particularly from 1997 to 2016. Finally, discussed and classify the challenges and some prospects for future improvements in this space.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Plants and fruits are the primary source of energy for the human as well as animals. Leafs of various plants and herbs are useful to mankind due to their medicinal attributes. Countries like Asia and Africa where over 50% of population depends on agriculture production for employment, export earnings and food security [79]. It is assessed that 30 to 40% of crops are lost each year through the production chain [26]. Losses from diseases also have an important economic impact, causing a drop in income for crop producers, higher amounts for consumers and distributors. A lot of studies have been carried out under changed environmental conditions, in different locations, to estimate the losses occur due to different diseases.
In 1980, it was assessed that the annual loss due to plant diseases in U.S.A was about four billion dollars while in India it was probably even more than that [65]. Pre and post-harvest fungal diseases crop losses annually exceed $200 billion euros and over $600 million are annually spent on fungicides [28] in the United States alone. In 2007, the loss within the jurisdiction of Georgia was about $539.74 million, where about $185 million was expensed to control the diseases, and the rest was use to pacify the damage caused by the diseases. So plant disease resistance and managing are crucial to the consistent production of food [13, 49]. Less than 25% of Malawi farmers attained self-sufficiency in maize in years (2000–2001) [21]. According to the study in the Georgia it is estimated that in the year 2010, approximately $701.2 million plants diseases losses considered including control costs. The cost of crops estimate was approximately $4236.51 million, resultant in a 16.5 total percent disease loss across all crops included. In India, the annual estimated losses due to nematodes have been assessed out to be about Rs. 242.1 billion.
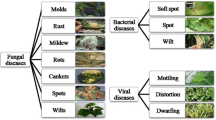
The above mentioned illustrations validate that various parts of the world where a large proportion of the population is reliant on a single or a few crops, is at risk, since a crop failure due to devastating disease can lead them to starvation [47]. Thus there is a need to evaluate the quality of plant so as to identify any disease in them that can be a potential danger to the life of the plant, thereby contributing towards economical, biological, sociological and ecological losses. Plant diseases can be broadly classified either infectious or noninfectious. The demand for high level of safety and superior quality in agricultural products is of prime concern. The foundation of quality assessment is basically dependent upon features of leafs such as its appearance, cracks, texture, and surface where human alertness could be easily fooled. Figure 1 demonstrates the general structure of categorization of plants diseases.
The demand for high level of safety and superior quality in agricultural products is of prime concern. The different methods such as thermography [19, 47], immunofluorescence techniques [83], fluorescence imaging [44], gas chromatography techniques [24], chain reactions [17] and DNA/RNA based affinity biosensor [23] etc. have been often used for quality evaluation of leafs. The problem of the above mentioned techniques were inefficiency, inconsistency and broadly prolonged. Therefore, consideration for an automatic and precise along with inexpensive and efficient technique to identify plant disease is of great realistic significance.
By now, studies show that the image processing methods work as effective tools for the identification and classification of plants diseases. Major advances have been made to improve reliability, precision and accuracy of image analysis for detecting and classifying plant diseases over the last 25 years. It is stated that more than 1000 papers are published each year in the growing fields of computer vision and image processing. The important doctrine and standards remain the same but calculations and testing became more accurate with the application of computer vision technology. The image analysis technique is used to distinguish the objects from the background, thereby isolate quantifiable information which is used in various control systems of decision making. In fact, about 60% to 70% of disease appears on leafs only. So, we have interest in the plant leaf rather than whole plant. This paper represents the latest growths and improvements of the computer and image processing methods in the strata of plant leaf diseases detection, identification with an emphasis on image processing in a significant way.
2 Leaf diseases and symptoms
The quality of leafs defines the degree of excellence or a state of being free from defects, deficits, and substantial variations. It depends on many factors such as sowing seeds, temperature, transplanting, and harvesting etc. [25, 31]. Plant leaf diseases can be mostly categorized based on their nature of their key causal agent (i.e. infectious and noninfectious. Infectious leaf diseases are instigated by a pathogenic organism such as a bacterium, mycoplasma, fungus, virus, nematode, viroid etc. An infectious agent is proficient of reproducing within or on its host and scattering from one susceptible host to another host. On the other hand, noninfectious plant diseases owe their origin to critical growing conditions, disadvantageous relationships between moisture and oxygen, excesses of temperature, toxic constituents in the soil or atmosphere, and deficiency of an essential mineral. Noninfectious causal agents are not contagious and not proficient of reproducing within a host. Table 1 briefly represents the mostly occurring leafs disease and their symptoms and descriptions [16, 75].
3 Challenges of image processing
Computer vision technology has been studied and broadly used in agriculture applications to recognize and classify plants leaf diseases. In the presented study we emphasize on latest studies that reflect image processing methods contribution in the detection of plants leaf diseases and their classification under various field conditions. Figure 2 represents a methodology structure for detection of diseases and their classification techniques. This survey is structured as follows: Section 4, describes an assessment of classification and identification approaches, a comparison of segmentation technique based on recent studies. Section 5, briefly discuss an overall remaining challenges, limitations, and discussion. Section 6, explains the conclusion and future scope of mentioned techniques in literature.
4 Literature review
Pathologists focus on diseases in different parts of the plant like roots, kernel, stem and leaf. As discussed earlier, the present paper concentrates, particularly on leafs. A large amount of the information on the identification of disease and classification field can be found in the various papers [11, 27, 40, 69]. It usually includes preprocessing stage, selection of segmentation techniques followed by the identification and classification stage using selection of proper techniques. The work is divided into two subsections. The first sub-section emphasizes on the contribution of researchers with respect to the technical explanations considered in their algorithm and a summarizing table which contains the information regarding technical culture. In the subsequent sub-sections, a concluding segment is presented with remarks on the contribution of researchers. Certain features common in mostly used methods are offered in this section: the images are acquired via consumer level cameras in a suitable laboratory situation, and the format used for the images are RGB, CIELAB etc. All the methods apply some kind of preprocessing to avoid the noise, segmentation process to select the region of interest (disease) and so on. Figure 3 represents the basic set up of leaf diseases detection and their classification. Therefore, unless stated otherwise, these are the situations under which the described methods operate. Also, practically all the methods mentioned in this paper apply some kind of preprocessing to clean up the images but we are not focusing much on these techniques.
4.1 Disease detection analysis with color and texture characteristics
This section represents the review of various techniques used for the identification of diseases using texture and color characteristics.
Pydipati et al. [63] represents the color co-occurrence method (CCM) to determine texture based hue, saturation and intensity (HSI) color features in the combination with statistical classification algorithms to classify diseased and normal citrus leafs. In the first stage, features are extracted using CCM by considering the four different categories of leafs. Further, SAS statistical analysis is used to evaluate the potential classification accuracies with reducing the variable sets and using color features individually. However, best results were attained using combined HIS features. The authors explicated that hue and saturation features are influenced by low lighting conditions and such methods prove to be the best under controlled laboratory lightning conditions. The most significant challenge is to used this method under natural lightning conditions.
A. Camargo et al. [18] presented image processing method to identify the plant diseases visual symptoms from the exploration of colored images. They transformed an RGB color space of the diseased leaf into H, I3a and I3b transformation. The I3a and I3b conversions are generally developed from an adjustment of the original I1 I2 I3 color transformations to meet the requirements. By analyzing the scattering of the intensities in a histogram, transformed images are segmented. After post processing, it checked the accuracy by manually comparing the segmented images of plant leafs with automatically segmented images. The authors concluded that this technique is suitable to identify a diseased segment even when the segment is represented by an extensive range of various intensities.
Haiguag Wang et al. [82] described a method for the identification of diseases in grape and wheat leafs. Firstly, captured images were transformed from RGB color space to XYZ color space and then again transformed from XYZ color space to L*a*b color spaces. Squared Euclidean distance (D) as a similarity distance is used to evaluate the color difference in a*b, a two dimensional data space. Total 4 shape features, 21 color features and 25 texture features are extracted to distinguish various diseases. Furthermore, for reducing dimensions in the featured data processing, Principal component analysis (PCA) is performed. Later on, different neural network techniques with different parameters including Back Propagation (BP) networks, Generalized Regression Networks (GRNNs) and Probabilistic Neural Networks (PNNs), Radial Basis Function (RBF), were used to classify and identify wheat and grape leaf diseases respectively. The author confirmed that this technique provides a fast and precise recognition of plant diseases. Table 2 describes the various color space advantages and disadvantages.
Qinghai He et al. [32] proposed the diseases identification method for cotton leaf in which three color models are used for extracting the disease. Firstly, images are captured and to eliminate the influence of noise, spatial non-linear and frequency domain filtering is used. Afterwards, the edges of background image are highlighted by using histogram equalization process and then the contrast of the image is raised. The enhanced images are converted into the RGB, HIS, and YCbCr color model. The percentage of damage (γ) is selected as a feature to evaluate the amount of damage due to leaf diseases or pests. After applying different color models, comparative results were obtained. The comparison of the results show YCbCr color format measured as the best color model for identifying the injured color leaf image. The author validated that the projected algorithm fails to handle the random noise interference and leaf shadow in case of outdoor conditions.
Auzi Asfarian et al. [8] presented the work on paddy diseases identification using the fractal descriptors based on Fourier transforms with texture analysis. Here, injuries (lesions) are collected manually and then each of these lesion images were transformed to HSV color space. Histogram equalization is performed for reducing the lightning effects after extracting the saturation components. Conclusively, from each lesion, the fractal descriptors were extracted and then given to PNN classifier for concluding the classification. 5-fold cross validation used to split the training and testing data for verification and validation. The author stated that this method, if it is combined with other features has a prospective to be used as one of the possible features, especially when two diseases comparatively involved have the same color.
Pradnya Ravindra Narvekar et al. [58] discussed the effective way of disease detection in grape leafs using SGDM (spatial gray level dependence matrices) method. They used four classes of leafs under different disease conditions (i.e. Black rot, downy mildew, powdery mildew and normal). After color transformation of RGB to HSI, the H component is considered for further analysis. The S and I components were throw down since they do not provide any useful information. Based on the specified and varying threshold, green pixels were masked using Otsu method. In the next step the pixels with zero red, green and blue color values were entirely removed. Later on, features based on the color and texture were extracted and classified in different disease category. The authors concluded that the proposed work indicates the valuable approach with little computational efforts but with the use of hybrid algorithms, the recognition rate of classification can be increased.
Vinita Tajane et al. [78] suggested CBIR (content base image retrieval) method for identifying medical plants leaf diseases using canny edge detection algorithm and histogram analysis. In the first stage, CBIR smooth the image to eliminate the noise and finds the image gradient to highlight regions with high spatial derivatives. In the next stage medical plant leaf diseases were detected based on its edge features. Firstly, leaf images are transformed into gray scale images and edge histogram is calculated. The area and color features of the image are evaluated using histogram for the overall image. Based on their research, they evaluate color histogram for each red, green and blue color region individually to check the intensity of each color pixels in the sample image for identification of healthy and infected sample. The author concluded that Canny edge detection algorithm is an effective method for distinguishing edges of healthy and infected plants and it also filters useless information and preserve significant structural properties in an image.
Juan F. Molina et al. [54] presented a color based strategy to detect an early blight disease or any kind of infection on tomato leaf lets. The proposed method based on the categorization of tomato leafs with the help of color descriptors. Color characterization is done using color structure descriptors (i.e. color histogram by quantifies the amount of significant color) using Hue-Max-Min method, scalable color descriptor (i.e. leading spatial color distribution) and color layout descriptors (color pattern variation) using YCbCr color space transform. After calculation of all these feature or descriptor values, a novel strategy based on nested leaf one out cross validation method is used to achieve better classification ratio. The individual descriptor configuration evaluation performed using an inner loop permit, while the outer loop measures performance assessment between different descriptors. The author concluded that color structure descriptor provided better accuracy than other methods. Table 3 represents the summarization of color and texture technique for diseases detection.
4.2 Disease detection analysis with thresholding and clustering parameters
This section characterizes the review of various techniques used for diseases identification using thresholding and clustering parameters.
Tucker and Chakraborty [81] focused on identifying and detecting diseases in oat and sunflower leafs. Firstly images were segmented with varying threshold w.r.t the disease being measured (blight or rust). Then, using clustering, subsequent pixels were connected into clusters for signifying the diseased regions. Based on the features of the lesions, they are segregated into the proper categories. The authors observed good results but due to inappropriate illumination during the capturing of the images, some errors exist.
D.G. Sena Jr. et al. [73] proposed work to develop a procedure for identifying damaged maize plant leaf diseases at simplified lightning conditions. Damaged and healthy maize plant leaf images were captured in three different light intensities and in eight different locations. The proposed algorithm is divided into two stages: preprocessing and the image analysis. During the first stage the original RGB images were transformed to 256 grey level images by applying the additional green index and rescaling the pixel values to create binary images. Then, the monochrome images were threshold by the iterative method. In the second stage, the images were sectioned into blocks and classified as healthy or damaged based on the number of objects originated in each block. The author concluded that the proposed algorithm perform well and gives good accuracy.
Shen Weizheng et al. [84] have implemented an image processing based technique for analyzing the leaf spot disease. They investigated all the influencing issues that existed in the process of segmentation. Leaf regions were segmented using Otsu thresholding. After color transformation from RGB color system to the HSI color system, H element is selected for segmentation of the leaf spot to reduce the disruption of lightning changes. Further, Sobel operator is considered in order of observing the edges of the leaf disease spots. Finally, grading is achieved by assessing the quotient of the diseased region and leaf areas. The author indicates that this method, to grade plant leaf spot diseases is fast and accurate.
Kurniawati et al. [45] defined a technique to detect and label three different kinds of diseases namely brown spot blast and narrow brown spot that influence paddy crops. The healthy and diseased region segmentation is accomplished by means of thresholding using Otsu’s and local entropy. Color, texture and shape features are extracted to decide the diseases that best fits the characteristics of the selected region. The characteristics of image consisting of boundary color, spot color, lesion type and broken paddy leaf color from paddy leaf images. The best accuracy achieved is by using local entropy threshold. The final classification is done by discriminating between the selected regions according to the corresponding predefined set of rules. The author concluded that the Otsu thresholding method is unable to accomplish segmentation task correctly to different intensity standards and less susceptible to illumination.
Sekulska- Nalewajko and Goclawski et al. [72] presented a method to detect disease signs in cucumber and pumpkin leafs. Firstly after capturing, the images were converted from RGB to HSV color space format. After thresholding the leaf area, the brightness section is discarded. Then, Fuzzy c means clustering algorithm is applied in hue-saturation space in order to group the pixels into clusters. These clusters are then combined with the interactive phase into two final clusters, where one of them decides the searched disease areas ultimately, the author justified that the present approach gives good classification results. However, this approach requires too many processes to achieve the desired results.
Zulkifli Bin Husin et al. [35] discuss the effective method used for evaluating primary detection of chili disease using leaf features inspection. Now days, from time to time the chemicals are applied to the plants without seeing the prerequisite of each plant life cycle. This technique will confirm that the chemicals are applied only when the plants are noticed to be instigated with the diseases. Firstly, leaf images are captured under controlled field condition and preprocessing is done to achieve improved image information. Features are extracted using color information for differentiating between healthy and non-healthy leafs. The resultant color pixels were gathered to achieve different groups of colors in the image to distinguish the disease affected area of leaf. In the end, histogram graphs are evaluated to measure the healthiness of leafs. The author concluded that the proposed method is fast and efficient in recognition of plant chili disease.
Revathi et al. [67] defined two segments to categorize the affected part of the leaf using proposed homogeneous pixel counting technique for cotton diseases detection (HPCCDD) technique. Initially, sobel edge detection method is used to recognize the diseased segments of the leaf spot and distinguish the diseased boundary. Then evaluate the entered pixels of opposed neighboring one, two, three pixels of clarity of edges. The Homogeneity based edge detector precedes the result of any edge detector and splits it by the average range of the part. The average range of an area is calculated by convolving the part with a mask containing all the ones. Finally, extracted edge features are used in the classification process to classify the disease spots. The author testified that the proposed method provides good accuracy than other existing algorithms.
Mokhled S. Al-Tarawneh et al. [5] suggested a method to detect diseases in olive leafs. The proposed method combines auto cropping technique with fuzzy c means clustering to identify the disease. After, color transformation of RGB to CIELab color space, the region of interest (diseased region) is cropped using automatic polygon to segment the leaf. The polygon cropping follows the edging contour of entire image to define the masked polygon points of diseased segments. At the end, classification is done using fuzzy c-means clustering for statistical usage to define the defect and severity. The severity percentage is calculated based on the classification of detected diseased and total leaf area. The author actualized that the experimental results of FCM algorithm with polygon auto-cropping segmentation shows encouraging accuracy prospects. Table 4 represents the various advantages and disadvantages of the segmentation techniques.
D. Zhihua et al. [92] discussed cotton mite diseases recognition approach using color features and thresholding methods. Proposed technique is distributed into three steps: 1) diseases spots and stems extraction of the green plants. 2) Special features detection in gray histogram and transform the segmented image into 8-bit gray scale image using thresholding. Histograms illustrated two points in two different gray scale portions as disease spotted portions with values 255 and grayscale values less than 120 depicted as non-disease spotted portions. The black pixels signified the corresponding disease spotted part taking mite disease spots, stems and white pixels representing non-disease spotted areas. 3) Segmentation of binary images using area thresholding and comparing the areas with spots to the stems. The author concluded that the mite and stem disease spotted portions are almost similar in color therefore it is a difficult task to compare the areas with spots on the stems.
Krishnan et al. [42] defined approach to analyze shade leafs disease (i.e. bacterial scotch). Firstly, shade tree leafs images were attained under controlled lightning conditions, followed by clustering based approach. K-means clustering approaches utilize distinct clusters and accomplish different cluster centers to achieve different clusters for defining region of interest. Final clustering is achieved by subtracting reference images from base images to form convoluted clusters from original images. The authors concluded that the proposed algorithm gives high accuracy with less operational time. But, the classification of algorithm can be improved by updating the clusters through repetition procedure. The clustering algorithms built on limitations and certain relaxation algorithms could also be improved to provide accuracy to the proposed scheme.
Eric Hitiman et al. [33] explained a technique for leaf injury detection and severity assessment of coffee leaf. Further, Gaussian kernel is used to suppress the noise and image color space is transformed into CIELAB color space. After increasing the contrast image with Look up Table (LUT) based gamma correction using average pixel intensity. Later, the boundary features of the threshold image are detected using canny edge detector and joined to modify the overall structure of the object. Using background removal method based on luminance and color, only leaf area (foreground) is extracted and the background free leaf image is processed with YUV color space (i.e. V channel) to maximize leaf injury detection. Finally, leaf damage severity is assessed w.r.t to percentage of pixel distribution of the damaged and healthy leaf. The author consummated that the proposed method is fast and avoids the defoliation. Moreover, the proposed method can handle all the images (i.e. destroyed and color changes) and provides good accuracy rate. Table 5 represents the summarization of thresholding and clustering techniques for diseases detection.
Jiangsheng Gui et al. [29] proposed a novel method for the identification of diseases of soybean leaf by means of salient regions using color image with complex background. Based on idea of Itti method and low level features of luminance and color, salient regions are detected. Then w.r.t combined experience threshold and K-means algorithm salient regions can be precisely extracted from the images of diseased soybean leafs based on salient maps. In the last, segmentation using threshold is performed on R component of RGB space. Morphology algorithm is used to fill small holes and eliminate the insignificant small sized regions of the non-diseased area to correct the diseased segment. The author finally conferred that the proposed technique using salient regions can rapidly and precisely detect the segment of diseases in complicated farmland backgrounds.
K. W. V Sanjaya et al. [70] proposed system to measure and predict orchid leaf healthiness and diseases. Gaussian operator and histogram equalization method is used for removing noise and enhancing the image. After, enhancing the image, we acquire the orchid leaf object by removing the background noise in the image. Next, we input RGB image and it is transformed into the HSV color model and apply thresholds for hue, saturation and value color bands separately. Threshold values are manually selected by observing the histograms of three color bands for segmenting the green and yellow objects. After segmentation, image shape and color features are extracted to detect orchid leaf diseases. The extracted geometric features of the leaf image are used to predict the orchid species and the color features are used to predict the healthiness of the orchid plant. In order to make predictions, decision trees method is used. The author concluded that proposed system accuracy is reliable but the system can identify only three orchid species and three orchid diseases.
Sourabh Shrivastva et al. [74] presented a technique to automatically identify and estimate the level of disease severity in soybean plant foliar. The concerned method converts the RGB image into the Y, Cb, Cr channels and then segments through simple thresholding method. The authors used various novel parameters such as disease level parameter, severity index, diseases severity index and infected area to automatically measure the disease level severity. The author highlighted that it is a low cost method and has the potential for the extensive usability in field conditions. Moreover, the performance of this method might be improved by using unconventional background separation approach to distinct the leaf object from a complex background.
Amar Kumar Dey et al. [76] considered image processing procedures to identify leaf rot disease by classifying the color feature of the rotted leaf region of betel vine. After analyzing different color spaces, HVS gives promising results where the hue component provides the strong remark of rotted leaf area. Afterwards, using Otsu thresholding method rotted area is segmented using various threshold values and area of rotted portion is considered as the feature set. A leaf disease severity scale can be prepared by calculating the proportion of diseased area. Using frequency of precise amounts of pesticide application, disease severity volume can be regulated, which moderates the pesticide cost used for management. The author concluded that the results presented promising results of this automatic vision based system with easy validation.
4.3 Diseases classification using artificial neural networks
This section describes the review of various techniques used for classification of diseases using Artificial Neural Networks (ANN).
Kuo-YI Huang et al. [34] explained an approach using artificial neural networks for recognition of Phalaenopsis seedling diseases with texture and color analysis. Lesion areas were segmented using underlining principle to differentiate background (pot), object (leafs) and exponentially transform a variable parameter to enhance the image and to evaluate texture features of the lesion area Gray Level Co-occurrence matrix (GLCM) used. After that, tri-color mean values of lesion areas are employed to classify diseases. The back propagation neural network classifier is then used to categorize various defects. Finally, authors were able to distinguish and categorize visible lesion areas but they were incapable to examine the infected area on the enclosed blades. The recognition and classification system can be also useful for the development of flowers and plant leafs in greenhouse.
Noor Ezan Abdullah et al. [1] classified the diseases in rubber tree leafs using multilayer perception neural networks. After the preprocessing by using median filter, the color features were extracted using RGB distribution indices to calculate the region of interest (i.e. lesion spots). The Principal Component Analysis (PCA) is employed on the progression values of each image. The improved artificial neural networks designed were based on optimized dominant pixel (mean) of RGB and normalized PCA data. The optimized models were then evaluated and validated through analysis of various performance indexes. According to authors, the proposed method is found to be the best models for identify bird eye spot and otrichum diseases. However, the effectiveness of this classification can be increased providing a better processor and high resolution camera is employed.
Santanu Phadikar et al. [60] defined the framework to identify the diseases of rice plant leafs using zooming algorithm and neural networks. The input RGB color space images are converted into HIS color space images and the infected regions are extracted using entropy based thresholding method. Then, the features are extracted using zooming algorithm and are classified using Self Organizing Map (SOM) neural network where train images are obtained by extracting features of the infected parts of the leaf while four different types of images are applied for testing purposes. The authors explained that it is a simple and computationally efficient technique, which results in a satisfactory classification of results. However, it has been observed that the transformation of the image in the frequency domain does not produce a better classification parallel to the original image.
Dheeb Al Bashish et al. [12] suggested a structure for classification and identification of diseases such as early scorch, ashen mold, cottony mold, late scorch and tiny whiteness etc. Subsequently color transformations of RGB into HIS color space, the images are clustered with K-means clustering approach using Squared Euclidean distance. This method segregates the leaf image into four clusters and evaluates the color and texture features, SGDM matrices are created for each pixel map of the image but only for H and S component. Statistical examination tasks are accomplished to select the best features for reducing the feature redundancy. Then for final classification, neural network classifier is used. It s found that the proposed method can expressively support precise and automatic recognition of diseases in leafs. However, due to invariability in lighting conditions some samples of diseased leafs were misclassified. An example of the output of K-Means clustering for a leaf infected with early scorch disease is shown in Fig. 4.
H. Al. Hiary et al. [4] shed light on an automatic disease detection method based on K-means segmentation process. In the proposed method, the pixels were categorized to K number of classes on the basis of a set of features, which masked the green pixel values obtained using the Otsu’s thresholding technique. Features are extracted using color co-occurrence matrix to analyse affected leafs texture. The leaf spots are considered as an indicator of crop diseases. In the end, the classification of diseases is evaluated using artificial neural networks. The author concluded that this method requires small computation of time and computational effectiveness.
Wanrat Abdullakasim et al. [2] elucidated an image analysis method for recognition of brown leaf spot disease. Various color descriptors or color indices i.e. RGB and HIS descriptors were used to recognize different regions of leaf based on its color. The range of color indices of RGB and HSI varies from 0 to 1. Then, ANN is used to classify or differentiate between an infected region and a healthy region. Brier score is employed to estimate the recognition capability of the ANN with more hidden layers. It is found that this algorithm properly identifies 79.23% diseased leafs and 89.92% healthy leafs, but this study also includes misclassification of infected leafs. This method can be improved by combining the lightning conditions, effects of infection phases and proper segmentation.
Kai et al. [39] suggested a technique to recognize diseases in maize leafs based on neural networks. Firstly, the images were transformed to the YCbCr color illustration to evaluate Cb and Cr components because they are less affected by brightness. In this two dimensional plane, the lesion area is relatively concentrated in accordance with Gaussian distribution. After that texture features are evaluated using SGLCM matrix. Furthermore, the back propagation based neural networks with sigmoid function using 3 hidden layers are used to classify various diseases. The exploratory results proclaimed that the accuracy is very high. The author concluded that a neural network error value is very low, weight design is reasonably correct and overall performance is good.
D.S. Guru et al. [30] discussed innovative idea for extracting lesion areas from tobacco leaf using neural networks. Contrast stretching transformation with an adjustable parameter and morphological operations are used to segment lesion areas. R channel based statistical texture features (i.e. statistical moments, uniformity and entropy) are extracted from lesion area to identify and analyze the diseased category. Based on extracted texture features, Probabilistic Neural Network (PNN) is used to categorize tobacco seedling leaf diseases as anthracnose and frog-eye spots. The author concluded that the first order statistical texture features perform better than Gray level co-occurrence matrix features. Classification accuracy can be improved with combination of different color and texture features.
Tushar H Jaware et al. [37] explained an effective and precise method for classification and recognition of leaf diseases. The suggested procedure is validated on five diseases; early scorch, cottony mold, late scorch, ashen mold and tiny whiteness. Primarily after the color transformation, pattern of input RGB color space to HIS format, K-means clustering technique is used for segment diseased region. Furthermore the red, green and blue zero pixel values and boundary pixels of the diseased leafs were completely removed and masked the green pixels only. Next, the diseased clusters were transformed back from HIS format to RGB format and SGDM matrices were generated for each pixel map of the image and extracted the features for further classification. Lastly, the features were trained using a pre-trained neural network. The results showed that the suggested system can be effectively distinguished and the diseases are categorized.
Weidan Zhang et al. [90] proposed the new method to identify the six varieties of jujube leaf diseases i.e. jujube fruit rust disease, jujube rust, jujube white rot, jujube anthracnose, ascochyta spot of jujube, jujube witches broom based on color, texture, morphological features and classified using neural networks. Firstly, after image acquisition nine colors, eleven texture and five morphological features are extracted corresponding to the red, green and blue plane of the image. Best twelve features using step wise discriminant analysis (STEPDISC) and four principal components using Principal Component Analysis (PCA) are selected from the original 24 features space. Furthermore, cross validation is performed for each feature set space. In the last stage, a two layer tan sigmoid model using 12 parameters diagnose the jujube diseases. The authors concluded that the detection precision of jujube white rot disease is the highest, due to big diseases spots. The high similarity structure index between jujube diseases decreases the classification accuracy.
Pranjali Vinayak Keskar et al. [41] suggested leaf disease analysis and recognition method for the assessment of injured leafs and classifying the category of diseases. To enhance the quality of acquired images, image enhancement methods were applied. The proposed model is involved with four stages: first stage includes transformation of RGB to the HIS color space, analyzing the histogram and intensity adjustment by applying the thresholding. Further next stage, contains adaption of fuzzy feature algorithm to segment image with adjustable parameter to fit the application in concern. In the third stage, features are extracted using component labeling based on color, size and shape of the diseased spots. The fourth stage is classification, which includes artificial neural networks to categorize disease category. The author concluded that the proposed model clearly differentiates between healthy and diseased leafs with maximum accuracy rate.
S. Sannakki et al. [71] proposed the organization model to classify grape leaf diseases using neural networks. Initially, the grape leaf image with complex background is captured and the background is removed using green color pixel masking and the noise is removed with the help of anisotropic diffusion up to 5 iterations to preserve the infected part information. K-means clustering is used for segmentation and the textural information from diseased part of the segmented image is calculated from GLCM matrix for nine features. The extracted features are used by Feed forward BPNN for its classification. The author concluded that the maximum accuracy is attained using the hue features only.
Suhaili Beeran Kutty et al. [46] discussed the method for the classification of downy mildew and anthracnose diseases of watermelon leaf using neural network study with respect to RGB color component mean values. Few samples of the infected leafs are assembled and taken with digital camera with detailed calibration process under controlled lightning conditions. Using identified region of interest, RGB color pixel indices are extracted. The suggested classification technique considers the procedure for categorization of disease neural network pattern recognition toolbox in MATLAB and statistical package for the social sciences (SPSS). The authors concluded that the proposed system provides satisfactory results but the CMYK, HSV and HIS can be used as an input in order to increase the efficiency.
Kholis Majid et al. [48] has established a portable presentation for paddy malady diseases identification system, using Probabilistic neural system and fuzzy entropy classifier that keeps running on versatile Android working system. It recognizes specific brown spot, leaf blast, tungro and bacterial leaf blight. After capturing images significant features are extracted, using fuzzy entropy with membership functions for extracting the membership of brightness levels. After feature extraction by using the extracted features, probabilistic neural networks method is used for further classification. The authors concluded that PNN approach provides optimal result, but the tungro disease has the lowermost accuracy of recognition, because certain features of tungro diseases are alike to bacterial leaf blight diseases. But proposed method accuracy can be increased by increasing the dataset.
Revathi et al. [68] demonstrated a new method for the detection of cotton diseases using improved PSO feature selection technique which implements skew divergence method with parameters like variances, texture, color and edge to excerpt the required features. Color variance feature is evaluated using color histogram and color descriptor. The Sobel and Canny edge detection method is used to extract shape Skew divergence feature and texture feature is measured by texture descriptor and Gabor filter. The cross information gains depth, forward neural network (CIGDFNN) supports to distinguish and classify cotton leaf spot diseases such as root rot, leaf blight, micro nutrient, verticilium wilt, bacterial blight and fusarium wilt precisely diminishing the error rate.
Sachin B. Jagtap et al. [36] presented an integrated image analyzer with in a diagnostic expert system model which detected diseases based on color, size, and shape. Histogram analysis is used to increase contrast of image by adjusting its intensity for image enhancement. After image enhancement and color transformation from RGB to HIS color space fuzzy c means clustering is used for identifying region of interest by setting all the important parameters, such as feature of dataset, optimal no. of clusters and degree of fuzziness. Finally, the author effectuated that due to the integration of this proposed system diagnosis, the accuracy will increase. Additionally, the proposed system focuses on specific identification disorder which can be further extended to include more disorders. Such extension of the system is carried out in such a way that it will be capable of detecting and identifying abnormalities on the other parts of plants also e.g. fruit, stem and root.
J.W. Orillo et al. [59] projected a technique to detect rice plant diseases such as bacterial leaf blight causing brown spot on the leafs using Otsu thresholding method and neural networks. At the first stage, after transformation of RGB to HSV color space, diseased area of the leaf is evaluated using Otsu thresholding method as in a gray level histogram, which has been resulting from the perspective of discriminant analysis. After segmentation, there are four features namely, (a) segment enclosed by the disease on the leaf (b) R, G, and B means value of the disease (c) R, G, and B standard deviation values (d) H, S and V mean values of the disease. In the last stage, back propagation neural network is used to evaluate the accuracy and performance of proposed method. The author finally verified that representative lesions of the diseased leaf are recognized accurately. Table 6 represents the summarization of neural networks techniques used for disease classification.
4.4 Diseases classification using Naives Bayes classifier
This section describes the review of various techniques used for the classification of diseases using Naives Bayes classifier.
Aduwo et al. [3] presented an automated vision based analysis system to detect cassava mosaic disease, based on color and shape features and classify using different classifiers. The procedure begins with the capturing of the leaf images with a standard digital camera. Then, three techniques were used to attain representative feature data from the leaf images of the health plants. For first dataset, to extract color features normalized histogram of the HSV color of hue pixels are evaluated. While shape features are extracted using SURF and SIFT methods for two other dataset of cassava leafs. The image is either classified as healthy or diseased, based on other methods like a k-nearest neighbor classifier (KNN), Naive Bayes and SVM. A comparison of the different classifiers is done and results for the three main datasets were produced and Naives Bayes provided the best results. The author concluded that the study of the color histograms by the use of divergence procedures has the ability to give good classification of the performance by using histograms, because they are more naturally represented as distributions. The extra information in the augmented feature sets lead to better generalization of the classifiers using them.
Dhiman Mondal et al. [55] proposed a technique to detect yellow vain mosaic virus disease in okra leaf with combination of k means and Naive Bayes classifier. Initially twenty three features are evaluated using gray image and converted into the size of invariant features by representing different features range into a standardized feature range. Using gray level co-occurrence, matrix texture features are extracted in the second phase. After feature extraction, Pearson correlation coefficient method is used to identify dominant features. Feature values analogous to dominant feature set are designated to create a feature matric and clustered using K-means algorithm. The final classification is done using Naive Bayes classifier. The author concluded that the proposed method, identifies 87% diseases correctly and classification rate can be increased using more appropriate features. Table 7 represents the summarization of Naives Bayes used for disease classification.
4.5 Diseases recognition and classification using fuzzy logic analysis
This section describes the review of various techniques used for diseases classification using Fuzzy classifiers.
Azmi et al. [15] presents an orchid diseases detection system using combination of image processing and fuzzy logic. The proposed scheme comprises of two phases; in the first phase, the leafs are segmented using Otsu thresholding methods. After segmentation the area and the number of the diseased spots is calculated. In the second phase extracted diseased spots and area are used as the input of fuzzy logic system. The number of diseased spots for each leaf is stored in a matrix of number during the batch processing of images. Based on these two features, fuzzy system determines the diseases category using nine ‘if-else’ rules based on inputs using Mandani method. The defuzzification is done using the center of maximum method, the mean of maximum method and the center of area method. The author concluded that this method did not provide the actual number of diseases spots. They detect more number of diseased spots as compared to the actual one.
Billah et al. [14] suggested a model for recognizing tea leaf diseases, which uses color wavelet features and adaptive neuro fuzzy inference system. After processing, color wavelet features based on the combined information from both color and texture fields of the image with the second order statistical representation of the wavelets transform are extracted. The extracted features classified by Adaptive Neuro Fuzzy Inference System (ANFIS) along with the three types of disease categories. The author concluded that the proposed techniques gives promising results and can recognize the diseases accurately. The color wavelet analysis performs better than other feature extraction methodologies.
K. Muthukannan et al. [56] defined a fuzzy rule-based technique using color features for the disease classification of tomato leaf. Firstly, gradient operator is used for preprocessing to suppress noise or small fluctuation in the image. Two important features i.e. mean and standard deviation are extracted from cropped image with different image size samples. In the last, Fuzzy Inference System using fuzzy rules with selected color features classified the image region as healthy, slightly healthy and highly affected disease portion of the plant leaf. Where, orange color indicates healthy portion of the image, yellow and red color indicated the diseased region of the leaf.
The author concluded the performance of fuzzy rule based classification is satisfactory. The experimental results show the proposed method can detect leaf diseases with little computational effort. Table 8 represents the summarization of fuzzy logic analysis used for disease classification.
4.6 Diseases detection and classification using PSO
This section describes the review of technique used for diseases classification using PSO.
Zhang et al. [91] proposed an improved PSO algorithm and neural networks to recognize and diagnose maize leaf diseases. Image is enhanced using the histogram equalization method and de-noising is realized using image filters. Color, texture and shape features based on HSI (hue-saturation-intensity) color components are extracted. An improved PSO algorithm based on opposition learning method is proposed to decrease the prospect that the exploration for particle swarm falls into the local optima so as to achieve a more optimal solution. To achieve high convergence speed, more compatible particle obtained to continuously optimize the global area to increase neural network analytical model accuracy. The authors concluded that traditional neural networks have slow convergent speed, easy getting into local minimum and low rate of correct motion pattern recognition. The results show more effectiveness than existing ones.
Muthukannan et al. [57] stated the importance of particle swarm optimization based segmentation of images for plants leaf diseases identification. After data set collection, Gaussian filter is used to remove the unwanted information of speckles. The results of preprocessing are analysed using PSNR and Max error parameters. The segmentation is achieved using binary PSO to select the best number of clusters and the centers of the selected clusters are refined by k-means clustering. The PSO is employed for allocating each pixel to a cluster. After segmentation, hybrid feature parameters based on texture, color and shape using GLCM matrices are extracted on the basis of which leaf diseases are classified. According to authors, hybrid feature extraction approach is useful for plant leaf disease classification in terms of reduction of misclassification and also increases the correct prediction classification accuracy. Table 9 represents the summarization of PSO techniques for disease classification.
4.7 Diseases detection and classification using membership function
This section describes the review of various techniques used for diseases classification using membership function.
Anthonys et al. [6] presented framework to recognize paddy diseases using membership functions. The color images captured by digital camera in the laboratory with dark background are used to avoid the effect of environmental factors. The images are then transformed into CIE XYZ color space and using appropriate threshold value, images were segmented and edges detected using sobel method. Texture and shape features are extracted by considering the parameters like area, roundness, shape complexity etc. Using these features, membership functions are calculated and defined for each class of disease for disease recognition and classification using nearest neighbor. The author concluded that proposed work gives satisfactory results with less recognition time but noise affects the performance. Table 10 represents the summarization of membership technique used for disease classification.
4.8 Diseases classification using combination of classifiers
This section describes the review of various techniques used for diseases classification using combination of classifiers.
Tian et al. [80] proposed a system that utilizes stacked generalization structure to combine the classification decisions attained from three kinds of support vector machines (SVM) classifiers rather than using single SVM classifiers to identify wheat leaf disease. The leaf region is segmented by using simple threshold method. Color, texture and shape features are extracted and used as training set for three corresponding SVM classifiers. Later on, different extracted feature sets are classified by the classifiers in low level and mid-level categories, which are partly described by the symptom of crop diseases according to the knowledge of plant pathology. Then the mid-level features are extracted from mid-categories produced from low-level classifiers. Finally high-level SVM trained and correct errors made by the color, texture and shape features. The author concluded that compared with other classifiers for wheat leaf diseases recognition, the proposed approach can obtains better success rate of recognition.
El Massi et al. [50] proposed an approach using serial arrangement of two neural networks classifiers. Firstly, images are transformed from RGB to LAB color space. Then images are segmented using k means clustering to extract the lesion region. Based on color, first classifier evaluates difference between classes. The damages having a same or an adjacent color are considered in the same class. Then according to the shape and texture features, second classifier is used to find difference between the classes. The method is verified on four categories of diseases, including two categories of pest insects (i.e. Leaf miners and the caterpillar Tuta absoluta), and the symptoms of two fungal diseases (i.e. internal powdery mildew and downy mildew). The author concluded that the proposed approach using serial classifiers arrangement method is stimulating and can resolve the difficulties of the individual classifiers.
Es-saady et al. [22] presented a system on serial grouping of two SVM classifiers. Firstly, images are transformed from RGB to LAB color space. Then images are segmented using k means clustering to extract the lesion region. Based on color first classifier, the difference between the classes is evaluated. The damages having a same or an adjacent color are considered in the same class. Then, according to the shape and texture features, second classifier is used to evaluate the difference between classes. The technique is verified on four classes, with the damages of six classes of plant leafs diseases. The author concluded that proposed system using serial classifiers combination is interesting, and can resolve the difficulties of the individual classifiers and can be improved using relevant features. Table 11 represents the summarization of combinations of classifiers used for disease classification.
4.9 Diseases classification using orthogonal locally discriminant projection algorithm (OPDPA)
This section describes the review of technique used for diseases classification using orthogonal locally discriminant projection algorithm.
Zhang et al. [87] presented orthogonal locally discriminant projection algorithm to classify the maize plant diseases. The original leaf images are pre-processed and then converted from RGB color space to HSI color space. After that, the green pixels are masked and removed to obtain the non-green component to achieve the useful segment. Finally, every processed image is symbolized as a point in the vector space. Segmentation is done using gray level selection thresholding method. Afterwards, the training data is constructed from the transformation matrix and is assign to low-dimensional feature subspace which is orthogonal locally discriminant projection with respect to the test data points. The nearest neighbor graph weights are adjusted according to their reliability between two nodes. The final results prove that the proposed method is achievable and effective. The effectiveness of kernel space in the OLDPA need to be more examined. Table 12 represents the summarization of technique used for diseases classification using orthogonal locally discriminant projection algorithm.
4.10 Diseases classification using minimum path evaluation theory
This section describes the review of various techniques used for diseases classification minimum path evaluation theory.
Pixia et al. [62] suggested a novel scheme based on minimum distance to identify the cucumber leaf diseases. The leaf images are preprocessed using median filtering, image smoothing and segmented using the color range of different diseases to attain lesion segment. The features are extracted using morphological, texture and color feature extraction method. Thirteen important features are extracted and normalized. Finally, the classification is done calculating the shortest distance path method to evaluate the distinguishing value of disease samples and an average center distance among diseases. As the recognized diseases are more similar with standard diseases, the distance is assumed to be less. The authors concluded that the characteristics based on the minimum path evaluation are very effective. Table 13 represents the summarization of minimum path evaluation theory used for disease classification.
4.11 Diseases detection and classification using fractal dimensions
This section describes the review of various techniques used for diseases classification with fractal dimension and chaos theory.
Da-Ke et al. [85] proposed technique to recognize leafminer infected leaf based on fractal dimensions. Two different kernels i.e. polynomial based kernel function and radial based kernel functions are considered for classifying models. Fractal dimension of the damaged cucumber leafs are applied to the threshold method and support vector machine for reorganization and classification of leafs. Where, fractal dimension provides a quantitative index of the roughness diseased leaf image. Finally, radial kernel based SVM excels to the polynomial–based kernel. The author concluded the recognition and precision are depends on the input vectors. The threshold method is good only for linear problems.
V. Surendrababu et al. [77] defined a method for distinguishing rice leaf disease with fractal dimension and chaos theory. The study of an unhealthy leaf is carried out with respect to its fractal dimension, image pattern, especially box-counting ratio calculation, and chaos to recognize the unhealthy outline self-similarity to reconstruct the fractal. The fractal pattern for the unhealthy leaf at the concluding phase will have the similar pattern during the preliminary phase for each category of rice leaf disease because of self-similarity. The author concluded that the proposed method evaluating the initial information for the progress of an early detection system or for emerging information based expert system. Table 14 represents the summarization of the fractal dimension, and chaos theory used for disease classification.
4.12 Diseases classification and identification using support vector machine (SVM)
This segment describes the review of various techniques used for diseases cataloging using Support Vector Machine (SVM).
Meunkaewjinda et al. [51] proposed a self-organizing feature map to distinguish the color images of the grape leafs. Before extraction color of grape leafs, anisotropic diffusion method is used to maintain the information of affected pixels. The resulting color pixels are assembled by the modified unsupervised self-organizing feature map and a genetic algorithm to optimize and obtain group of different colors in the image. In the next phase, Gabor filter is applied on segmented images to analyze diseases more efficiently. Subsequently, the support vector machine is applied to categorize grape leaf diseases. The authors concluded that the appearance of grape leaf disease features using proposed method can achieve a very efficient quality of classification, but there are certain restrictions with respect to the background of the image, regarding extraction of indistinct color pixels.
Youwen et al. [86] suggested a technique to recognize two diseases namely powdery mildew and downy mildew that can apparent in the cucumber leafs. Statistic pattern recognition approach is used to segment healthy and diseased regions of leafs. To differentiate diseased varieties color, texture and features are extracted. Using all the features, SVM performs the final classification using multiple kernel functions such as linear, polynomial, radial basis and sigmoid function. The authors concluded that using radial basis kernel function with only one shape feature, SVM can recognize cucumber diseases accurately. While with the linear kernel function, combination of two feature sets (texture and shape) provides good performance. SVM has more diseases recognition accuracy and high speed as compared to BP artificial neural network in same environment.
Jian et al. [38] explained the method to identify the diseases in cucumber leafs using SVM with a polynomial function, radial basis function and sigmoid kernel function. The classifier is trained using the above mentioned kernel functions and results were compared to decide the best kernel function for identification of diseases. The author summarized that the radial basis function is the best method to classify leafs diseases and promises good accuracy to identify diseases but this method is preferred when the leafs are affected by a single disease. This method cannot diagnose accurately when two or more diseases occur simultaneously.
Asraf et al. [9] introduced the method to detect the symptoms of the oil palm leafs nutrient disease using support vector machine along with polynomial kernel with hard margin, polynomial kernel with soft margin, and linear kernel. Color and histogram based texture features (based on RGB color and gray level co-occurrence matrix) are extracted and separated into numerous sub-features. SVM classifier is then used for classification using all 27 extracted features using different kernel functions. After, classification, a polynomial kernel with soft margin is more proficient of accurately classifying nutrient disease according to its class as compared to other methods. The author finally deduced that more accurate results can be attained using proper kernel trick function.
Lu et al. [20] proposed a novel technique to recognize the maize leaf diseases, based on fuzzy least square vector machine (FLSVM) algorithm. Using YCbCr color space, spatial GLCM is used to calculate texture characteristics of maize leaf. After feature extraction, fuzzy least square vector machine is used for categorization. Where, the sample mean is calculated with respect to the center of each class. Then according to the distance between center (sample class) and sample, it calculates the initial membership function using fuzzy K nearest neighbor method. It requires membership function to be accurately reflected the distribution characteristics of the samples. The author concluded that proposed model provides encouraging results. It gives smaller errors in the identification process when the samples number is too small. The developed FLSVM also gives prediction uncertainty.
Arivazhagan et al. [7] discussed a system for automatic plant disease detection, which comprised of four stages. In the first stage, a color conversion space is created for the RGB image and then using specific threshold value green pixels are masked, removed and monitored by the segmentation process. Using co-occurrence matrix texture features are calculated for useful segments. Finally, the useful features are handed over to SVM classifier where classifier gain is obtained by the minimum distance criterion. The results specify that the proposed technique can distinguish and classify leaf diseases with a minimalistic computational effort. The author envisioned improving the disease identification rate along with color and texture features for any given input conditions.
Zhou et al. [93] explained an approach which focused on cercospora leaf spot identification in sugar beet using support vector machine with hybrid algorithms of template matching. The technique divided in three stages. Firstly, to discriminate leaf parts from the soiled background for automatic collection of initial sub-templates a plant segmentation index of G-R is introduced. Secondly, a robust template matching technique is implemented for constant monitoring of foliar translation, dynamic object examining and disease development. At last, SVM used for disease classification using color features. Ultimately the author analyzed that the segmentation process is not suitable for other Dicot family plant.
Ratnasari et al. [66] explained a model to identify the sugarcane leaf diseases and recognize severity of spot disease based on segmentation spot. Severity measurement is accomplished only on sample data consisting of regular sized leafs. Disease identification is achieved through segmentation of diseases spot using thresholding of a* component from L*a*b* color space. After segmentation using GLCM, texture and color features are extracted. In the end, SVM classifies the various sugarcane leaf diseases (i.e. ring, rust and yellow spot) using extracted features through the use of different kernels such as polynomial with third order, quadratic, radial basis function (RBF) and linear function. The author effectuated that the proposed model displays more accuracy in classifying sugarcane leaf disease with small average error severity calculation. Linear kernel function gives better results than others. On the other hand, due to limitations of segmentation techniques, lesion cannot be identified accurately.
Mokhtar et al. [53] applied Gabor wavelet transform technique to extract various diseases in tomato leaf. Various relevant features along with SVM having different kernel functions are used to identify and recognize different kind of disease that infects the tomato plant. Initially, a wavelet based feature technique is used to identify an optimum feature subset. Finally, SVM classifiers with different kernel functions such as the Cauchy kernel, Invmult Kernel and Laplacian Kernel were employed to identify and detect tomato leaf infected with powdery mildew or early blight. Finally, it is concluded that the Cauchy and Laplacian kernel functions provides good accuracy but the choice of optimum factors of kernel functions is still one of the critical issue.
Zhang et al. [89] explained a new methodology to advance the recognition rate of diseases in cucumber leaf using singular value decomposition. Watershed algorithm is used for segment spot from leaf images in the first phase. In the second phase, every spot is distributed into little blocks, and combining features of global local singular values are extracted from each block by SVD. In the third phase, main point vectors are assembled and their dimensionalities are adjusted. Finally, SVM classifier is used to identify the category of the unknown disease leaf image. The author concluded that presented method achieve good result. The main limitation of the proposed method is that it needs more computation efforts to evaluate the singular values and advanced color features can improve a better recognition results.
Qin et al. [64] investigated a system for the analysis and classification of the four categories of alfalfa leaf diseases. Using artificial cutting from every captured diseases image, a sub image with single or multiple typical diseases are achieved. Every sub images are transformed into HSV color space and L*a*b color space. In each pixel in the sub-image, (a*) component value and the (b*) component value were regarded as the color features of the pixel. All pixels in the image were clustered into ten classes. After all pixels in a sub-image are clustered into ten classes using a clustering algorithm, the mean of the H components of all pixels in each class is calculated. Then using twelve lesion segmentation methods combined with clustering techniques (i.e. fuzzy C-means clustering K-means clustering and K-median clustering) and supervised classification algorithms (i.e. Naive Bayes algorithm logistic regression analysis, linear discriminant analysis and regression tree) used to segment the sub images. The pixels in the class with the minimum mean were treated as typical lesion pixels, and the pixels in the seven classes with the largest means were treated as typical healthy pixels. From the lesion images based on color, shape and texture total 129 feature are extracted. Using ReliefF, IR and correlation based feature selection methods useful features are selected. Finally diseases recognized and classified using three supervised learning models using support vector machine, random forest and K-nearest neighbor. Evaluations of the classification results of the learning models are evaluated. The author concluded that the SVM model using most significant 45 features is the optimal model. Table 15 represents the summarization of support vector machine techniques used for disease classification.
4.13 Diseases classification using discriminant analysis
This section describes the review of various techniques used for diseases classification using discriminant analysis.
Bandi et al. [10] suggested a technique to classify four classes of citrus leaf diseases i.e. normal, greasy spot, melanose and scab. Based on the color co-occurrence method texture features are evaluated for each citrus leaf sample. After extracting features, they are classified with leaf age condition using Naive Bayes classifier (NBC), Linear Discriminate Analysis (LDA) classifier¸ k-Nearest Neighbor (kNN) and Random Forest Tree Algorithm classifier (RFT). Eventually using earphone operative characteristics contour all the classifiers are compared. The author concluded that Normal leafs and Greasy spot leafs can be classified easily from the other classes of leafs. Whereas, Melanose diseased leafs classification rate is less. The LDA outperforms than other classifiers.
Kruse et al. [43] studied the classification of each pixel as injured or healthy from images of clove leafs through the extraction of color and texture based information features. The four classification approaches were evaluated as Fit to pattern model approach (FPM) combined with T statistics, linear discriminant analysis (LDA), K means clustering and residual sum of squares for classifying and calculating leaf surface injury. The predicted leaf pixel classification is then compared with manually segmented images. The ground truth (binary mask) is used to evaluate pixel classification accuracy. It is concluded that an LDA classifier performed well as compare to mentioned three methodologies in pixel identification owing to its higher accuracy, precision and less computational time. It is determined that the simple feature vector with only color information is sufficient for leaf pixel injury classification. The spatial information does not increased the computation time significantly. Table 16 represents the summarization of discriminant analysis techniques used for disease classification.
4.14 Diseases classification using AdaBoost algorithm
This section describes the review of various techniques used for diseases classification using AdaBoost algorithm.
Min Zhang et al. [88] explained a new method using zone based local and global features to identify citrus canker from citrus leaf images. In the first phase, an improved AdaBoost algorithm is used to evaluate the most imperative features (citrus lesions) after extracting the lesions from their background. In the second phase, for combining texture and color information about lesion, a special citrus canker feature descriptor is proposed. To disclose the spatial properties of citrus canker, Local Binary pattern descriptors are used. In the last phase, to identify canker lesion, two-level hierarchical recognition structure is established. The author compared the performance of the suggested method with human experts and concluded that the accuracy of the presented approach is almost similar.
K. Jagan Mohan et al. [52] proposed a system to identify and classify paddy leaf diseases i.e. brown spot disease, leaf blast disease and bacterial blight disease. The proposed work is divided into two parts. Firstly, diseases are identified using HAAR features and classified using AdaBoost classifier to locate the disease affected portion of the paddy plant and provides identification rate 83.33%. Secondly different diseases are recognized using SIFT (Scale Invariant Feature Transform) feature extraction method to extract local features of image and classified using k-NN (k-Nearest Neighbors) and SVM (Support Vector Machine) classifiers with recognition rate 91.10% and 93.33%. The author concluded that this approach can detect the disease at an early stage and thus can minimize the loss of production. Table 17 represents the summarization of AdaBoost algorithm used for disease classification.
4.15 Diseases classification using rule set theory
This section describes the review of various techniques used for diseases classification using rule set theory.
Phadikar et al. [61] proposed a method based on infected region features to classify different categories of rice. To separate the infected segment, Fermi energy segmentation method is used. Using novel algorithm, shape, position and color features, diseases symptoms are extracted. Change in shape, position and color of diseases spot defined as features to classify diseases with respect to boundary of the leaf. Genetic algorithm is used to detect the shape of the infected segment which estimates the structure of the leaf region. The disease spot is separated into different blocks and then settled as a quad tree at different labels. Consequently, the position of infection is determined. Binary illustration of each block decreases computational complication reasonably. To minimize the loss of information and complexity, significant features are selected using rough set theory. Finally, a rule base classifier using selected features has been assembled that cover all rice leaf diseases. The author concluded that proposed algorithm gives superior outcomes as compare to traditional classifiers. It involves lesser computational complexity. Table 18 represents the summarization of rules based theory used for disease classification.
5 Discussion
Precise and well-timed identification and classification of diseases is helpful to improve the growth. Diseases can be identified by naked eye observation using continuous monitoring experience level. But, it results in high cost as well as time consuming. It has been shown that the image processing proves to be an effective tool for identification and classification of plant diseases, wherein the digital camera functions as a better substitute for the human eye and human brain is superseded by a learning and optimization algorithm. To overcome the difficulties of manual process, several techniques based on computer vision are developed in recent years to identify and recognize disease of agriculture and horticulture crops. The above literature review explained various methods for identification and classification for different plant leaf diseases. Even though the importance of the subject of identifying leaf diseases using digital image processing is tremendous and this has been studied from approximately 25 years, but the advances achieved seem to be a little timid. Some facts lead to this conclusion:
-
a)
Setup conditions are too firm: The presence of noise in captured images makes the quality analysis very difficult. Another constraint is environment conditions for capturing images in controlled lighting conditions. The implementation of image processing techniques in real world application is problematic for number of issues. Existing techniques exhibit high accuracy while examined under controlled laboratory lighting conditions. However, their use in outdoor condition is problematic, since the accuracy in such conditions is drop dramatically. Robust calibration is required to reduce the effect of illumination, angle of camera, capturing device and distance between objects. The other significant challenge arises due to the inherent variability of color under natural lighting conditions.
-
b)
Lack of technical knowledge or Selection of image processing schemes: The choice of the image-processing techniques as well as the classification strategies is crucial for the effective performance of any computer vision system. Assessment of segmentation and classification techniques cause problems like oversized sample set, over fitting etc. In some cases, authors do not provide required technical information.
-
c)
Processing Speed: Processing speed of the detection and classification technique is an important issue as we need to process the huge amount of data. Many feature extraction and selection techniques are proposed to reduce the dimensions of the obtained data firstly and then analyzed for further processing.
-
d)
Significance of universal approach: It is difficult to develop a universal system which has the capability to detect and recognize every type of plant leaf disease. Most of the techniques or systems proposed by authors are able to deal with only one species of plant diseases. Therefore, there is a need to develop a universal system for the detection of plant leaf diseases.
-
e)
Complexity of images: Due to complexity of considered image, it is somehow challenging to separate the desired leaf region from the complex image background.
-
f)
Reliability of data (images): The additional concern is related to the unavailability of complete and reliable information of data. The device classes i.e. lighting set up, camera angle, distance etc. that have been used to produce the results are not fully known. Many of the authors do not provide their testing and training data information, which is the main concern during examine validation of results. It is necessary to provide different reliable measures.
-
g)
Image quality: For better diseases detection and recognition, high quality images dataset are recommended. The lighting setup must be in proper position because it also can affect the image captured
We concluded that the authors should devote more time in learning and understanding of the tools which they intend to use. Apparently, some authors provide high quality manuscripts, in which they define their work clearly with precise device classes, measurable parameters and used technique and tools.
6 Conclusion and future scope
This paper highlighted the concepts and techniques used by various researchers to identify and classify diseases, challenging issues and problems. The ultimate goal is to limit the impact of plant diseases on agricultural production using image processing techniques. Moreover, it is important to understand the correlation between the diseases symptoms and its impacts on yield. The large number of agriculture and horticulture applications based on the detection and classification of plant leaf diseases make it difficult for someone to prospect all important concepts present in the literature. This is a reason of missing potential solutions for problematic issues. The authors should implement more novel algorithms and understand more concepts about tools to achieve better results. They should provide more reliable results by considering the accuracy and quality parameters, which are needed in this highly competitive and changing industry. A brief discussion of popular detection and classification techniques is provided along with possibilities of extensions. The key findings of our literature can also be investigated and analyzed for the other pattern recognition problems. Based on our key findings from the previous studies, following future aspects can be considered for further research.
-
a)
In future, the existing algorithms can also be utilized in outdoor conditions along with the combination of leaf front and leaf backs into a common dataset.
-
b)
Further research can be devoted to model the detection and recognition techniques under mixed lighting conditions. It is of great importance to confirm that the accuracy of classification can be maintained under these conditions or not.
-
c)
An unexplored combination of feature extraction, selection and learning methods can also be used to increase the effectiveness of detection and classification techniques.
-
d)
The future work can also be dedicated to the automatic estimation of severity of the detected diseases.
-
e)
The instant solutions can be made available to the farmers by designing the mobile based applications.
-
f)
Online solutions related to plants diseases can be provided by using web portals.
-
g)
It is also required to increase the number of data for training and testing purposes to achieve better accuracy.
-
h)
Existing work can also be extended to achieve high speed and accuracy by developing the advanced algorithms.
In this review, as there are so many methods are proposed and implemented to identify, recognize and classify plant diseases using digital imaging and classification techniques. The paper presents an overview of technical concepts used in the existing methods related to the objective of the paper. The plant diseases detection and recognition area of research is not limited to above-mentioned future scope.
References
Abdullah NE, Rahim AA, Hashim H, Kamal K (2007) Classification of rubber tree leaf diseases using multilayer perceptron neural network. In: Fifth student conference on research and development (SCORed), Selangor, 11-12 December. pp. 1–6
Abdullakasim W, Powbunthorn K, Unartngam J, Takigawa T (2011) An images analysis technique for recognition of brown leaf spot disease in cassava. Journal of Agricultural Machinery Science 7(2):165–169
Aduwo JR, Mwebaze E, Quinn JA (2010) Automated vision-based diagnosis of cassava mosaic disease. In: Industrial Conference on Data Mining-Workshops, pp. 114–122
Al-Hiary H, Bani-Ahmad S, Reyalat M, Braik M, ALRahamneh Z (2011) Fast and accurate detection and classification of plant diseases. Int J Comput Appl 17(1):31–38
Al-Tarawneh MS (2013) An empirical investigation of olive leaf spot disease using auto-cropping segmentation and fuzzy c-means classification. World Appl Sci J 23(9):1207–1211
Anthonys G, Wickramarachchi N (2009) An image recognition system for crop disease identification of paddy fields in Sri Lanka. In: Fourth IEEE international conference on industrial and information systems (ICIIS 2009), Sri Lanka, 28-31 December. pp. 403–407
Arivazhagan S, Shebiah RN, Ananthi S, Varthini SV (2013) Detection of unhealthy region of plant leafs and classification of plant leaf diseases using texture features. Agric Eng Int CIGR J 15(1):211–217
Asfarian A, Herdiyani Y, Rauf A, Mutaqin KH (2013) Paddy diseases identification with texture analysis using fractal descriptors based on Fourier spectrum. In: International conference on computer, control, informatics and its applications (IC3INA), Jakarta, 19-21 November. pp. 77–81
Asraf HM, Nooritawati MT, Rizam MS (2012) A comparative study in kernel-based support vector machine of oil palm leafs nutrient disease. Procedia Engineering 41(2012):1353–1359
Bandi SR, A Varadharajan, Chinnasamy A (2013) Performance evaluation of various statistical classifiers in detecting the diseased citrus leaf. Int J Eng Sci Technol 5(2):298–307
Barbedo JGA (2013) Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus 2(1):660
Bashish DA, Braik M, Bani-Ahmad S (2010) A framework for detection and classification of plant leaf and stem diseases. In: IEEE International Conference on Signal and Image Processing, pp.113–118
Bentley JW, Boa E, Danielsen S, Franco P, Antezana O, Villarroel B, Rodríguez H, Ferrrufino J, Franco J, Pereira R, Herbas J (2009) Plant health clinics in Bolivia 2000-2009: operations and preliminary results. Food Sec 1(3):371–386
Billah M, Miah MBA, Hanifa A, Amin R (2015) Adaptive neuro fuzzy inference system based tea leaf disease recognition using color wavelet. Commun Appl Electron 3(5):1–4
Bin MohamadAzmi MT, Isa NM (2013) Orchid disease detection using image processing and fuzzy logic. In: International conference on electrical, electronics and system engineering (ICEESE), Kuala Lumpur, 4-5 December. pp.37–42
Bos L (1970) Symptoms of virus diseases in plants, 2nd edn. VADA, Wageningen
Cai HY, Caswell JL, Prescott JF (2014) Non-culture molecular techniques for diagnosis of bacterial disease in animals: a diagnostic laboratory perspective. Vet Pathol 51(2):341–350
Camargo A, Smith JS (2009) An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosyst Eng 102(1):9–21
Chaerle L, Leinonen I, Jones HG, Van Der Straeten D (2007) Monitoring and screening plant populations with combined thermal and chlorophyll fluorescence imaging. J Exp Bot 58(4):773–784
Cunlou L, Gao S, Zhou Z (2013) Maize disease recognition via fuzzy least square support vector machine. J Inf Comput Sci 8(4):316–320
Devereux S (2009) Why does famine persist in Africa? Food Sec 1(1):25
Es-saady Y, El Massi I, El Yassa M, Mammass D, Benazoun A (2016) Automatic recognition of plant leaf diseases based on serial combination of two SVM classifiers. In: International Conference on Electrical and Information Technologies, pp.561–566
Eun AJC, Huang L, Chew FT, Li SFY, Wong SM (2002) Detection of two orchid viruses using quartz crystal microbalance (QCM) immunosensors. J Virol Methods 99(1):71–79
Fang Y, Umasankar Y, Ramasamy RP (2014) Electrochemical detection of p-ethylguaiacol, a fungi infected fruit volatile using metal oxide nanoparticles. Analyst 139(15):3804–3810
Figueiredo AC, Barroso JG, Pedro LG, Scheffer JJ (2008) Factors affecting secondary metabolite production in plants: volatile components and essential oils. Flavour Fragance J 23(4):213–226
Flood J (2010) The importance of plant health to food security. Food Sec 2(3):215–231
Ghaiwat, SN, Arora N (2014) Detection and classification of plant leaf diseases using image processing techniques: a review. International Journal of Recent Advances in Engineering and Technology 2347–2812
González-Fernandez R, Prats E, Jorrín-Novo JV (2010) Proteomics of plant pathogenic fungi. J Biomed Biotechnol 2010:1–36
Gui J, Hao L, Zhang Q, Bao X (2015) A new method for soybean leaf disease detection based on modified salient regions. International Journal of Multimedia and Ubiquitous Engineering 10(6):45–52
Guru DS, Mallikarjuna PB, Manjunath S (2011) Segmentation and classification of tobacco seedling diseases. In: Fourth Annual ACM Bangalore Conference, Bangalore, March 25-26
Haferkamp MR (1988) Environmental factors affecting plant productivity. Achieving efficient use of rangeland resources. Montana State University Agricultural Experiment Station, Bozeman, pp 27–36
He Q, Ma B, Qu D, Zhang Q, Hou X, Zhao J (2013) Cotton pests and diseases detection based on image processing. Indonesian Journal of Electrical Engineering and Computer Science 11(6):3445–3450
Hitimana E, Gwun O (2014) Automatic estimation of live coffee leaf infection based on image processing techniques. In: Second international conference on signal, image processing and pattern recognition (SIPP), Sydney, 24 Febuary
Huang K-Y (2007) Application of artificial neural network for detecting Phalaenopsis seedling diseases using color and texture features. Comput Electron Agric 57:3–11
Husin ZB, Shakaff AY, Aziz AH, Farook RB (2012) Feasibility study on plant chili disease detection using image processing techniques. In: Third IEEE International Conference on Intelligent Systems, Modeling and Simulation (ISMS), Kota Kinabalu, 8-10 Febuary. pp. 291–296
Jagtap SB, Shailesh MH (2014) Agricultural plant leaf disease detection and diagnosis using image processing based on morphological feature extraction. IOSR J VLSI Sig Proc 4(5):24–30
Jaware TH, Badgujar RD, Patil PG (2012) Crop disease detection using image segmentation. World J Sci Technol 2(4):190–194
Jian Z, Wei Z (2010) Support vector machine for recognition of cucumber leaf diseases. In: Second IEEE international conference on advanced computer control, vol 5. IEEE, Shenyang, pp. 264–266
Kai S, Zhikun L, Hang S, Chunhong G (2011) A research of maize disease image recognition of corn based on BP networks. In: IEEE Third International Conference on Measuring Technology and Mechatronics Automation, pp.246–249
Kavya, RM, Gowda A, Bharathi PT, Virupakshaiah HK (2016) Image processing techniques based plant disease detection. Int J Adv Found Res Computer: Pragmatic Rev 3(8):1–8
Keskar PV, Masare SM, Kadam MS, Deoghare SM (2013) Leaf disease detection and diagnosis. Int J Emerg Trends Electr Electron 2(2):28–31
Krishnan M, Sumithra MG (2013) A novel algorithm for detecting bacterial leaf scorch (BLS) of shade trees using image processing. In: Eleventh IEEE malaysia international conference on communications (MICC), Kuala Lumpur, 26-28 November. pp. 474–478
Kruse OM, Prats-Montalbán JM, Indahl UG, Kvaal K, Ferrer A, Futsaether CM (2014) Pixel classification methods for identifying and quantifying leaf surface injury from digital images. Comput Electron Agric 108:155–165
Kuckenberg J, Tartachnyk I, Noga G (2009) Temporal and spatial changes of chlorophyll fluorescence as a basis for early and precise detection of leaf rust and powdery mildew infections in wheat leafs. Precis Agric 10(1):34–44
Kurniawati NN, Abdullah SNHS, Abdullah S, Abdullah S (2009) Investigation on image processing techniques for diagnosing paddy diseases. In: IEEE International conference of soft computing and pattern recognition (SOCPAR 09), Malacca, 4-7 December. pp. 272–277
Kutty SB, Abdullah NE, Hashim H, Kusim AS, Yaakub TN, Yunus PN, Rahman MF (2013) Classification of watermelon leaf diseases using neural network analysis. In: IEEE business engineering and industrial applications colloquium (BEIAC 2013), Langkawi, 7-9 April. pp. 459–464
Mahlein AK (2016) Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis 100(2):241–251
Majid K, Herdiyeni Y, Rauf A (2013) I-PEDIA: Mobile application for paddy disease identification using fuzzy entropy and probabilistic neural network. In: IEEE international conference on advanced computer science and information systems (ICACSIS), Bali, 28-29 September. pp. 403–406
Martinez A (2007) Georgia plant disease loss estimates. The University of Georgia Cooperative Extension Bulletin. University of Georgia, Athens
Massi IE, Saddy YE, Yassa ME, Mammass D, Benazon A (2015) Serial combination of two classifiers for automatic recognition of the damages and symptoms on plant leaf. In : IEEE Third World Conference on Complex Systems, pp.1–6
Meunkaewjinda A, Kumsawat P, Attakitmongcol K, Srikaew A (2008) Grape leaf disease detection from color imagery using hybrid intelligent system. In: Fifth IEEE International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology, pp.513–516
Mohan KJ, Balasubramanian M, Palanivel S (2016) Detection and recognition of diseases from Paddy Plant leaf images. Int J Comput Appl 141(12):34–41
Mokhtar U, Ali MA, Hassenian AE, Hefny H (2015) Tomato leafs diseases detection approach based on support vector machines. In: Eleventh International IEEE, Computer Engineering Conference, pp.246–250
Molina JF, Gil R, Bojaca C, Gomez F, Franco H (2014) Automatic detection of early blight infection on tomato crops using a color based classification strategy. In: XIX IEEE Symposium on Image, Signal Processing and Artificial Vision, pp. 1–5
Mondal D, Kole DK (2015) Detection and classification technique of yellow vein mosaic virus disease in okra leaf images using leaf vein extraction and Naive Bayesian classifier. In: IEEE international conference on soft computing techniques and implementations (ICSCTI), Faridabad, 8-10 October. pp. 166–171
Muthukannan K, Latha P (2014) Fuzzy inference system based unhealthy region classification in plant leaf image. Int J Comput Inf Eng 8(11):2103–2107
Muthukannan K, Latha P (2015) A PSO model for disease pattern detection on leaf surfaces. Image Anal Stereol 34:209–216
Narvekar PR, Kumbhar MM, Patil SN (2014) Grape leaf diseases detection & analysis using SGDM matrix method. International Journal of Innovative Research in Computer and Communication Engineering 2(3):3365–3372
Orillo JW, Cruz JD, Agapito L, Satimbre PL, Valenzuela I (2014) Identification of diseases in rice plant (Oryza Sativa) using back propagation artificial neural network. In: IEEE International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment and Management, pp.1–6
Phadikar S, Sil J (2008) Rice disease identification using pattern recognition techniques. In: 11th international conference on computer and information technology (ICCIT 2008), Khulna, 24-27 December. pp. 420–423
Phadikar S, Jaya S, Das AS (2013) Rice diseases classification using feature selection and rule generation techniques. Comput Electron Agric 90:76–85
Pixia D, Xiangdong W (2013) Recognition of greenhouse cucumber disease based on image processing technology. Open J Appl Sci 3:27–31
Pydipati R, Burks TF, Lee WS (2006) Identification of citrus disease using color texture features and discriminant analysis. Comput Electron Agric 52(1):49–59
Qin F, Liu D, Sun B, Ruan L, Ma Z, Wang H (2016) Identification of alfalfa leaf diseases using image recognition technology. PLoS One 11(12):e0168274
Rangaswami G, Mahadevan A (1998) Diseases of crop plants in India, 4th edn. PHI Learning Pvt. Ltd, New Delhi
Ratnasari EK, Mentari M, Dewi RK, Ginardi RH (2014) Sugarcane leaf disease detection and severity estimation based on segmented spots image. In: IEEE International Conference on Information, Communication Technology and System, pp.93–98
Revathi P, Hemalatha M (2012) Classification of cotton leaf spot diseases using image processing edge detection techniques. In: IEEE international conference on emerging trends in science, engineering and technology (INCOSET), Tiruchirappalli, 13-14 December. pp. 169–173
Revathi P, Hemalatha M (2014) Cotton leaf spot diseases detection utilizing feature selection with skew divergence method. International Journal of scientific engineering and technology 3(1):22–30
Sabrol H, Kumar S (2015) Recent studies of image and soft computing techniques for plant disease recognition and classification. Int J Comput Appl 126(1):44–55
Sanjaya KWV, Vijesekara HM, Wickramasinghe IM, Amalraj CR (2015) Orchid classification, disease identification and healthiness prediction system. International Journal of Scientific and Technology Research 4(3):215–220
Sannakki SS, Rajpurohit VS, Nargund VB, Kulkarni P (2013) Diagnosis and classification of grape leaf diseases using neural networks.In: Fourth International Conference on Computing, Communications and Networking Technologies, pp.1–5
Sekulska-Nalewajko J, Goclawski J (2011) A semi-automatic method for the discrimination of diseased regions in detached leaf images using fuzzy c-means clustering. In: Seventh IEEE international conference on perspective technologies and methods in MEMS design (MEMSTECH), Polyana, 11-14 May. pp. 172–175
Sena Jr DG, Pinto FAC, Queiroz DM, Viana PA (2003) Fall armyworm damaged maize plant identification using digital images. Biosyst Eng 85(4):449–454
Shrivastava S, Singh SK, Hooda DS (2015) Color sensing and image processing-based automatic soybean plant foliar disease severity detection and estimation. Multimed Tools Appl 74(24):11467–11484
Singh DV (1950) Introductory plant pathology. Trans Br Mycol Soc 33:154–160
Deya AK, Sharmaa M, Meshram MR (2016) Image processing based leaf rot disease, detection of betel vine (Piper BetleL.). In: International conference on computational modeling and security. Procedia Comput Sci 85:748–754
Surendrababu V, PS C, Umapathy E (2014) Detection of rice leaf diseases using chaos and fractal dimension in image processing. International Journal on Computer Science and Engineering 6(1):69
Tajane V, Janwe NJ (2014) Medicinal plants disease identification using canny edge detection algorithm histogram analysis and CBIR. Int J Adv Res Comput Sci Soft Eng 4(6):530–536
Thresh JM (2003) The impact of plant virus diseases in developing countries. Virus and virus like diseases of major crops in developing countries. Springer, Netherlands, pp 1–30
Tian Y, Zhao C, Lu S, Guo X (2012) SVM-based multiple classifier system for recognition of wheat leaf diseases. In: IEEE World Automation Conference, pp.189–193
Tucker CC, Chakraborty S (1997) Quantitative assessment of lesion characteristics and disease severity using digital image processing. J Phytopathol 145(7):273–278
Wang H, Li G, Ma Z, Li X (2012) Image recognition of plant diseases based on principal component analysis and neural networks. In: IEEE eighth international conference on natural computation (ICNC), Chongqing, 29-31 May. pp. 246–251
Ward E, Foster SJ, Fraaije BA, Mccartney HA (2004) Plant pathogen diagnostics: immunological and nucleic acid-based approaches. Ann Appl Biol 145(1):1–16
Weizheng S, Yachun W, Zhanliang C, Hongda W (2008) Grading method of leaf spot disease based on image processing. In: IEEE International Conference on Computer Science and Software Engineering 6: 491–494
Wu D, Xie C, Ma C (2008) The SVM classification leafminer-infected leaves based on fractal dimension. In: IEEE conference on cybernetics and intelligent systems, Chengdu, 21-24 September. pp. 147–151
Youwen T, Tianlai Li, Yan N (2008) The recognition of cucumber disease based on image processing and support vector machine. In: IEEE Conference on Image and Signal Processing, pp. 262–267
Zhang S, Zhang C (2013) Orthogonal locally discriminant projection for classification of plant leaf diseases. In: Ninth IEEE international conference on computational intelligence and security, Leshan, 14-15 December. pp. 241–245
Zhang M, Meng Q (2011) Automatic citrus canker detection from leaf images captured in field. Pattern Recogn Lett 32:2036–2046
Zhang S, Wang Z (2016) Cucumber disease recognition based on global-local singular value decomposition. Neurocomputing 205:341–348
Zhang W, Guifa T, Chunshan W (2013) Identification of jujube trees diseases using neural network. Optik International Journal for Light and Electron Optics 124(11):1034–1037
Zhang Z, Li Y, Wang F, He X (2014) A particle swarm optimization algorithm for neural networks in recognition of maize leaf diseases. Sensors & Transducers 166(3):181
Zhihua D, Huan W, Yinmao S, Yunpeng W (2013) Image segmentation method for cotton mite diseases based on color feature and area thresholding. J Theor Appl Inf Technol 48(1):527–533
Zhou R, Kaneko S, Tanaka F, Kayamori M, Shimizu M (2013) Matching-based Cercospora leaf spot detection in sugar beet. In: Seventh International Conference on Agricultural, Biotechnology, Biological and Biosystems Engineering, pp.204–2020
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dhingra, G., Kumar, V. & Joshi, H.D. Study of digital image processing techniques for leaf disease detection and classification. Multimed Tools Appl 77, 19951–20000 (2018). https://doi.org/10.1007/s11042-017-5445-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-5445-8