Abstract
Groundnut (Arachis hypogaea L.) is an important oilseed crop grown in semi-arid tropics where it experiences moisture stress at different stages of growth resulting in reduced growth and productivity. In this study, we report that the stress tolerance of groundnut can be improved by overexpression of stress-specific transcription factor through transgenic approach. In silico electronic-northern analysis of AtNAC2 showed increased expression under different abiotic stresses. The transcript levels of a homolog of AtNAC2 gene were upregulated under different drought regimes in groundnut. Groundnut transgenics overexpressing AtNAC2 showed enhanced tolerance to drought and salinity with improved yield under water-limited conditions. The study demonstrates that AtNAC2 is a potential candidate gene to improve stress tolerance by transgenic approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In semi-arid tropics, high temperature and reduced soil moisture levels affects crop growth and productivity. The impending climate change characterized by an increase in frequency and severity of drought and elevated temperature has accentuated negative impact on the peanut productivity (Bhatnagar et al. 2008; Akcay et al. 2010). Plants have evolved diverse adaptive strategies to cope with water-deficit conditions like drought avoidance, drought escape, and drought tolerance. Drought tolerance phenomenon is complex and polygenic and is based on several inherent and acquired traits. In this context, developing superior genotype having better adaptation to drought stress has relevance. Under water-deficit conditions, the cellular level of tolerance mechanisms which bring about alteration in cell metabolism for plant adaptation, assumes significance. The molecular mechanism of stress perception and signal transduction has been fairly well understood in recent years in plants like Arabidopsis, rice, and maize (Shinozaki and Dennis 2003). Several stress-related proteins including transcription factors (TFs) have been identified, including those encoding key enzymes in abscisic acid biosynthesis (Nambara and Marion-Poll 2005), proteins involved in cellular dehydration (Yao et al. 2011), cellular protective enzymes (Puckette et al. 2007), and proteins involved in cell signaling (Zhu 2002).
Transcription factors control large number of genes by up- or down-regulating the target genes by binding to the respective cis motifs present in the promoter sequences of the genes (Babitha et al. 2013). TFs such as AP2-ERF, DREBs, CBF, bZIP, zinc-finger, MYB, and NAC [NAM (No Apical Meristem), ATAF1, 2 (Arabidopsis thaliana Transcription Activation Factor 1, 2), CUC2 (Cup-Shaped Cotyledon 2)] are directly or indirectly involved in the regulation of plant defense and stress response (Mukhopadhyay et al. 2004; Yanhui et al. 2006).
NAC proteins constitute one of the largest families of plant-specific TFs, and the family is present in a wide range of land plants. NAC TFs were first described a decade ago (Riechmann et al. 2002). Proteins of this family are characterized by a highly conserved DNA-binding domain, known as the NAC domain, in the N-terminal region. The C-terminal region of NAC proteins, which usually contains a transcriptional activation domain, is highly diversified both in length and sequence (Ooka et al. 2003). About 135 and 163 members of this family have been identified in Arabidopsis and rice, respectively (Fang et al. 2008). NAC proteins play an important role in diverse processes, including plant developmental processes such as pattern formation in embryos and flowers, formation of secondary walls, leaf senescence (Mitsuda and Ohme-Takagi 2008), and lateral root development (He et al. 2005). NAC TFs are also involved in plant response to various biotic and abiotic stresses such as pathogens, drought, cold, salinity, low-oxygen, and extreme temperature (Puranik et al. 2012). In Arabidopsis, three NAC members, ANAC019, ANAC055 and ANAC072 bind to the promoter region of ERD1 and improved the tolerance to drought stress (Tran et al. 2004). AtNAC2 is involved in plant responses to hormone signals such as ABA, 1-aminocyclopropane-1-carboxylic acid, and α-naphthalene acetic acid (He et al. 2005). Another NAC gene identified from Hordeum vulgare L. i.e. NAC6 showed enhanced pathogen resistance in barley and Arabidopsis (Jenson et al. 2007). In addition, many NACs have been characterized in rice, and SNAC1 is involved in drought stress tolerance at anthesis stage (Hu et al. 2006). Overexpression of root-specific NAC, OsNAC10 improves drought tolerance and yield in rice (Jeong et al. 2010). SNAC2/OsNAC6, OsNAC045, and OsNAC063 were shown to enhance tolerance to multiple abiotic stresses (Zheng et al. 2009). Transcription factors TaNAC4 and TaNAC8 are found to be involved in both biotic and abiotic stress responses in wheat (Xia et al. 2010a, b). Tobacco transgenics overexpressing NAC1 from Eleusine coracana L. showed enhanced tolerance to several abiotic stresses (Ramegowda et al. 2012).
The emphasis of this study was to assess the functional role of AtNAC2 (AT5G39610) in groundnut for abiotic stresses. The transgenics developed using this gene showed improved phenotypic characters, besides showing tolerance to moisture stress, salinity with improved yield under water-limited conditions.
Materials and methods
Plant material and stress imposition
The expression pattern of AhNAC2-2 a groundnut homolog of AtNAC2 was studied in leaves of groundnut plants exposed to different soil moisture status. Moisture stress treatments were imposed to 4-week-old groundnut seedlings (cv. K-134) by maintaining soil water status at 100, 60, and 40 % field capacity (FC) in pot by gravimetric approach (Govind et al. 2009). The leaf tissues were collected and stored in −70 °C for expression analysis.
Construction of plant transformation vector with AtNAC2 and maintenance of plants
The AtNAC2 gene also known as ANAC092 was obtained from the RIKEN Centre Japan (AT5G39610). The 1.2-kb cDNA fragment of AtNAC2 was subcloned into pTZ57R/T and subsequently the fragment was released using KpnI and SalI (MBI Fermentas) and ligated into pBinAR binary vector. The pBinAR:AtNAC2 plasmid was mobilized to Agrobacterium strain LBA4404 by thr electroporation method and the positive clones were identified using colony PCR analysis with gene-specific primers. Agrobacterium harboring the recombinant binary vector (pBinAR:AtNAC2) was grown in LB medium containing 50 μg/ml kanamycin overnight at 28 °C. The bacterial cells were later resuspended in Winan’s AB medium (pH 5.2) and grown for 18 h. Wounded tobacco leaf extract was added to the suspension before infection. Groundnut cv. K-134 was used to develop AtNAC2 transgenic groundnut plants by Agrobacterium-mediated in planta transformation method (Rohini and Sankara Rao 2000).
Isolation of genomic DNA and total RNA for molecular analysis
Genomic DNA from composite samples was isolated by CTAB method (Doyle and Doyle 1990). The PCR analysis was carried out with gene specific and npt II primers using 94 °C for 5 min initial denaturation, 94 °C for 1 min, 58 °C for 1 min, 72 °C for 1 min of 30 cycles, and final extension at 72 °C for 5 min PCR conditions using the following primers: FP-TAGGTACCCTCTTCTCAAAAACCCTTC and RP-GCGTCGACTTTAACTTCGTGATATCTTTAGAC.
Further to carrying out RT-PCR analysis, total RNA was extracted from a few plants of the T3 generation by using the protocol described by Datta et al. (1989). All care was taken to use RNase-free materials for RNA work according to the instructions (Sambrook et al. 1989). cDNA was prepared using 5 μg of total RNA and reverse-transcribed to make single-stranded cDNA in a 20-μl reaction mix consisting of 25 units M-MuLV reverse transcriptase (MBI Fermentas), 1× MMuLV-RT buffer, 40 pmol oligo dT primer, and 10 μM dNTP mix. Reverse transcription was performed at 42 °C for 1 h, and RNA was reverse-transcribed to cDNA immediately after isolation and stored at −20 °C. Then, 1 μl of the cDNA mix was used as template for PCR amplification. To study the expression of AtNAC2 homolog gene in groundnut, the plants were exposed to 60 and 40 % FC conditions. The RT-PCR analysis was carried out using FP-CGGCGAGAAAGAGTGGTATT and RP-GTTTTCACTCCCTTGGG.
Physiological analysis of transgenic groundnut plants
Rate of water loss, specific leaf area (SLA) and relative water content
The rate of water loss was estimated using method explained by Mao et al. (2012).
SLA of leaves was calculated by measuring leaf area (LA) using an area meter, and subsequently dry weight (DW) was recorded. Then SLA was calculated,
Relative water content (RWC) of leaf discs was measured according to Barrs and Weatherly (1962).
Chlorophyll stability index
One set of leaf discs from wild-type and transgenic plants were incubated in acetone:DMSO (1:1) solution (10 ml) for 72 h as control. Another set of leaf discs were incubated in water for 30 min at 60 °C to give heat stress and were then transferred to acetone:DMSO (1:1) solution (10 ml) for 72 h. Absorbance was recorded at 645, 653, and 663 nm, using UV–Vis spectrophotometer (UV 2450; Shimadzu). Chlorophyll content was estimated by substituting the absorbance values in the following formulae:
-
Chlorophyll a: 12.7 (A 663) − 2.69(A 645)V/weight × 1,000
-
Chlorophyll b: 22.9 (A 645) − 4.68(A 663)V/weight × 1,000
-
Total chlorophyll (mg g FW−1) = (chlorophyll a + chlorophyll b)
-
Percentage of reduction over control (R) = (control-stressed)/control × 100
-
Chlorophyll stability index (X) = 100 – R.
Membrane integrity
Percent leakage of solutes reflecting loss of membrane integrity was quantified in both control and stressed leaf discs. The leaflets from plants grown under contained field conditions were collected and allowed for live wilting in laboratory conditions. One set allowed for 4 h (mild stress) live wilting and another set for 12 h (severe stress). After that, respective leaf discs were incubated in 25 ml of water with continuous shaking for 2 h and initial electrical conductivity (EC) was taken using EC-TDS analyzer (ELICO-CM183). Then, the leaf discs were boiled for 30 min and final EC was taken (Hoekstra et al. 2001). The cell leakage was computed by using following formula,
Yield analysis
The plants were harvested 120 days after sowing (DAS) and several yield parameters such as plant height, pod weight, and total dry matter (TDM) were recorded. The obtained values were subjected to statistical analysis using one-way ANOVA followed by Duncan’s Multiple Range Test (DMRT) using MSTATC software.
Results
In silico analysis of AtNAC2
Stress inducibility of AtNAC2 expression was analyzed by electronic-northern (e-northern) analysis using Arabidopsis Botany Array Tool (http://bbc.botany.utoronto.ca). The microarray dataset showed that AtNAC2 was up-regulated under drought (twofold), oxidative stress and salt stress (Fig. 1a–c). Further, AtNAC2 showed a homology with NAC family genes from different crop species, to an extent of 49 % in Brachypodium distachyon, 54 % in Brassica napus, 55 % in Arachis hypogaea, 48 % in Medicago sativa, 54 % in Nicotiana tabacum, 50 % in Hordeum vulgare, 55 % in Gossypium hirsutum, 50 % in Triticum aestivum, 51 % in Saccharum officinarum, 58 % in Oryza sativa, and 69 % in Glycine max (supplemental Fig. 1). There are two groundnut genes homologous to AtNAC2 gene, namely AhNAC2 (AC142833; Liu et al. 2011) and a gene identified as ACG49995. The nearest neighbor analysis of phylogenetic view shows that AtNAC2 has 55 % identity with ACG49995 and 54 % with AhNAC2. From this result, we refer to this homologous gene (ACG49995) as AhNAC2-2 and AhNAC2 (Liu et al. 2011) was cited here as AhNAC2-1.
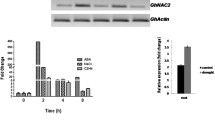
In silico analysis of AtNAC2 and expression analysis of AhNAC2-2. e-Northern expression data of AtNAC2 under different stresses like salinity (a), drought (b), and oxidative stress (c). RT-PCR analysis of AhNAC2-2 (AtNAC2 homolog) in groundnut at different field capacity (FC) levels (60 and 40 %) (d)
Expression analysis of AtNAC2 in groundnuts
To study the expression pattern of AtNAC2 homolog, i.e. AhNAC2-2 in groundnut leaves to moisture stress, 4-week-old plants were exposed to gradual soil moisture stress of 60 and 40 % FC by gravimetric approach (Govind et al. 2009). The transcript levels were significantly increased at 60 % FC moisture levels (Fig. 1d). This confirms the stress-inducible nature of AhNAC2-2.
Further to study the functional relevance of AtNAC2, an overexpression cassette was developed by subcloning AtNAC2 into pBinAR binary vector under CaMV 35S promoter having neomycin-phosphotransferase gene (npt II) as a plant selection marker (Fig. 2a). The binary vector was mobilized into Agrobacterium strain LBA4404 and subsequently the gene construct was transferred to groundnut cv. K-134 using in planta transformation method.
Analysis of T1 transformants
In planta transformation results in the production of many chimeras in the T0 generation and, hence, T1 generation seeds were screened using GRID PCR analysis (Ramu et al. 2012). PCR analysis using 35S CaMV promoter forward primer and gene-specific reverse primer indicated that a few plants were positive. The PCR-positive plants were advanced to next generation to study the stress response. Under normal growth conditions, the transgenics showed round and thicker leaves compared to wild-type (Fig. 2b). Transgenic and wild-type plants raised in pots were subjected to moderate drought stress by withholding irrigation for 72 h to reduce the soil water status to 60 % FC and analyzed for membrane integrity in leaf tissue. The transgenic plants showed relatively less membrane damage compared to wild-type (Fig. 2c). Based on the molecular and physiological data, 29 superior plants were advanced to the next generation.
Analysis of T2 transformants
Seeds from 29 T1 transgenic lines were raised in containers with 10 kg soil + farmyard manure (FYM) mixture at 3:1 ratio and grown into T2 plants. After 120 days from sowing, selected plants were harvested, and growth and yield parameters like plant height, branch number per plant, filled pods, unfilled pods, and dry pod weight per plant were recorded. The transgenic lines showed improved growth and productivity (Table 1). Out of 29 T2 transgenics, 8 lines showed higher pod weight and pod number. The PCR analyses of lines further confirmed the integration of AtNAC2 gene in groundnut genome (Fig. 3a, b).
AtNAC2 transgenic lines showed improved tolerance to abiotic stress
To assess the stress resistance capacity of AtNAC2 transgenics and wild-type plants, excised leaves from 35-day-old plants were collected and subjected to a few drought-screening assays in laboratory conditions like chlorophyll stability index (CSI), SLA, RWC, and relative water loss (RWL). The transgenic lines showed significantly higher CSI, RWC, and reduced RWL than wild-type plants. Whereas, in the vcase of SLA, there was not much difference between wild-type and transgenic lines (Fig. 4a–d).
Similarly, excised leaves were used to assess the electrolyte leakage by exposing to desiccation stress in laboratory condition by air drying for 4 h (mild stress) and 12 h (severe stress). Transgenic plants showed an average of 42.47 % leakage under mild stress and 49.35 % under severe stress, whereas wild-type had shown 46.24 % under mild stress and 68.47 % under severe stress condition respectively (Table 2a).
Further, the chlorophyll content in transgenic under mild stress was 1.93 mg g FW−1 as against 1.4 mg g FW−1 in wild-type (Table 2b).
To study the relative salinity tolerance capacity of transgenics, the leaf discs were floated on 300 mM NaCl solution for 72 h and then chlorophyll content was measured. Transgenic lines showed an average chlorophyll content of 2.20 and 1.35 mg g FW−1 under control and NaCl stress conditions, respectively, whereas wild-type had 1.93 and 0.97 mg g FW−1, respectively (Table 2b).
These results indicate improved growth and yield characteristics in the transgenics compared to wild-type. The TDM was significantly improved in transgenics in both T2 (Table 1) and T3 generation (Table 3). A positive correlation between TDM and pod weight (Fig. 4e) signifies the importance of the transgene.
Expression analysis of AtNAC2 in groundnut transgenics
To analyze the transcript levels of AtNAC2, four PCR positive lines (Fig. 5a, b) with higher TDM (Table 3) were selected for RT-PCR analysis. Total RNA was isolated and cDNA was synthesized and amplified with npt II and AtNAC2 gene specific primers. The transcript levels of these genes were significantly higher in transgenic lines, whereas wild-type plant did not show any expression of either of the genes (Fig. 5c).
Discussion
Plants exposed to desiccation stress alter cell metabolism for adaptation by expression of several stress-responsive genes. Many stress-responsive genes are up-regulated and some are down-regulated in many species and several stress-specific TFs have been well studied (Babitha et al. 2013). The NAC family TFs are a large class of TFs and they play diverse roles in the development and responses to environmental stimuli (Puranik et al. 2012; Ramegowda et al. 2012). Only a few stress-specific NAC proteins have been characterized (Nakashima et al. 2012) for their relevance in imparting tolerance to abiotic stresses. Over 140 putative NAC or NAC-like genes have been identified in rice and classified into different groups based on trait specificity (Fang et al. 2008). Phylogenetic analysis of deduced amino acid sequences of AtNAC2 with other closest accessions of NAC proteins showed that this gene belongs to the NAC family TF (supplemental Fig. 1) which are reported to have a stress-responsive nature. AtNAC2 is also stress responsive as confirmed by e-northern analysis (Fig. 1a–c). AtNAC2 and other NAC proteins have been found to be transcriptional activators (Aida et al. 1997; Fujita et al. 2004; Tran et al. 2004; Xie et al. 2000). The overexpression of AtNAC2 in Arabidopsis showed improved tolerance to salt stress and have a role in lateral root development and involved in ethylene and auxin signaling (He et al. 2005). The AhNAC2-2 gene (a homolog to AtNAC2) also showed the stress inducibility under moisture stress condition in groundnut (Fig. 1d). Previously, it was shown that AhNAC2-1 protein binds to specific NAC response elements (NACRE) in vitro and play a role in ABA signaling (Liu et al. 2011).
Overexpression of Triticum aestivum NAC2 in Arabidopsis was shown to enhance primary root growth, early flowering, reduced water loss, higher leaf water status, higher cell membrane stability, better osmotic adjustment, and increased biomass under stress (Mao et al. 2012). Similarly, AtNAC2 groundnut transgenics showed reduced rate of water loss and maintained higher RWC as compared to wild-type (Fig. 4c, d). This clearly indicates that transgenics have higher ability to retain water and maintain higher leaf water status. Maintenance of higher chlorophyll content which reflects the stay-green nature of AtNAC2 transgenics (Fig. 4a) (Table 2b) under stressful condition is one of the important parameters which result in maintenance of higher canopy photosynthetic rate during stress. The transgenics also showed reduced membrane damage under stress (Table 2a). Maintaining better membrane integrity will help in functional stability of macromolecules (Levitt 1980). Similar stress responses were noticed in transgenics overexpressing SNAC2 and AhNAC2-1 in rice and Arabidopsis, respectively (Hu et al. 2008; Liu et al. 2011).
Several transgenic plants with relevant regulatory genes have been developed in different crop species for abiotic stress conditions (Karaba et al. 2007; Bhatnagar et al. 2008; Anami et al. 2009; Yoshida et al. 2010; Jeong et al. 2010). In this context, our data on overexpression of AtNAC2 in groundnut strengthens the idea of using regulatory genes as candidate genes to improve stress tolerance of crops.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Akcay UC, Ercan O, Kavas M, Yildiz L, Yilmaz OA, Yucel M (2010) Drought-induced oxidative damage and antioxidant responses in peanut (Arachis hypogaea L.) seedlings. Plant Growth Regul 61:21–28
Anami S, De Block M, Machuka J, Van LM (2009) Molecular improvement of tropical maize for drought stress tolerance in sub-Saharan Africa. Crit Rev Plant Sci 28:16–35
Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja NK, Udayakumar M (2013) Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res 22:327–341
Barrs HO, Weatherly PE (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15:413–428
Bhatnagar MP, Vadez V, Sharma KK (2008) Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep 27:411–424
Datta SK, Patel H, Berry D (1989) Extraction and purification of RNA from crop plants. J Exp Bot 165:1252
Doyle JJ, Doyle JL (1990) Isolation of DNA from fresh tissue. Focus 12:13–15
Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 28:547–563
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Govind G, Harshavardhan VT, Patricia JK, Dhanalakshmi R, Senthil-Kumar M, Sreenivasulu N, Udayakumar M (2009) Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut. Mol Genet Genomics 281:591–605
He X, Mu R, Cao W, Zhang Z, Zhang J, Chen S (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signalling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hoekstra PA, Golovina EA, Buitink J (2001) Mechanisms of plant desiccations tolerance. Trends Plant Sci 6:431–438
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Jenson MK, Rung JH, Gregerson PL, Gjetting T, Fugslang AT, Hansen M (2007) The HvNAC6 transcription factor: a positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol Biol 65:141–152
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martine N, Krishnan A, Nataraja KN, Udayakumar M, Pereira A (2007) Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc Natl Acad Sci USA 104:15270–15275
Levitt J (1980) Responses of plants to environmental stresses: water, radiation, salt and other stresses, vol 2. Academic, New York, pp 3–211
Liu X, Hong L, Li XY, Yao Y, Hu B, Li L (2011) Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci Biotechnol Biochem 75:443–450
Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63:2933–2946
Mitsuda N, Ohme-Takagi M (2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56:768–778
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314
Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–242
Puckette MC, Weng H, Mahalingam R (2007) Physiological and biochemical responses to acute ozone-induced oxidative stress in Medicago truncatula. Plant Physiol Biochem 45:70–79
Puranik S, Sahu PP, Srivastava SP, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:1360–1385
Ramegowda V, Senthil-Kumar M, Nataraja NK, Reddy KM, Mysore SK, Udayakumar M (2012) Expression of a finger millet transcription factor, EcNAC1, in tobacco confers abiotic stress-tolerance. PLoS ONE 7(7):e40397
Ramu SV, Rohini S, Keshavareddy G, Gowri Neelima M, Shanmugam NB, Kumar ARV, Sarangi SK, Ananda Kumar P, Udayakumar M (2012) Expression of a synthetic cry1AcF gene in transgenic Pigeon pea confers resistance to Helicoverpa armigera. J Appl Entomol 136(9):675–687
Riechmann JL, Reuber L, Jiang Z, Keddie L, Adam O, Pineda O, Ratcliffe R, Samaha R, Creelman R, Pilgrim P, Broun J, Zhang Z, Ghandehari B, Sherman K, Yu L (2002) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Rohini VK, Sankara Rao K (2000) Transformation of peanut (Arachis hypogaea L.): a non-tissue culture based approach for generating transgenic plants. Plant Sci 150:41–49
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. Cold Spring Harbor Laboratory Press, Plain view
Shinozaki K, Dennis ES (2003) Cell signalling and gene regulation global analyses of signal transduction and gene expression profiles. Curr Opin Plant Biol 6:405–409
Tran LS, Nakashima K, Sakuma Y, Simpson D, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Xia N, Zhang G, Liu X, Deng L, Cai G, Zhang Y, Wang X, Zhao J, Huang L, Kang Z (2010a) Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol Biol Rep 37:3703–3712
Xia N, Zhang G, Sun YF, Zhu L, Xu LS, Chen XM, Liu B, Yu YT, Wang XJ, Huang LL, Kang ZS (2010b) TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol Mol Plant Pathol 74:394–402
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S, Li Z, Xiaohui D, Jingchu L, Xing-Wang D, Zhangliang C, Hongya G, Li-Jia Q (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124
Yao D, Zhang X, Zhao X, Liu C, Wang C, Zhang Z, Zhang C, Wei Q, Wang Q, Yan H, Li F, Su Z (2011) Transcriptase analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics 98:47–55
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685
Zheng X, Chen B, Lu G, Han B (2009) Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun 379:985–989
Zhu J (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgments
Authors acknowledge the financial support from Department of Biotechnology, Centre of Excellence programme support (BT/01/COE/05/03), Indian Council of Agricultural Research—Niche area of Excellence programme (F. No. 10-(6)/2005 EP&D) and Department of Science and Technology—Fund for Improvement of Science and Technology (SR/FST/LSI-051/2002).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patil, M., Ramu, S.V., Jathish, P. et al. Overexpression of AtNAC2 (ANAC092) in groundnut (Arachis hypogaea L.) improves abiotic stress tolerance. Plant Biotechnol Rep 8, 161–169 (2014). https://doi.org/10.1007/s11816-013-0305-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-013-0305-0