Abstract
Boraginaceae species, such as those from the genus Echium, contain high levels of the Δ6-desaturated γ-linolenic (18:3n-6) and octadecatetraenoic (18:4n-3) acids. These are unusual fatty acids among the plant kingdom that are gaining interest due to their benefits to human health. The potential utility of acyltransferases aimed at an increase in oil yield and fatty acid profiling has been reported. In this work, a gene encoding an acyl-CoA:diacylglycerol acyltransferase (DGAT, EC 2.3.1.20) was cloned from Echium pitardii. Genomic and cDNA sequences obtained revealed a gene structure composed of 16 exons, yielding a protein (EpDGAT) of 473 amino acids with high similarity to DGAT1 enzymes of plants. Protein features such as a predicted structure with a highly hydrophilic N-terminus followed by 10 transmembrane domains, as well as the presence of diverse specific signatures, also indicate that EpDGAT belongs to the DGAT1 family. indeed. DGAT activity of the protein encoded by EpDGAT was confirmed by heterologous expression of the full-length cDNA in a yeast mutant (H1246) defective in the synthesis of triacylglycerols. Fatty acid composition of the triacylglycerols synthesized by EpDGAT in H1246 yeast cultures supplemented with polyunsaturated fatty acids suggest a substrate preference for the trienoic fatty acids α-linolenic acid (18:3n-3) and γ-linolenic acid over the dienoic linoleic acid (18:2n-6). Site-directed mutagenesis has revealed the presence of a critical residue (P178 in EpDGAT) within a reported thiolase signature for binding of acyl-enzyme intermediates that might be involved in the active site of the enzyme. Transcript analysis for EpDGAT shows an ubiquitous expression of the gene which is increased in leaves during senescence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant oils have become important renewable resources as biofuel and for human consumption [1–3]. Nowadays there is a growing demand, mainly due to an increase in the biodiesel market specially in Europe [1]. Consequently, any improvement in seed oil production is of interest, and great efforts are being made with this aim within the biotechnology field.
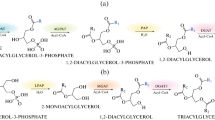
Plant oil is mostly composed of triacylglycerol (TAG), the main storage lipid. TAG is usually synthesized by sequential incorporation of acyl groups through the glycerol-3-phosphate (G3P) pathway, also known as the Kennedy pathway [4–6]. Briefly, G3P is first acylated by the action of the acyl-CoA:glycerol-3-phosphate acyltransferase (GPAT; EC 2.3.1.15), followed by a second acylation step catalyzed by the acyl-CoA:lysophosphatidate acyltransferase (LPAT; also called acyl-CoA:acyl-glycerol-3-phosphate acyltransferase, AGPAT; EC 2.3.1.51). The phosphatidic acid obtained is then dephosphorylated by a phosphatidate phosphatase (PAP; EC 3.1.3.4) to generate diacylglycerol (DAG) which is finally used as substrate for the acyl-CoA:diacylglycerol acyltransferase (DGAT; EC 3.2.1.20) to produce TAG [5, 6]. GPAT and LPAT are acyltransferases common to TAG and membrane-lipid biosynthesis while DGAT catalyze the only step which is committed to TAG biosynthesis [4, 5, 7].
Two alternative pathways for the synthesis of TAG have also been described involving the phospholipid:diaylglycerol acyltransferase (PDAT) and DAG:DAG transacylase (DGTA) enzymes, respectively [5, 8]. It has been suggested that PDAT has a role in directing unusual fatty acids (such as ricinoleic acid) to TAG, thus avoiding their incorporation into polar lipids and possible disturbances of membrane functions [9, 10]. The presence of PDAT in plants lacking such unusual fatty acids indicates that it may play a different role in lipid biosynthesis, though this function remains still unknown [11, 12]. Nevertheless, current evidence strongly suggests that PDAT is not a major determinant of TAG production in seed plants [9, 12–14]. With regard to DGTA, involvement in remodeling of TAG was proposed [15] although it does not seem to affect the net biosynthesis [8, 13].
There is growing evidence supporting a major contribution of DGAT to TAG synthesis in seed plants [14, 16]. The AS11 Arabidopsis mutant, bearing reduced DGAT activity, showed a 75% reduction in seed lipids [17]. Conversely, DGAT over-expression determines a net increase in seed oil content in Arabidopsis [18]. Additional evidence is also available from studies of soybean [19], oil-seed rape [20], olive [21] and maize [3]. In agreement with this, it is considered that DGAT catalyzes a rate limiting step within the TAG biosynthesis pathway [5, 18, 20, 22, 23]. Consequently, DGAT is also regarded as being a key enzyme, from a biotechnological point of view, in order to increase oil content in oleaginous species [7, 23–27].
Another area of active research is the biosynthesis of oils containing particular fatty acid profiles. In this regard, the importance of acyltransferases for the production of ‘designer oils’ in genetically engineered plants has been emphasized [14, 24]. Though initial studies on DGAT activity had suggested a wide substrate utilization by this enzyme [24], later works indicated that the DGAT specificity is species-dependent. While DGAT from plant species like saffron or peanut showed wide acyl utilization [28], in others such as spinach, maize [28], castor bean [29], Arabidopsis [17], Garcinia indica [30], Vernonia galamensis and Stokesia laevis [31], this enzyme exhibited some acyl preference. This behavior opens the possibility of using appropriate DGAT genes to engineer TAG fatty acid profiles.
The first eukaryotic DGAT gene was cloned from the mouse, based on similarity to the acyl-CoA:cholesterol acyltransferase (ACAT) enzyme [32]. A similar gene was later cloned from Arabidopsis [33–35] and other plants species [36–41], all of them encoding proteins related to ACAT. A different family of enzymes with DGAT activity was uncovered after cloning of the DGAT gene from Mortierella rammaniana [42], and finding of similar genes in different plant species [13, 41]. Members of this group share homology with a broader family of genes that transfer acyl groups from coenzyme A to neutral lipids including monoacylglycerol, diacylglycerol, and fatty alcohol species [43]. Thus, two evolutionary unrelated DGAT families are present in plants, encoding membrane bound proteins, that have been named as DGAT1 (ACAT-related) and DGAT2 [23]. Recently, a new DGAT gene was reported encoding a soluble cytosolic enzyme [44]. This protein is closely related to bacterial bifunctional DGAT/wax ester synthetase, thus representing a third unrelated group named as DGAT3 [14].
Experimental evidence indicates that DGAT1 and DGAT2 are the major isoenzymes acting in the biosynthesis of TAG [14], while the contribution of DGAT3 seems to be just marginal [44]. The importance of DGAT1 for the synthesis of TAG in the seed has been well documented in Arabidopsis thaliana [12, 17, 18, 34, 35, 45, 46] and more recently in maize [3]. On the other hand, DGAT2 has proven to be essential in determining both TAG profile and TAG content in Vernicia fordii [41]. Paradoxically, in that work, DGAT2 was shown to be less active in vitro than DGAT1, and yeast transformed with DGAT1 were more efficient in producing TAG than that transformed with DGAT2 [41]. Similar studies performed in castor bean (Ricinus communis) suggested that DGAT2 was the main enzyme for TAG synthesis in seeds [13] and metabolic engineering of Arabidopsis with both castor bean fatty acid hydroxylase 12 and DGAT2 resulted in a significant increase in ricinoleic acid in the seed oil [47]. However, another study on R. communis has shown a DGAT1 activity pattern matching in time to that of TAG accumulation [48]. Moreover, an extensive search for DGAT2 was unsuccessful in Tropaelum majus suggesting that DGAT1 may be the sole DGAT in this plant species [7], and Arabidopsis DGAT2 showed no detectable activity in yeast complementation assays [41]. Therefore, the question about the relative contribution to TAG synthesis by DGAT1 versus DGAT2 is far from being answered [14, 23]. In this regard, it has been suggested that both DGAT isoenzymes may play distinct roles in different tissues and plant species [41, 48].
In this work, we report on the cloning of the DGAT1 gene from Echium pitardii A. Chev. ex D. Bramwell (Boraginaeae). Boraginaceae species such as those from genus Echium are characterized by the accumulation of high levels of the Δ6-desaturated fatty acids, γ-linolenic (18:3n-6, GLA) and octadecatetraenoic (18:4n-3) acids, that are unusual among the plant kingdom. More specifically, Macaronesian species of Echium such as E. pitardii are considered among the richest sources of GLA found in nature, reaching 28% of total fatty acids in the seeds of E. gentianoides [49]. E. pitardii was chosen for our work since its herbaceous habit makes it appropriate for the study in the laboratory [50]. The highest content of GLA (19% for E. pitardii) is found in the seed, but it is also accumulated, although at a lower level, in other organs of the plant such as the leaves, roots and stem [50].
Molecular characterization of the Echium DGAT was performed in our study that included confirmation of the DGAT activity by heterologous expression in a yeast system.
Experimental Procedure
Biological Material
Seeds of E. pitardii A. Chev. ex D. Bramwell (= E. lancerottense Lems et Holz) were collected from plants located in their natural habitat at Lanzarote (Canary Islands). Seedlings (6–8 leaves stage) were grown at 25°C, under the controlled conditions of growth cabinets with a 16 h light/8 h dark photoperiod and 70% relative humidity. Leaf material from seedlings was used as a DNA source, while the different tissues of E. pitardii utilized for RNA extraction and Northern blot analysis were sampled from mature plants cultivated in a greenhouse.
The wild type yeast strain INVSc1 (purchased from Invitrogen), and the H1246 mutant strain (Matα yor245c::KanMX4 lro1::TRP1 are1::HIS3 are2::LEU2 ADE2 ura3) [51], kindly provided by Dr. Stymne (Swedish University of Agricultural Sciences, Uppsala) were used to assay DGAT activities by heterologous expression.
Cloning of the DGAT1 Gene of E. pitardii
Cloning of the EpDGAT gene was achieved by RT-PCR amplification of a partial cDNA sequence, followed by bi-directional walking through inverse PCR (IPCR) on genomic DNA. Briefly, a cDNA was synthesized from 5 μg of total RNA obtained from developing flowers of E. pitardii (RNeasy Plant Mini Kit, QIAGEN) by employing the kit “SuperScript First-Strand Synthesis System for RT-PCR” (Invitrogen), and following the manufacturer's instructions. RT-PCR amplification on the cDNA was done using the degenerated oligonucleotide primers DAG1-Up (5′-ATTATCGARAAYYTIATGAARTAYGG-3′) and DAG2-Down (5′-GCRTTCCACCARTCYTTRTARAAYTC-3′) designed against the DGAT conserved motifs IIENLMKYG and EFYKDWWNA, respectively. The reaction was performed using a proofreading polymerase (AccuTaq, Sigma) and a program consisting of a denaturation step of 2 min at 94°C, followed by 40 cycles of 15 s at 94ºC, 30 s at 45ºC and 75 s at 72ºC, ending with a 5 min step at 72ºC. The resulting fragment (about 800 bp, spanning the central coding region) was cloned in the vector pGEM-T-Easy® (Promega), and several clones were sequenced that resulted in their being all identical. Starting with this partial sequence, a total of five successive IPCR rounds were carried out following the method reported in [52] with minor modifications [53]. DNA clones were sequenced on both strands using a Perkin-Elmer ABI-310 DNA automated sequencer, and the BigDye® Terminator v3.1 chemistry. About 7.5 Kb of genomic sequence were assembled which included the whole coding sequence and about 700 bp of the 5′-region upstream the ATG. A whole cDNA was obtained by RT-PCR as described before on mRNA from developing fruits, using the flanking primers DAG2-Up (5′-CATAGGTACCATGGCAATATGGGAGTCGCCGGA-3′) and DAG2-Down (5′-CATACTCGAGTCAGTTTGCATTAACTTTTCTATTCAAGAC-3′) which contained suitable restriction sites for cloning in the pYES2 vector and a single nucleotide change involving the second codon, ACA (Thr) → GCA (Ala), to conform the Kozak consensus and maximize expression in yeast. The cDNA and genomic sequences were deposited in the GenBank under the accession numbers FJ226588 and FJ226589, respectively.
Cladistic Analysis
Alignment of DGAT protein sequences was achieved using the program Clustal X v.1.7 [54] using the default settings, and further refined by visual inspection. The alignment output was used to generate a cladogram based on the minimum evolution method [55], as implemented in the MEGA package v3.1 [56]. The Poisson model was used together with the pairwise deletion of gaps option, and confidence of the tree branches was checked by bootstrap generated from 1,000 replicates. Rooting of the tree was accomplished by using the plant cytosolic DGAT3 sequences as outgroup. For sequences selected in Fig. 1, the alignment was visualized using the Boxshade v. 3.21 software.
Sequence comparison of EpDGAT with related DGAT1 enzymes from higher plants. The amino acid sequence of EpDGAT (GenBank accession no. FJ226588) was aligned, using the software ClustalX v1.7 together with those of characterized DGAT1 type I DGAT from Olea europaea (acc. no. AAS01606), Nicotiana tabacum (acc. no. AAF19345), Euonymus alatus (acc. no. AAV31083), Ricinus communis (acc. no. AAR11479), Arabidopsis thaliana (acc. no. AAF19262), and Vernicia fordii (acc. no. ABC94471). The Boxshade program is used to highlight the homology between protein sequences. Shading is applied when there is agreement for a fraction of sequences above 0.5. Amino acids identical to EpDGAT are enclosed in black boxes while similar residues are in grey. Gaps introduced for maximum alignment are represented by dashes. Transmembrane domains inferred from the TMpred software [62] are marked by horizontal solid bars. Conserved motifs or putative signatures (see text for details) are boxed, such as the N-terminal basic motif, the Acyl-CoA binding signature, and DAG-binding and putative ER retrieval motifs (ER-DIR). The region containing a conserved leucine repeat (L) coinciding with a thiolase acyl-enzyme intermediate binding signature is also marked (pointed arrow) besides previously described critical Pro and Ser residues which are marked by asterisks. Position of the Pro residue in EpDGAT which was analyzed in this work by site-directed mutagenesis is indicated by an arrow
Southern and Northern Blot Analysis
Genomic DNA was isolated from Echium seedlings by a CTAB-based extraction procedure [57]. DNA (about 5 μg) was restricted with the appropriate restriction enzymes, separated on a 0.8% agarose gel, and transferred by capillarity onto Hybond® N+ nylon membranes (Amersham). Filters were UV-crosslinked, pre-hybridized at 42ºC for 5 h in the 50% formamide/high SDS buffer recommended by the DIG manufacturer (Boehringer-Mannheim), and hybridized at the same temperature and same buffer solution (stringent conditions), containing the digoxigenin-labeled DGAT specific probe. High stringency washes were performed twice at 65ºC during 15 min in buffer containing 0.1× SSC, 0.1% SDS, and the luminogenic substrate CSPD® was used for the detection, following the instructions provided with the DIG detection kit. Images were obtained by exposure of Biomax® ML films (Kodak) for 10–25 min and final developing by standard procedures. The DGAT probe was obtained by random primed labeling from a cDNA fragment spanning 512 bp of the 5′-coding sequence.
Total RNA was extracted from different tissues of Echium plants, using the ConcertTM Plant RNA Reagent (Invitrogen) following the protocol provided by the manufacturer. About 10 μg per lane of total RNA was loaded onto an agarose/formaldehyde gel, electrophoretically separated, and transferred to Hybond®-N+ membranes. Filters were hybridized at 50°C (stringent conditions) as described for Southern analysis, and using the same DGAT specific probe. Stringent washes, accomplished at 68ºC, and detection of the DIG-labelled probe were as indicated before.
Heterologous Expression of EpDGAT in Yeast
The whole EpDGAT coding sequence was transcriptionally fused to the GAL1 inducible promoter of the pYES2® expression vector (Stratagene), and the resulting plasmid used to transform Saccharomyces cerevisiae (INVSc1 or H1246 strains) according to the LiAcO method [58]. Cultures were grown at 28°C in standard minimal medium supplemented with the auxotrophic requirement of the strain plus 1% (w/v) raffinose, and expression was further induced on a 0.4 OD600 culture by the addition of galactose 2% (w/v). Incubation under inductive conditions was prolonged for 48 h at the same temperature. Supplementation of cultures with LNA, ALA and GLA in some experiments was carried out with 0.5 mM of each, in the presence of 0.1% Tween-40 in the induction medium. Yeast cells were collected by centrifugation, further washed with 1.3% NaCl, and the resulting biomass subjected to lyophylization and pulverization in a mortar. The material was stored at −25°C until processed for lipid analysis.
Lipid Analysis
Fatty acid composition of the different materials (lipid extract, lipid fractions, etc.) was analyzed by GC of methyl esters, as previously described [59] and using heptadecanoic acid as the internal standard.
Total lipids were extracted from 200 mg of yeast lyophylized biomass as described elsewhere [60]. Particular care was taken with the procedure to minimize the action of endogenous lipases and subsequent liberation of free fatty acids. The lipid extract was dried in a rotary evaporator under an argon stream and then resolubilized in 2 ml of CHCl3. The lipid extract was fractionated by column chromatography (CC) on a silica gel cartridge (Sep-Pack Classic, Waters) accordingly to [60] with some modifications. Briefly, after cartridge equilibration with CHCl3, the lipid extract was adsorbed into the silica gel cartridge and lipid fractions were sequentially eluted with 30 ml of CHCl3 (neutral lipids, NL) and then 30 ml of MeOH (polar lipids, PL). Lipid fractions were dried in a rotary evaporator as described above and resuspended in 2 ml CHCl3.
Neutral lipid classes were separated by one-dimensional TLC using silica gel plates (Macherey Nagel). Plates were activated in an oven at 120°C for 2 h before use. Solvents employed were: petrol–Et2O–HOAc (80:20:1) [61]. NL classes were visualized by iodine vapor, marked with a pencil and immediately scrapped-off and analyzed individually by GC as previously described. NL classes were identified by co-chromatography with authentic standards. The areas corresponding to NL classes were always scrapped-off and processed even when they were not visible (i.e. SE, TAG, and MAG from the H1246 yeast mutant strain).
The complete process, from lipid extraction to TLC separation, was repeated three times for each yeast biomass. As a result of the procedures employed, all lipid data refer exclusively to saponifiable matter (i.e. acyl-lipids); non-saponifiable matter was therefore excluded from quantitation. This is especially relevant for NL because they always contain non-saponifiable lipids (e.g. sterols).
Site-Directed Mutagenesis of EpDGAT
Site directed mutagenesis was performed directly on the pYES2 plasmid containing the EpDGAT cDNA, using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene), following manufacturer instructions and the oligonucleotides Forward (5′-CCAGTAATTGTCATCCTCAGATGTGATAG/TCGCCGTTCTATCAG-3′) and Reverse (5′-CTGATAGAACGGCGC/ATATCACATCTGAGGATGACAATTACTGG-3′). Primers were degenerated to introduce nucleotide substitutions corresponding to codons for Ser or Ile at position 178 of EpDGAT. Mutations were checked by fully sequencing of the cDNA and GAL-1 promoter regions. Mutated plasmids were introduced in H1246 yeast cells and DGAT activity was assayed by analysis of lipid fractions (PL and NL) and NL classes from galactose-induced cultures, as described above.
Results and Discussion
Cloning and Molecular Characterization of the DGAT1 Gene from E. pitardii
A genomic sequence (about 8.4 Kb; GenBank accession no. FJ226589) containing the whole coding region for EpDGAT was obtained by PCR using degenerated primers against conserved motifs in other previously cloned DGATs, followed by progressive walking through inverse PCR, as indicated in “Experimental Procedure”. This also allowed the design of flanking primers to obtain a cDNA (GenBank accession no. FJ226588) by RT-PCR on mRNA from developing fruits (see “Experimental Procedure”).
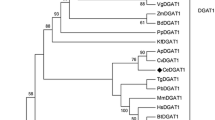
The polypeptide deduced from the longest ORF in the cDNA (Fig. 1) contains 473 amino acids (MW 54.8 kDa) and shows a high similarity to DGAT1 of higher plants like Olea europaea (87% similarity; 78% identity) and Nicotiana tabacum (80% similarity; 70% identity). Cladistic analysis of EpDGAT against other DGATs, covering the three families described in plants (membrane bound DGAT1 and DGAT2, and cytosolic DGAT3) [23], groups EpDGAT with members of the DGAT1 family (Fig. 2). According to this, EpDGAT also share distinctive characteristics of DGAT1. A protein structure is recognized (Fig. 1) composed of a strongly hydrophilic N-terminal region of about 50 amino acids in EpDGAT, followed by a long hydrophobic stretch (ca. 400 residues) with a predicted arrange of 10 transmembrane domains (TMpred software; [62]) that are likely to anchor the protein to the endoplasmic reticulum (ER) membrane. Little conservation is found for the amino terminal domain, which is unusually short in the Echium protein, except for the characteristic basic repeat [36] consisting of three arginine residues in EpDGAT. Also relevant is the presence of the reported acyl-CoA binding signature R61–G79 [18, 37] close to residues (R94–N99) that have been involved in the active site, as well as a DAG/phorbol ester binding motif [18, 35]. A previously reported leucine zipper motif [23, 36] is also found overlapping with a putative thiolase acyl-enzyme intermediate binding motif reported for the Arabidopsis and Tropaeolum DGAT1 [7, 35]. This sequence contains an invariant proline residue (P169 in EpDGAT, Fig. 1) that has been shown critical for DGAT1 activity [7]. Within the same domain it is also remarkable the presence of a proline residue at position 178 of EpDGAT instead of the extremely conserved serine present in other DGAT1 (Figs. 1, 7a). The relevance of the Pro replacement in EpDGAT was assessed in this work by site directed mutagenesis (see results below). A C-terminal YYHDV motif conforming the putative ER retrieval motif is also present in the Echium DGAT similarly to other DGAT1 proteins of plants [13].
Minimum evolution tree showing relationships among EpDGAT and diverse DGAT enzymes from higher plants. Amino acid sequences for plant DGAT enzymes covering the different types described (membrane bound DGAT1 and DGAT2, and cytosolic DGAT3) were obtained from the GenBank, aligned with that of EpDGAT (marked by an arrow) as indicated in Fig. 1, and the resulting matrix analyzed using the minimum evolution method, as described in the “Experimental Procedure”. Branches with a bootstrap value below 50% were collapsed, and higher values represented on the corresponding nodes. The tree was rooted using the cytosolic DGAT3 protein from Arachis hipogaea (acc. no. AAX62735), and putative orthologues of Oryza sativa (acc. no. AAS98422) and Arabidopsis thaliana (acc. no. AAK06873). DGAT2 were from Vernicia fordii (acc. no. ABC94473), Ricinus communis (acc. no. AAY16324), Arabidopsis thaliana (acc. no. AAK32844), and two related sequences from Oryza sativa [acc. nos. BAD33251 (1) and BAD07792 (2)]. DGAT1 were chosen from Nicotiana tabacum (acc. no. AAF19345), Perilla frutescens (acc. no. AAG23696), Olea europaea (acc. no. AAS01606), Euonymus alatus (acc. no. AAV31083), Glycine max [acc. nos. AAS78662 (1) and BAE93461 (2)], Lotus japonicus (acc. no. AAW51456), Jatropha curcas (acc. no. ABB84383), Vernicia fordii (acc. no. ABC94471), Ricinus communis (acc. no. AAR11479), Arabidopsis thaliana (acc. no. AAF19262), Brassica napus [acc. nos. AF251794 (1), AF164434 (2), and AF155224 (3)], Brassica juncea [acc. nos. AAY40784 (1), and AAY40785 (2)], Tropaeolum majus (acc. no. AAM03340), and Oryza sativa [acc. nos. AAW47581 (1), BAF19794 (2), BAF16783 (3), and AAU10815 (4)]
Gene Structure and Genomic Organization of EpDGAT
A comparison between the genomic and cDNA sequences allows the identification of 15 introns interrupting the coding region (Fig. 3a, GenBank accession no. FJ226589). The same structure is also shared by DGAT1 genes of dicot plant species such as Arabidopsis [35], N. tabacum [36], and V. fordii [41]. An exception to this rule are the DGAT genes of the legumes Lotus and Glycine, with only 14 introns, a difference that is likely due to combination of the last two exons [40]. When sequences around the inferred splicing sites are analyzed (Table 1 in Supplementary data) the usual GT pair in all donor splice sites is found, although a non-consensus acceptor splice sequence is observed in the third intron of the EpDGAT gene whose sequence (GG) differ from the common AG. This particular deviation has also been observed in other organisms and has been illustrated in the case of A. thaliana (http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_nonconsensus_splice_sites.shtml).
a Genomic structure of EpDGAT together with a restriction map showing relevant endonuclease sites (Hae, HaeIII; E1, EcoRI; EV, EcoRV; HIII, HindIII; V, VspI) for Southern analysis. Position of the cDNA probe used for hybridization experiments is represented correlating with exons covered in the genomic DNA. b Analysis of the genomic organization of EpDGAT by Southern blot. Genomic DNA of E. pitardii was restricted with the different enzymes indicated above each lane and analyzed as indicated in “Experimental Procedure”, using a digoxigenine labeled cDNA as a probe (see upper panel). Marker sizes (Kbp) are indicated. c Expression analysis of EpDGAT by Northern blot in different tissues of E. pitardii. Equivalent amounts of total RNA (10 μg) from roots (R) developing (Ld) or mature (Lm) leaves, floral stem (S) developing flowers (Fl) and developing fruits (Fr), collected from adult plants, were subjected to electrophoresis in an agarose/formaldehyde gel, run for 1 h at 110 V, blotted and hybridized with the EpDGAT-specific probe (see above) under highly stringency conditions, as indicated in “Experimental Procedure”. The ethidium bromide staining of the gel is also shown as a loading control
Genomic organization of the EpDGAT gene was investigated by Southern-blot on genomic DNA restricted with different enzymes under highly stringency conditions (Fig. 3b). The pattern obtained is in agreement to that expected from the known genomic sequence and the cDNA probe used in the analysis (Fig. 3a, b). This also indicates that the DGAT1 gene is represented by a single copy in the Echium genome. This is also suggested by the finding of identical sequences for the different cDNA clones obtained in the initial RT amplification using primers against highly conserved motifs (see “Experimental Procedure”).
Expression Analysis of EpDGAT
The transcript level of EpDGAT was determined by Northern blot on total RNA. An ubiquitous expression is observed among the different organs of the plant (Fig. 3c). This generalized expression pattern is also found for DGAT1 genes of other plants such as Arabidopsis, Brassica and soybean [33, 35, 40, 45, 63], where expression is not restricted to typical oil-accumulating organs like seeds. As in the case of Echium, relatively high transcript levels are also present in flowers, stem, roots, and leaves, which may indicate a more generalized function in the plant. Low expression levels, though comparable to those of developing seeds, are found in the leaves of Ricinus [13] and tung tree [41], where a second DGAT enzyme (DGAT2) is also present with a location predominantly in the seed. However, a seed-specific pattern of DGAT1 has been reported in the case of Tropaeolum [7]. It seems likely that differences may exist in the contribution of the DGAT isozymes and metabolic pathways to the synthesis of TAG in the diverse organs, among the plant species considered.
Interestingly, the mRNA level was remarkably higher in the old leaves of Echium than in young developing leaves. Up-regulation of the DGAT1 gene during senescence has been also reported in Arabidopsis and soybean [40, 63], and a role in sequestering fatty acids mobilized from plastid galactolipids into TAG has been proposed [63]. Related to this, is the presence of two ethylene responsive motifs (ERE) in the 5′-regulatory region of EpDGAT located at −350 and −380 as a part of a wider direct repeat CACCTATATTTCAAA (see GenBank accession no. FJ226589). Two direct ERE motifs are also found in the promoter of the Arabidopsis DGAT1 gene that have been related with seed maturation [45]. However, a possible involvement of the ERE motifs in the regulation of DGAT1 during the leaf senescence remains unknown, but it is also likely given the involvement of ethylene in the senescence of vegetative tissues [64, 65].
The Protein Encoded by EpDGAT shows TAG Biosynthetic Activity
To assess DGAT activity of the EpDGAT product, a complementation assay was carried out using the H1246 strain of S. cerevisiae [51]. This is a dga1 lro1 are1 are2 quadruple mutant that is defective in the DAG1 and LRO1 genes which are responsible for the TAG synthesis in the yeast, besides two other genes, ARE1 and ARE2, with overlapping acyl-CoA:sterol acyltransferase (ASAT) activities, thus rendering the yeast unable to synthesize both triacylglycerol (TAG) and steryl ester (SE) [51]. Yeast transformation was performed with a pYES2 expression plasmid containing the EpDGAT gene, or the empty vector as a control. The lipid content (as total fatty acids, TFA) from galactose induced cultures was determined from the lipid extract and the lipid extract was fractionated into polar lipids (PL) and neutral lipids (NL) as indicated in “Experimental Procedure”. As shown in Fig. 4a, nearly a fourfold increase (from 11 to 40 mg/g) in the NL fraction is obtained relative to the control, when the yeast mutant is transformed with EpDGAT, while a reduction in the PL was found. Expressed as percentages, this results in an increment of the NL from 22 to 66% and a parallel decrease of the PL from 78 to 34%. Similarly, expression of EpDGAT in a wild type yeast strain (INVSc1) also results in a clear increase of the NL fraction (Fig. 4a). The higher PL content in the H1246 mutant as compared to the same cells transformed with EpDGAT or with the wild type strain may be attributed to the higher availability of the DAG, an intermediary that is also employed for the synthesis of major phospholipids in yeast [66].
Synthesis of TAG directed by heterologous expression of EpDGAT in yeast cells. a Fatty acids content of the NL (black bars) and PL (grey bars) fractions from the defective H1246 or wild type INVSc1 yeast strains transformed with the empty expression vector (pYES2) or the same plasmid containing the Echium DGAT gene (EpDGAT). Lipids were extracted from the induced yeast cells and processed as indicated in the “Experimental Procedure” to obtain the NL and PL fractions. Fatty acids of acyl-lipids in these fractions were quantitated by GC analysis of methyl esters, as described in the “Experimental Procedure”, and expressed relative to the dry wt biomass. Mean values (n = 3) are represented together with their SE. b Lipid analysis by TLC of the NL fraction obtained from H1246 cells transformed with the empty vector (pYES2) or with the plasmid containing EpDGAT. Lipids were visualized by iodine staining. Predominant acyl-lipid classes in the NL fractions are indicated: triacylglycerol (TAG), steryl esters (SE), free fatty acids (FFA) and diacylglycerols (DAG). In our chromatography system DAG overlap with sterols (ST), and estimation of the DAG amount is therefore not possible from direct visualization of the band intensity
The NL fraction was further analyzed by resolving the lipid classes by TLC, and each saponifiable NL class was quantitated by GC (see “Experimental Procedure”). As expected for the mutant strain H1246 [51] neither SE nor TAG was detected in the NL fraction of the pYES2 control, where the predominant NL classes were free fatty acids (FFA) (73%) and DAG (27%) (Fig. 4b). Conversely, TLC plates of NL reveal the appearance of a prominent band corresponding to TAG in the yeast transformed with EpDGAT that is lacking in the pYES2 control (Fig. 4b). Most of NL of yeast expressing EpDGAT correspond to TAG (91%), with FFA in much lower amount (9%), and undetectable levels of DAG and SE. These results indicate that EpDGAT encodes a protein with TAG biosynthetic activity, what together with sequence identity suggests that it is a DGAT1 enzyme.
Analysis of the Acyl-CoA Preference of EpDGAT
Echium pitardii accumulates substantial amounts of GLA, an ‘unusual’ PUFA in higher plants. Since it has been reported that DGAT enzymes from plant species with ‘unusual’ fatty acids (e.g. ricinoleic, vernolic, eleostearic acids) exhibit preference for substrates containing its ‘unusual’ fatty acid, it may be hypothesized that EpDGAT could exhibit preference for some PUFAs. In this regard, we devised an “in vivo” experiment in yeast to get some hint on the possible acyl-CoA preference of EpDGAT. We compared the fatty acid profiles of the H1246 yeast mutant transformed with the EpDGAT gene with that containing the empty vector. Since the yeast does not synthesize PUFAs we supplemented the culture with equimolar amounts of LNA, GLA, and ALA together. As will be shown, these exogenous fatty acids are efficiently incorporated to the different cellular lipids. Similarly to the previous experiment, the lipid extract from the induced cultures was fractionated into PL and NL. As represented in Fig. 5a, the amount of PL fraction did not change significantly while a considerable increase was observed for the NL which increased from 14 mg/g in the pYES2 control to 129 mg/g in the yeast expressing EpDGAT. Overall these results are similar to those obtained in the experiment without added PUFAs, though a greater increase of the NL suggests a limitation of the FA supply for the synthesis of TAG in the non supplemented yeast, at least under our experimental conditions. Lipid class composition in the NL fraction reveals that, as expected, TAG are the main component (76%) when the yeast are transformed with EpDAGAT (Fig. 5b). When fatty acid composition of the NL was determined (Fig. 5c) differences were observed among them, with a lower proportion of C16 fatty acids (16:0 and 16:1n-7) in EpDGAT expressing cells, relative to the control, while 18:0 is increased and 18:1n-9 remains unchanged. With regard to the three PUFAs, exogenously provided, the trienoic acids, GLA and ALA, increased while LNA decreased in EpDGAT transformed yeast relative to the control (Fig. 5c). These differences were statistically significant as shown by ANOVA and non-parametric tests.
Lipid synthesis directed by EpDGAT in H1246 yeast supplemented with PUFAs. a Fatty acids content of the NL (black bars) and PL (grey bars) fractions from the defective H1246 yeast strain transformed with the empty expression vector (pYES2) or the same plasmid containing the Echium DGAT gene (EpDGAT), and cultivated in the presence of equimolar amounts of LNA, ALA and GLA. Lipids were analyzed as in Fig. 4a. b Contributions of individual fatty acids in the NL fractions of H1246 yeast cells transformed with pYES2 alone or the plasmid containing EpDGAT from cultures supplemented with PUFAs (see above). Values are expressed as percentage over total fatty acids in the NL fraction. Mean values (n = 3) are represented together with their SE. Significance of the differences was checked by ANOVA and non-parametric tests (ns non significant; *P < 0.05; **P < 0.01; ***P < 0.001)
When PUFA contributions to the different lipids are compared (Fig. 6) several observations are remarkable. The three PUFAs are efficiently incorporated into the different lipids if we compare them to the endogenous fatty acids of the yeast. Nevertheless, GLA is incorporated in all lipids to a lower extent than LNA and ALA (Fig. 6), even though they are supplied to cultures in equimolar amounts. This indicates that, in the yeast, GLA is discriminated against by some of the yeast enzymes (acyl-CoA synthetases and/or acyltransferases) involved up to the synthesis of DAG. Thus, a LNA:GLA:ALA ratio of 1:0.25:0.93 in the total lipid extract (TL), and 1:0.36:1 in the NL fraction are obtained for the pYES2 control (Fig. 6a). However, when cells are transformed with EpDGAT, a remarkable change in the ratio of PUFAs is observed both in TL (1:0.57:1.56) and NL (1:0.69:1.86), so that LNA contribution is reduced relative to GLA and ALA, thus indicating that these two PUFAs are being favored in their incorporation to the TAG by the Echium DGAT (Fig. 6a) as compared to the rest of acyl-lipids. This suggestion is also supported by comparison of the PUFA composition of lipid classes in the NL fraction of EpDGAT transformed yeast (Fig. 6b). A PUFA ratio of 1:0.77:2.08 was obtained for TAG while, in DAG and FFA, the ratios were 1:0.40:1 and 1:0.41:0.76, respectively, fairly similar to that of the NL fraction in control cells. In other words, GLA and ALA increased from DAG to TAG while LNA decreased (Fig. 6b). These differences in PUFA percentages between DAG and TAG were statistically significant (P < 0.05) by ANOVA and non-parametric tests. Since in the H1246 cells the synthesis of TAG comes exclusively from the acylation of DAG via EpDGAT it seems likely that ALA and GLA are being preferentially selected against LNA in their incorporation to DAG by EpDGAT, thus increasing the proportion of the trienoic acids in TAG relative to that in the DAG substrate.
PUFAs composition in different lipid fractions of H1246 yeast cells transformed with the empty vector (pYES2) or the vector containing EpDAGAT (see experiment in Fig. 5). LNA, GLA and ALA contributions are represented for the total lipids in the biomass (TL in panel a), the NL (in panel a), and individual NL classes (TAG, DAG and FFA in panel b). Values correspond to percentages on the total FAs in each fraction. Acyl-lipid composition of the NL fraction is represented as a percentage for each lipid class (panel c). Mean values (n = 3) are represented besides their SE
A number of reports on DGAT activity using microsomes from different species have indicated a preference for substrates containing particular acyl groups [23]. This is exemplified in plants like Cuphea and Ricinus, producing unusual and potentially toxic fatty acids such as lauric and ricinoleic acids, respectively [29]. In these cases it was proposed that fast conversion of DAG to TAG would act as a way to prevent incorporation of harmful fatty acids to membrane phospholipids [29]. Channeling of particular fatty acids into TAG has also been described in cocoa, for stearic acid [67], and rape, for erucic acid [68].
However, studies on substrate preference dealing with particular DGAT isoenzymes (DGAT1, DGAT2) are still scarce, and even a clear picture of their particular roles and distribution within the plant is lacking at the moment. It has been shown that DGAT1 of some plants like Ricinus [38] and Tropaeolum [7] favor the incorporation of their respective unusual ricinoleic and erucic fatty acids into TAG. However, in other species like Vernonia [69] and Vernicia [41], the DGAT1 enzyme does not show preference for vernolic and eleostearic acids, respectively, contrary to the results obtained with seed microsomes. In these cases the DGAT2 isoenzyme was proposed to be responsible for channeling of the unusual fatty acids. It seems therefore that substrate preference and isoenzyme contribution to the synthesis of TAG may be different in each plant.
There is little information available for Boraginaceae species. The analysis carried out with Borago seed microsomes indicated a strong selectivity of the DGAT activity by GLA-CoA [70]. Even though interpretation of our results must be cautious, data obtained for Echium DGAT do not show a strong preferential utilization of GLA-CoA. It is possible, as described above for other species, that additional activities (e.g. DGAT2) are involved in determining the fatty acid profile of TAG in Boraginaceae species. It should be also noticed that, contrary to Borago, the seeds of Echium accumulate high amounts of ALA [49], which is consistent with the observed preference of the Echium DGAT1 enzyme for this fatty acid.
Site Directed Mutagenesis of the Pro178 Residue of EpDGAT
As previously stated, a proline residue is present at position 178 of EpDGAT, a place where a serine is invariantly found for DGAT1 proteins of plants and animals (Fig. 7a). We have performed SDM on this residue in order to assess its importance for DGAT activity. In particular, the P178 was replaced either by serine (P → S), the common residue, or by isoleucine (P → I), and DGAT activity was recorded by yeast complementation of the defective H1246 strain, as described before. As shown in Fig. 7b, the P → S replacement does not have an appreciable effect on the synthesis of TAG, estimated by the NL content, which is predominantly composed by TAG, as it was shown previously. On the contrary, changing of the P178 to Ile produces a drastic reduction of the NL content thus showing the critical role of this residue for DGAT1 activity. As expected, none of the replacements had an appreciable effect on the PL content (Fig. 7).
Site-directed mutagenesis of EpDAGAT. a Protein alignment of the leucine repeat region showing the P169 residue in EpDAGAT which was replaced either by Ser (P→S) or Ile (P→I). Positions of other critical amino acids in the same region, revealed in previous studies, are marked by asterisks. b Fatty acids content of the NL (grey bars) and PL (white bars) fractions from the defective H1246 yeast strain transformed with the wild type EpDGAT gene (wt) and the two replacement mutants (P→S and P→L). Lipids were extracted from the induced yeast cells and processed as indicated in the “Experimental Procedure” to obtain the NL and PL fractions. Fatty acids of acyl-lipids in these fractions were quantitated by GC analysis of methyl esters and expressed as a percentage over the dry wt biomass. The experiment was performed three times using independent cultures, and the mean value is shown besides their SE
Different studies have underlined the importance of this region for DGAT1 activity. Thus, two critical positions have been identified (Fig. 7a), a serine residue (Ser199 in EpDGAT) which is essential for ACAT activity [71], and an extremely conserved proline (Pro169 in EpDGAT) present in diverse acyltransferases [72]. This Pro was demonstrated to act as a catalytic site in GPAT enzymes [72], and a recent work also proved its importance for DGAT1 activity [7]. As stated before the P178 is located between these two amino acids as a part of a sequence that contains a thiolase signature (PCDO0092) for binding of acyl-enzyme intermediates [7, 35]. The loss of activity produced by the P→I replacement suggests the possible involvement of this residue as a part of the DGAT1 active site. On the other hand, it was noticed that DGAT1 proteins of plants contain a regularly spaced leucine repeat overlapping the thiolase signature (Fig. 7a) which has been proposed to act as a leucine zipper in protein to protein interactions [37]. However, specialized leucine zipper prediction tools such as TRESPASSER [73] or 2ZIP [74] fail to recognize this structure, and it should also be noticed that the Leu repeat is absent from animal enzymes. The notion that this motif may not represent a leucine zipper is in agreement with our results since the presence in the EpDGAT of the Pro residue, a typical helix breaker, does not seem to affect enzyme activity which is similar to that of the Ser containing mutant.
The cloning and molecular characterization of the gene encoding the DGAT1 enzyme of Echium was achieved. Functional assay by heterologous expression in yeast show that the encoded protein promotes the synthesis of TAG, and increases the oil amount. We also report evidence that Echium DGAT1 catalyzes a preferential incorporation of ALA over LNA, and that it does not discriminate the GLA negatively. This is interesting regarding the possible utility of this gene to modify the fatty acid profile of transgenic plants in order to increase the contribution of trienoic fatty acids in oils, including the ‘unusual’ GLA.
Abbreviations
- ALA:
-
Alpha-linolenic acid
- cDNA:
-
Complementary DNA
- CTAB:
-
Cetyl trimethylammonium bromide
- DAG:
-
Diacylglycerol
- DGAT:
-
Acyl-CoA:diacylglycerol acyltransferase
- DIG:
-
Digoxigenin
- FFA:
-
Free fatty acids
- GC:
-
Gas chromatography
- GLA:
-
Gamma-linolenic acid
- IPCR:
-
Inverse PCR
- LNA:
-
Linoleic acid
- NL:
-
Neutral lipids
- PCR:
-
Polymerase chain reaction
- PL:
-
Polar lipids
- PUFA:
-
Polyunsaturated fatty acid
- RT-PCR:
-
Reverse transcriptase PCR
- SDS:
-
Sodium dodecylsulfate
- SE:
-
Steryl esters
- ST:
-
Sterols
- TAG:
-
Triacylglycerol
- TL:
-
Total lipids
- TLC:
-
Thin layer chromatography
References
Dyer JM, Stymne S, Green AG, Carlsson AS (2008) High-value oils from plants. Plant J 54:640–655
Durrett TP, Benning C, Ohlrogge J (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54:593–607
Zheng P, Allen WB, Roesler K, Williams ME et al (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40:367–372
Dircks L, Sul HS (1999) Acyltransferases of de novo glycerophospholipid biosynthesis. Prog Lipid Res 38:461–479
Lehner R, Kuksis A (1996) Biosynthesis of triacylglycerols. Prog Lipid Res 35:169–201
Coleman RA, Lee DP (2004) Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res 43:134–176
Xu J, Francis T, Mietkiewska E, Giblin EM et al (2008) Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol J 6:799–818
Stobart AK, Mancha M, Lenman M, Dahlqvist A, Stymne S (1997) Triacylglycerols are synthesized and utilized by transacylation reactions in microsomal preparations of developing safflower (Carthamus tinctorius L.) seeds. Planta 203:58–66
Dahlqvist A, Stahl U, Lenman M, Banas A et al (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97:6487–6492
Millar AA, Smith MA, Kunst L (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5:95–101
Banas A, Carlsson AS, Huang B, Lenman M et al (2005) Cellular sterol ester synthesis in plants is performed by an enzyme (phospholipid:sterol acyltransferase) different from the yeast and mammalian acyl-CoA:sterol acyltransferases. J Biol Chem 280:34626–34634
Mhaske V, Beldjilali K, Ohlrogge J, Pollard M (2005) Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol Biochem 43:413–417
Kroon JT, Wei W, Simon WJ, Slabas AR (2006) Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67:2541–2549
Cahoon EB, Shockey JM, Dietrich CR, Gidda SK et al (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10:236–244
Mancha M, Osorio J, Garces R, Ruso J et al (1994) New sunflower mutants with altered seed fatty acid composition. Prog Lipid Res 33:147–154
Bao X, Ohlrogge J (1999) Supply of fatty acid is one limiting factor in the accumulation of triacylglycerol in developing embryos. Plant Physiol 120:1057–1062
Katavic V, Reed DW, Taylor DC, Giblin EM et al (1995) Alteration of seed fatty acid composition by an ethyl methane sulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol 108:399–409
Jako C, Kumar A, Wei Y, Zou J et al (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126:861–874
Settlage SB, Kwanyuen P, Wilson RF (1998) Relation between diacylglycerol acyltransferase activity and oil concentration in soybean. J Am Oil Chem Soc 75:775–781
Perry HJ, Bligny R, Gout E, Harwood JL (1999) Changes in Kennedy pathway intermediates associated with increased triacylglycerol synthesis in oil-seed rape. Phytochemistry 52:799–804
Giannoulia K, Haralampidis K, Poghosyan Z, Murphy DJ, Hatzopoulos P (2000) Differential expression of diacylglycerol acyltransferase (DGAT) genes in olive tissues. Biochem Soc Trans 28:695–697
Triki S, Ben Hamida J, Mazliak P (2000) Diacylglycerol acyltransferase in maturing sunflower seeds. Biochem Soc Trans 28:689–692
Lung SC, Weselake RJ (2006) Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41:1073–1088
Frentzen M (1998) Acyltransferases from basic science to modified seed oils. Fett-Lipid 100:161–166
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4:12–21
Dyer JM, Mullen RT (2005) Development and potential of genetically engineered oilseeds. Seed Sci Res 15:255–267
Lardizabal KD, Effertz R, Levering CK, Mai JT et al (2008) Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 148:89–96
Ichihara K, Takahashi T, Fujii S (1988) Diacylglycerol acyltransferase in maturing safflower seeds: its influences on the fatty acid composition of triacylglycerol and on the rate of triacylglycerol synthesis. Biochim Biophys Acta 958:125–129
Vogel G, Browse J (1996) Cholinephosphotransferase and diacylglycerol acyltransferase (substrate specificities at a key branch point in seed lipid metabolism). Plant Physiol 110:923–931
Daniel J, Abraham L, Balaji K, Rajasekharan R (2003) Biosynthesis of stearate-rich triacylglycerol in developing embryos and microsomal membranes from immature seeds of Garcinia indica Chois. Curr Sci 85:363–370
Yu K, McCracken CT Jr, Li R, Hildebrand DF (2006) Diacylglycerol acyltransferases from Vernonia and Stokesia prefer substrates with vernolic acid. Lipids 41:557–566
Cases S, Smith SJ, Zheng YW, Myers HM et al (1998) Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA 95:13018–13023
Hobbs DH, Lu C, Hills MJ (1999) Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett 452:145–149
Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem 37:831–840
Zou J, Wei Y, Jako C, Kumar A et al (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19:645–653
Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H (2000) Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur J Biochem 267:85–96
Nykiforuk CL, Furukawa-Stoffer TL, Huff PW, Sarna M et al (2002) Characterization of cDNAs encoding diacylglycerol acyltransferase from cultures of Brassica napus and sucrose-mediated induction of enzyme biosynthesis. Biochim Biophys Acta 1580:95–109
He X, Turner C, Chen GQ, Lin JT, McKeon TA (2004) Cloning and characterization of a cDNA encoding diacylglycerol acyltransferase from castor bean. Lipids 39:311–318
Milcamps A, Tumaney AW, Paddock T, Pan DA et al (2005) Isolation of a gene encoding a 1, 2-diacylglycerol-sn-acetyl-CoA acetyltransferase from developing seeds of Euonymus alatus. J Biol Chem 280:5370–5377
Wang HW, Zhang JS, Gai JY, Chen SY (2006) Cloning and comparative analysis of the gene encoding diacylglycerol acyltransferase from wild type and cultivated soybean. Theor Appl Genet 112:1086–1097
Shockey JM, Gidda SK, Chapital DC, Kuan JC et al (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18:2294–2313
Lardizabal KD, Mai JT, Wagner NW, Wyrick A et al (2001) DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J Biol Chem 276:38862–38869
Turkish AR, Henneberry AL, Cromley D, Padamsee M et al (2005) Identification of two novel human acyl-CoA wax alcohol acyltransferases: members of the diacylglycerol acyltransferase 2 (DGAT2) gene superfamily. J Biol Chem 280:14755–14764
Saha S, Enugutti B, Rajakumari S, Rajasekharan R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141:1533–1543
Lu CL, de Noyer SB, Hobbs DH, Kang J et al (2003) Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol Biol 52:31–41
Zhang FY, Yang MF, Xu YN (2005) Silencing of DGAT1 in tobacco causes a reduction in seed oil content. Plant Sci 169:689–694
Burgal J, Shockey J, Lu C, Dyer J et al (2008) Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol J 6:819–831
Chen GQ, Turner C, He X, Nguyen T et al (2007) Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in castor bean (Ricinus communis L.). Lipids 42:263–274
Guil-Guerrero JL, Gomez-Mercado F, Rodriguez-Garcia I, Campra-Madrid P, Garcia-Maroto F (2001) Occurrence and characterization of oils rich in gamma-linolenic acid (III): the taxonomical value of the fatty acids in Echium (Boraginaceae). Phytochemistry 58:117–120
Garcia-Maroto F, Garrido-Cardenas JA, Rodriguez-Ruiz J, Vilches-Ferron M et al (2002) Cloning and molecular characterization of the Delta 6-desaturase from two Echium plant species: production of GLA by heterologous expression in yeast and tobacco. Lipids 37:417–426
Sandager L, Gustavsson MH, Stahl U, Dahlqvist A et al (2002) Storage lipid synthesis is non-essential in yeast. J Biol Chem 277:6478–6482
Ochman H, Ajioka JW, Garza D, Hartl DL (1990) Inverse polymerase chain reaction. Biotechnology 8:759–760
Garcia-Maroto F, Garrido-Cardenas JA, Michaelson LV, Napier JA, Alonso DL (2007) Cloning and molecular characterisation of a Delta(8)-sphingolipid-desaturase from Nicotiana tabacum closely related to Delta(6)-acyl-desaturases. Plant Mol Biol 64:241–250
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Rzhetsky A, Nei M (1992) Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J Mol Evol 35:367–375
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Taylor B, Powel A (1982) Isolation of plant DNA and RNA. Focus 4:4–6
Elble R (1992) A simple and efficient procedure for transformation of yeasts. BioTechniques 13:18–20
Rodriguez-Ruiz J, Belarbi EH, Sanchez JLG, Alonso DL (1998) Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol Tech 12:689–691
Alonso DL, Belarbi EL, Rodriguez-Ruiz J, Segura CI, Gimenez A (1998) Acyl lipids of three microalgae. Phytochemistry 47:1473–1481
Kates M (1988) Techniques of lipidology: isolation, analysis, and identification of lipids. Elsevier, Amsterdam
Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166
Kaup MT, Froese CD, Thompson JE (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129:1616–1626
Oh SA, Park JH, Lee GI, Paek KH et al (1997) Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J 12:527–535
John I, Drake R, Farrell A, Cooper W et al (1995) Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J 7:483–490
Athenstaedt K, Daum G (1999) Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem 266:1–16
Griffiths G, Harwood JL (1991) The regulation of triacylglycerol biosynthesis in cocoa (Theobroma cacao) L. Planta 184:279–284
Cao YZ, Huang AH (1987) Acyl coenzyme A preference of diacylglycerol acyltransferase from the maturing seeds of Cuphea, maize, rapeseed, and canola. Plant Physiol 84:762–765
Yu K, Li R, Hatanaka T, Hildebrand D (2008) Cloning and functional analysis of two type 1 diacylglycerol acyltransferases from Vernonia galamensis. Phytochemistry 69:1119–1127
Griffiths G, Stobart AK, Stymne S (1988) Delta 6- and delta 12-desaturase activities and phosphatidic acid formation in microsomal preparations from the developing cotyledons of common borage (Borago officinalis). Biochem J 252:641–647
Cao G, Goldstein JL, Brown MS (1996) Complementation of mutation in acyl-CoA:cholesterol acyltransferase (ACAT) fails to restore sterol regulation in ACAT-defective sterol-resistant hamster cells. J Biol Chem 271:14642–14648
Lewin TM, Wang P, Coleman RA (1999) Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38:5764–5771
Hirst JD, Vieth M, Skolnick J, Brooks CL 3rd (1996) Predicting leucine zipper structures from sequence. Protein Eng 9:657–662
Bornberg-Bauer E, Rivals E, Vingron M (1998) Computational approaches to identify leucine zippers. Nucleic Acids Res 26:2740–2746
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia y Tecnología (MCYT, AGL2005-01498/AGR) and Junta de Andalucía (P05-189-AGR). J.A. Garrido-Cárdenas and A. Mañas-Fernández were recipients of postgraduate fellowships from the MCYT and Junta de Andalucía, respectively. We are also grateful to J. Pérez-Parra and J.C. Gázquez for providing greenhouse facilities and technical assistance in plant culture at the “Estación Experimental Las Palmerillas (CAJAMAR)”. Dr. Stymne (Swedish University of Agricultural Sciences, Uppsala) kindly provided the mutant strain of S. cerevisiae H1246, and is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
11745_2009_3303_MOESM1_ESM.doc
Deduced donor and acceptor splice sites of the EpDGAT gene. Splicing sites were deduced from the comparison between genomic and cDNA sequences. Non-standard acceptor sequence on intron 3 is underlined. (DOC 27 kb)
About this article
Cite this article
Mañas-Fernández, A., Vilches-Ferrón, M., Garrido-Cárdenas, J.A. et al. Cloning and Molecular Characterization of the Acyl-CoA:Diacylglycerol Acyltransferase 1 (DGAT1) Gene from Echium . Lipids 44, 555–568 (2009). https://doi.org/10.1007/s11745-009-3303-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11745-009-3303-9