Abstract
Castor seed triacylglycerols (TAGs) contain 90% ricinoleate (12-hydroxy-oleate) which has numerous industrial applications. Due to the presence of the toxin ricin and potent allergenic 2S albumins in the seed, it is desirable to produce ricinoleate from temperate oilseeds. To identify regulatory genes or genes for enzymes that may up-regulate multiple activities or entire pathways leading to the ricinoleate and TAG synthesis, we have analyzed expression profiles of 12 castor genes involved in fatty acid and TAG synthesis using quantitative reverse transcription-polymerase chain reaction technology. A collection of castor seeds with well-defined developmental stages and morphologies was used to determine the levels of mRNA, ricinoleate and TAG. The synthesis of ricinoleate and TAG occurred when seeds progressed to stages of cellular endosperm development. Concomitantly, most of the genes increased their expression levels, but showed various temporal expression patterns and different maximum inductions ranging from 4- to 43,000-fold. Clustering analysis of the expression data indicated five gene groups with distinct temporal patterns. We identified genes involved in fatty acid biosynthesis and transport that fell into two related clusters with moderate flat-rise or concave-rise patterns, and others that were highly expressed during seed development that displayed either linear-rise or bell-shaped patterns. Castor diacylglycerol acyltransferase 1 was the only gene having a higher expression level in leaf and a declining pattern during cellular endosperm development. The relationships among gene expression, cellular endosperm development and ricinoleate/TAG accumulation are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The castor plant (Ricinus communis) is a perennial shrub mainly cultivated in tropical and subtropical areas of India, China and Brazil as an oilseed crop. Its seeds accumulate 60% oil in the form of triacylglycerol (TAG) that serves as a major energy reserve for seed germination and seedling growth. Castor oil is unique in that 90% of its fatty acid content is ricinoleate, 12-hydroxyoleic acid. The hydroxy group imparts unique chemical and physical properties that make castor oil a vital industrial raw material for numerous products such as cosmetics, paints, coatings, plastics and anti-fungal products [1]. Castor oil is also used as an additive to prevent fuels and lubricants used in aircraft engines from freezing at extremely low temperatures [1], and its replacement for sulfur-based lubricity components in petroleum diesel helps to reduce sulfur emissions [2].
One of the problems associated with castor seeds is that they contain the toxin ricin and hyper-allergenic 2S albumins that are detrimental to growers and processors. Therefore it is highly desirable to develop a safe source for castor oil production. In the past decade scientists have sought to identify key genes responsible for the ricinoleate synthesis in order to develop temperate oilseeds to produce ricinoleate. A castor gene (FAH) for the hydroxylase which is directly responsible for synthesis of ricinoleate was successfully isolated [3], but transgenic expression of the FAH in the model oilseed Arabidopsis produced only 17% ricinoleate [4], which was too low to be useful. There are many examples of transgenic production of other unusual fatty acids by over-expression of a key gene in oilseeds. With the exception of lauric acid and gamma-linolenic acid, the amount of desired fatty acids in the transgenic oilseed has been considerably lower than in the wild species from which the transgene was obtained [5]. These results suggest that expressing a single key gene required for unusual fatty acid biosynthesis in transgenic plants is insufficient to produce a large amount of unusual fatty acids in seed.
Current efforts on metabolic engineering of new oilseeds have been shifted towards searching for additional genes [6] or general transcription factors [7] that may up-regulate multiple activities or entire pathways leading to oil biosynthesis. Therefore, knowledge of the expression of multiple genes and their regulation during castor oil biosynthesis is needed to further understand the regulatory mechanisms controlling castor oil metabolism. In general, castor oil biosynthesis mostly follows the common biosynthetic pathways for fatty acid in the plastid as well as TAG in the endoplasmic reticulum (ER), although there is a modification step for ricinoleate formation in the latter pathway [8]. In this paper, we survey the available castor sequences in National Center for Biotechnology Information (http://www.ncbi.nlm.gov) to identify 12 genes that participate in different steps of the pathways leading to fatty acid and TAG synthesis. We characterize the expression profiles of these genes in developing castor seeds during the time-course of seed development and ricinoleate/TAG accumulation. Additionally, a leaf sample is included as vegetative control and for comparative studies. To further compare the temporal expression patterns among the genes, we perform a clustering analysis that reveals five groups with distinct temporal patterns. Our results provide not only the initial information on promoter activity for each gene, but also a first glimpse of the global patterns of gene expression and regulation, which are critical to metabolic engineering of transgenic oilseeds for safe castor oil production.
Experimental Procedures
Plant Material
Castor (R. communis L.) seeds, PI215769, were obtained from the USDA-Germplasm Resources Information Network, Southern Regional Plant Introduction Station (Griffin, GA). Plants were germinated and grown in a greenhouse at temperatures between 28 °C (day) and 18 °C (night), with supplemental metal halide lighting to provide a 15-h day length (1,000–1,250 μmol m−2 s−1). Mature female flowers were individually pollinated and tagged, and the tagging dates were recorded as 0 day after pollination (0 DAP). Capsules were harvested at 7-day intervals from 12 to 61 DAP. Dissected seeds were frozen immediately in liquid nitrogen and stored at −80 °C. Leaf tissue was collected from a fully expanded young leaf in a mature castor plant.
Isolation of Total RNA and Reverse Transcription
Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA). RNA pellets were dissolved in DEPC-treated water, quantified by absorbance at 260 nm and checked for quality by ethidium bromide gel electrophoresis. The total RNA samples were first treated with DNase I, and then reverse transcribed to first-strand cDNA using an Oligo(dT) primer and SUPERSCRIPT III (Invitrogen, Carlsbad, CA). Finished cDNA products contained 50 ng/μL reverse transcribed total RNA, and were stored at −80 °C.
Primer Design and Standard Curve Construction
Putative primers were designed according to the SYBR Green Design criteria incorporated in the Beacon Designer 4.0 software (Premier Biosoft International Palo Alto, CA). To ensure maximum specificity and efficiency during quantitative PCR, putative primer pairs were further tested for linearity of response by constructing standard curves on five or six serial tenfold dilutions. The templates used for the standard curve analysis were mixed cDNAs from 12 to 54 DAP samples with a starting concentration of 2.5 ng/μL. A standard reaction mixture (25 μL) contained 10 μL cDNA template, 1x iQ SYBR Green I Supermix (Bio-Rad, Hercules, CA) and 300 nM forward and reverse primers. The Bio-Rad iCycler iQ-system was used for all amplifications. The PCR protocol consisted of an initial denaturizing step of 95 °C for 3 min, followed by 40 repeats of 95 °C for 10 s and 55 °C for 30 s. A melt-curve protocol immediately followed the amplification with heating to 95 °C for 1 min and annealing at 50 °C for 1 min, followed by 80 repeats of heating for 10 s, starting at 50 °C with 0.5 °C increments. The Bio-Rad iQ iCycler System software produced standard curves by plotting the log10 of the starting quantity against the threshold cycle (C T). Correlation coefficients were determined from the standard regression formulas. Standard curves showing correlation coefficients of 0.99 or higher and PCR efficiencies between 90 and 110% were accepted. For each primer set, at least three independent standard curves were analyzed, and the average number was used in data analysis. PCR product specificity was confirmed by melting-curve analysis and by electrophoresis on 4% agarose gel to ensure that PCR reactions were free of primer dimers and non-specific amplicons. Information on optimized primer pairs is listed in Table 1.
Quantitative RT-PCR Data Analysis
The method of Pfaffl [9] was applied to calculate comparative expression levels between samples. The castor actin gene (a house-keeping gene, [10]) was used as internal reference to normalize the relative amount of mRNAs for all samples. For each selected gene, triplicate sets of PCR reaction samples including the actin controls, and duplicate negative controls (reaction samples without cDNA templates), were prepared and run in a 96-well plate. The SYBR fluorescence was analyzed by iQ iCycler software and the C T value for each sample was reported. The average C T from 26 DAP measurements were calibrated as 100, 1,000 or 105 copy numbers, and the relative copy numbers of a gene were averaged over triplicates. The PCR experiments were repeated for each plate to ensure that similar results could be obtained.
Data Clustering Analysis and Visualization
To partition the lipid genes into distinct groups such that genes assigned to the same cluster should have similar expression patterns, the quantitative expression data of each gene at various developmental stages were first normalized to the genes maximum expression (set at 100%) and then subjected to gene expression clustering analysis using the k-mean clustering method [11] provided within the software Expression Analyzer and DisplayER (EXPANDER, [12]). The k-mean clustering numbers, 4, 5 and 6 were tested (data not shown) and the clustering into five groups was selected because it gave the best scores for clustering quality. To view the expression patterns of clusters by a graph, the option of mean patterns with error bars operated by the EXPANDER was chosen that allows each cluster to be displayed in a separate panel with error bars representing standard deviations.
Chemicals
Ricinoleic acid methyl ester and cyclohexane were obtained from Sigma–Aldrich (St. Louis, MO). Nonadecanoic acid methyl ester and GLC-68 FAME standard mixture were obtained from Nu-Chek Prep, Inc. (Elysian, MN). Heptadecanoic acid methyl ester and anhydrous acetyl chloride were purchased from Alltech (Deerfield, IL) and butylated hydroxytoluene (BHT) was obtained from Spectrum Chemical MFG Corp. (Gardena, CA). Anhydrous sodium sulfate was purchased from J.T. Baker Inc. (Philipsburg, NJ). 2-Propanol, methanol, hexane and toluene were obtained from Fisher Scientific (Fair Lawn, NJ). Sodium chloride and potassium bicarbonate were obtained from Mallinckrodt Laboratory Chemicals (Philipsburg, NJ). Ethanol was purchased from AAPER Alcohol and Chemical Co. (Shelbyville, KY). The water used was double distilled, and all chemicals and solvents used were of reagent grade.
Lipid Extraction, Seed Dry Weight and Water Content Measurement
Castor seeds were peeled and thoroughly homogenized using a mortar and a pestle. Seed samples were weighed (seed fresh weight) into Eppendorf tubes and dried under vacuum centrifugation over night. The dried samples were weighed and the water content was calculated. Triplicate samples were prepared by accurately weighing 0.01 g of dried seed sample into 10 mL glass tubes. The lipids were extracted using 2 mL of hexane/2-propanol (8:2) containing 50 μg mL−1 of BHT. Internal standard (nonadecanoic acid methyl ester) was added and the extraction took place at 55 °C for 30 min with shaking every 10 min. The extracts were filtered and dried over sodium sulfate, and the solvent was evaporated under nitrogen. The lipid weight was determined gravimetrically. After addition of 0.5 mL toluene, the lipids were methylated for 1 h at 80 °C using methanolic hydrogen chloride (3%), as described by Christie [13]. The resulting FAMEs were dissolved in 10 mL of cyclohexane (0.01% BHT) for GC analysis. The non-lipid dry weight was obtained by subtracting the lipid and water weight from the seed fresh weight.
GC Analysis
Quantitative analysis was carried out by GC-FID using a Hewlett Packard 6890 GC system with split injection connected to a 7673 automatic liquid sampler, Agilent Technologies (Palo Alto, CA). Separation was achieved on a DB-WAX column (20 m × 0.12 mm i.d., 0.18 μm film thickness) purchased from J & W Scientific, Agilent Technologies. The injector and detector temperatures were 250 and 280 °C, respectively. The column temperature program was 100 °C for 1 min, 5 °C min−1 to 250 °C, and hold 1 min. Standard solutions of a mixture of FAMEs including methyl ricinoleate at three different concentrations in the range of 40–400 μg mL−1 for methyl ricinoleate and 5–150 μg mL−1 for the other FAMEs were used for generating standard calibration curves. 50 μL of methyl heptadecanoate (1 mg mL−1) was added as internal standard to each 1-mL aliquot of standard sample. One microlitre injections were used, and duplicate determinations were applied. Identification of peak components was achieved on a Hewlett Packard 5890 GC system connected to a 5970A mass selective detector, Agilent Technologies. Split injection was applied, and the same type of column and temperature program as described above was used. Comparison to mass spectra of known FAMEs was used for identification of each peak. In addition, double-bond locations for the unsaturated fatty acids were determined by interpreting spectra from picolinyl derivatives of free fatty acids (FFAs), employing the methodology described by Christie [13].
Results and Discussion
Changes in Water, Lipid and Ricinoleate Content During Seed Development
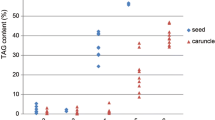
A mature castor seed contains a mass of endosperm that synthesizes and stores oil and protein as major reserves [14]. We have previously characterized the morphological changes of developing castor seeds and established a time-course for assessing endosperm development [15]. Castor endosperm development has a nuclear endosperm development pattern that starts with a free-nuclear stage and then progresses into cellularization and maturation. The whole course takes about 54 DAP. At 61 DAP and after, seeds are completely mature and begin desiccation [15]. During the first 19 DAP, castor seeds grew rapidly to full size ([15], Fig. 1d), and consisted mostly of water at 90% of seed fresh weight and small amounts of lipid at about 5% (Fig. 1a). The endosperm stayed at free-nuclear stage and did not enter cellular endosperm development (Fig. 1d). Between 26 and 40 DAP, the endosperm underwent cellularization, expanding and eventually filling the most of the seed at 40 DAP ([15], Fig. 1d). Then the cellular endosperm continued maturation up to 54 DAP without showing visible changes [15]. During the course of cellular endosperm development (26–54 DAP), the seeds gradually lost water and gained storage lipid to a maximum level of 60% at 54 DAP (Fig. 1a) and maintained the same high level thereafter.
To examine the ricinoleate content, total fatty acid represented by FAME and ricinoleate was measured in seeds at various developmental stages. As shown in Fig. 1b, the ricinoleate and total FAME showed accumulation patterns parallel to each other, and the increase of the FAME was attributed predominantly to the increase of ricinoleate. At early stages (12 and 19 DAP) in free-nuclear endospermic seeds, the ricinoleate was not detectable (Fig. 1c). However by 26 DAP when 30% of seed’s volume filled with cellular endosperm (Fig. 1d), the ricinoleate accumulated immediately to 9.2% of seed dry weight (Fig. 1b) and accounted for 77% of fatty acid content (Fig. 1c). During the remaining stages of cellular endosperm development, the ricinoleate kept increasing and reached plateaus of 55% of seed dry weight at 54 DAP and 90% of total FAMEs at 40 DAP (Fig. 1b, c). Besides ricinoleate, there were about seven minor fatty acids detected in seeds at all stages of the development, all of them accumulated at low background levels in a total amount of 2.8–6.5% of dry seed weight (data not shown). These fatty acids were probably components of structural lipids for maintaining the cell membrane and seed coat, in addition to being minor components of the oil. By using the same sets of developing seeds, similar results were observed by measuring changes in lipid classes, including acylglycerols, FFAs, phosphatidylcholine (PC) and phosphatidylethanolamine (PE) [16]. Before 19 DAP, there were considerable amounts of PC and PE present in seed lipid, and the acylglycerols contain less than 7% TAG. After 26 DAP, the relative amount of PC and PE dropped to negligible levels, and the fatty acids were almost exclusively in acylglycerols of which 77–89% was TAG [16]. It is known that ricinoleate accumulates almost exclusively as storage TAGs rather than as membrane lipids in castor seeds [8], therefore, the appearance of ricinoleate indicates the initiation of storage TAG synthesis. Taken together, the FAMEs measured in seeds before 19 DAP (Fig. 1b) would represent mostly fatty acids from membrane lipids, whereas, after 26 DAP, the FAME are mostly of TAG origin. As shown in Fig. 1, ricinoleate and TAG synthesis started at 26 DAP, coinciding with the beginning of endosperm cellularization. Our previous studies in castor showed increased expression of ricin [17] and 2S albumins [15] genes that also started at 26 DAP. These results all suggest that the time between 19 and 26 DAP is a critical switchover stage for synthesizing both oil and storage proteins in castor. Moreover, these results also demonstrate that our series of developing seeds allows us to identify the initial timing and temporal pattern of different biochemical and cellular activities during seed development and to draw accurate comparisons between experiments.
Selection of Lipid Genes and Optimized PCR Primers
In order to find the lipid genes for castor oil biosynthesis, we searched the Genbank nucleotide database (http://www.ncbi.nlm.gov) by using compound queries each consisting of R. communis and the nomenclature of genes involved in synthesis of fatty acid and TAG from a comprehensive lipid gene catalog [18], which lists 600 genes representing 210 cellular activities involved in various aspects of acyl lipid metabolic pathways including fatty acid and TAG biosynthesis from plant kingdom. We noticed that all hits were for cDNA clones from developing castor seeds, indicating their functional expression during seed lipid biosynthesis. In some cases, more than one accession exists because of multiple partial sequences submitted by different laboratories. We examined the candidate genes by multiple sequence alignment and selected the most complete sequence to represent the gene. The search also found a partial sequence (GenBank ID T15158) containing an enoyl-reductase domain of Type I fatty acid synthase (FAS). This sequence is tentatively designated as FASI-like gene in Table 1.
Quantitative PCR is one of the most sensitive and quantitative methods for measuring mRNA levels [19]. We optimized all assay conditions (“Experimental Procedures”), and the primer sets and PCR efficiencies are listed in Table 1. In addition, PCR products were designed to have similar sizes between 100 and 230 bp (Table 1). These optimized conditions allow simultaneous analysis of multiple genes on a 96-well plate and permit accurate comparison of relative copy numbers among genes.
General Expression Profiles of Lipid Genes and Lipid Gene Clusters
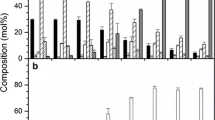
Using the quantitative RT-PCR technology, we examined the steady state mRNA levels of lipid genes in seed samples from various developmental stages. For convenience, these mRNA levels are referred to as ‘expression’ in this article. Expression profiles of the lipid genes are shown in Fig. 2. With the exception of the diacylglycerol acyltransferase 1 (DGAT1), we detected low background expression for the majority of lipid genes in leaf tissue and in young seeds at 12 and 19 DAP when endosperm was at the free-nuclear stage. When the seeds progressed to cellular endosperm development (26–54 DAP), the expression of the majority of lipid genes was induced to higher levels, displaying various temporal patterns; the maximum induction also varied dramatically, ranging from 4- to 43,000-fold (Fig. 2, Table 2). The results revealed that a major transcriptional activation of lipid gene expression occurred at the onset of cellular endosperm development, coinciding with the beginning of the storage TAG accumulation (Fig. 1b). It indicates a primary role of gene transcription in regulating castor oil biosynthesis.
Expression of lipid genes in developing seeds and leaf. Abbreviated names for the genes are described in Table 1. Each data point represents the mean ± SD of three replicates. DAP days after pollination. Values in parentheses indicate relative copy number
To examine relationships among the temporal expression patterns of the lipid genes, we performed clustering analysis (“Experimental Procedures”), which classified the lipid genes into five groups based on their pattern similarities. As expected, the DGAT1 itself formed a cluster showing a specific declining pattern (cluster 1 in Fig. 3a). The majority of the lipid genes showed various up-regulated patterns during seed development including flat-rise (cluster 2), concave-rise (cluster 3), bell-shaped (cluster 4) and linear-rise (cluster 5). The clusters are summarized in Fig. 3 and Table 2, together with their gene members and normalized mean pattern description, the changes in expression (maximum ratio) for each gene as well as the maximum ratio between the developing seed and mature leaf.
Expression patterns of lipid gene clusters during seed development. Abbreviated names are described in the Table 1. Each cluster is represented by the mean expression pattern over all the genes assigned to it. Error bars denote ±SD. DAP days after pollination
Expression Profiles of the Lipid Genes Involved in Fatty Acid Biosynthesis
According to our current knowledge of castor seed TAG synthesis, oleic acid is synthesized in the plastid and then exported to cytoplasm following the standard fatty acid biosynthesis pathway [20]. Oleic acid is activated to oleoyl-CoA in the cytoplasm and imported into ER with help of acyl-CoA binding protein (ACBP) for TAG synthesis [20]. We have examined seven castor lipid genes involved in oleic acid biosynthesis and transport and found they belong to either cluster 2 or cluster 3. Cluster 2 has a flat-rise pattern (Fig. 3b) with four members: acetyl-CoA carboxylase biotin carboxylase subunit (BC), 50 kDa β-ketoacyl-ACP synthase (KASB), enoyl-ACP reductase (EAR), and ACBP; while cluster 3 (Fig. 3c) includes three genes, acyl carrier protein (ACP), 46 kDa β-ketoacyl-ACP synthase (KASA), and stearoyl-acyl-carrier protein desaturase (SAD), all with a concave-rise pattern. Further comparisons among gene clusters reveal that cluster 2 and 3 share certain expression characteristics. First, the maximum induction ratios between seed and leaf in cluster 2 and 3 (4.7 to 28-fold) are much smaller than those in cluster 4 and 5 (18,288 to 252,230-fold). Second, during the time-course of seed development, cluster 2 and 3 have moderate maximum induction ratios (4 to 70-fold) compared to cluster 4 and 5 (940 to 43,083-fold). Third, the temporal patterns of cluster 2 and 3 are less dynamic than those of cluster 4 and 5; cluster 2 has a simple flat-rise, whereas cluster 3 has a quick rise that reaches its half-maximum at the beginning of cellular endosperm development, and increases slowly (<2 fold) during the rest of the seed development. As a consequence of these features, the levels of induction among clusters 2 and 3 at the onset of cellular endosperm development (26–33 DAP) are similar, ranging between 4 to 21 and 11 to 35 fold, respectively. These shared characteristics between clusters 2 and 3 suggest that common regulatory factors might be present to up-regulate the expression of these lipid genes in plastid. Moreover, the appearance of overall moderate expression and related patterns shared by clusters 2 and 3 would also suggest their house-keeping functions in cellular endosperm tissue. There is evidence that fatty acid biosynthesis is a house-keeping function and it undergoes coordinated regulation through gene expression to provide constant demands of fatty acids for membrane lipid biosynthesis, in addition to its involvement in storage TAG assembly. For instance, biochemical data show that individual enzymes required for fatty acid biosynthesis in the plastid may be retained as a functional unit, or a house-keeping metabolon [21]. Genes encoding components of the plastidic acetyl-CoA carboxylase in Arabidopsis [22] and the FAS in Brassica [23] are expressed at constant molar ratios both in different tissues of mature plants and throughout seed development. Furthermore, the microarray analysis of more than 100 Arabidopisis lipid genes demonstrates that 60% of them do not change in expression during the time-course of seed development, including some core enzymes of FAS, such as β-ketoacyl-ACP synthaseIII and isoforms of ACP [24]. In castor seed, cluster 2 and 3 may represent house-keeping genes and their boosted expressions obviously associated with the increasing demand of the oleic acid for TAG biosynthesis during cellular endosperm development. It is possible that cluster 2 and 3 genes also contribute to castor membrane lipid biosynthesis, because we have also observed low expression of cluster 2 and 3 in non-storage tissues such as leaf and young seeds at 12 and 19 DAP (Fig. 2). The low expression of cluster 2 and 3 in the vegetative tissues would correlate with the low demand for fatty acid in membrane lipid synthesis. Similar results were reported for the castor SAD gene expression using Northern blot analysis, showing highest expression in developing seed, moderate to low expression in leaf and low expression in root [25]. So probably there are two different mechanisms existing in castor to regulate the expression of genes involved in fatty acid biosynthesis, one at low levels for membrane function and the other at higher levels for reserve synthesis. Without castor whole genome information, other alternatives also exist, for example, isoforms of cluster 2 and 3 may contribute to fatty acid biosynthesis in different tissues or at different developmental stages.
Expression Profiles of Castor Oleosin Genes
The clustering analysis groups two oleosin genes into cluster 4 showing a bell-shaped pattern (Fig. 3d). Oleosins are oil body-associated proteins that cover and stabilize oil bodies during TAG accumulation [26]. In most plants oleosins are encoded by multi-gene families and some of them are co-expressed [26]. We have observed that castor oleosin1 and oleosin2 genes were co-expressed during seed development with maximum induction ratios of 24,172 and 27,150-fold, respectively (Fig. 2k, l; Table 2). The oleosin proteins have been shown to accumulate in equal amounts [27] or different amounts with the oleosin1 at a higher level [10]. The difference in relative protein level between oleosin1 and oleosin2 could be due to genotype discrepancy among castor cultivars and/or post-transcriptional regulation involvement in oleosin protein accumulation. However our time-course data showed peaks at 40 DAP, a time point when cellular endosperm reached its full expansion (Fig. 1d). The peak expression of oleosins is probably driven by the demand for newly synthesized oil bodies resulting from the growing ER membranes during cellular endosperm expansion.
Expression Profiles of Castor FAH and FASI-Like Genes
The FAH and FASI-like genes are grouped into cluster 5 and display a linear rise pattern during seed development (Fig. 3e). As the enzyme responsible for ricinoleate biosynthesis, the FAH hydroxylates the sn-2 oleoyl chain of oleoyl PC to form sn-2 ricinoleate-PC [8]. The expression of the FAH gene was reported to be high in seed and very low in leaf [3]. Our data on the FAH gene expression is consistent with the previous report, but with new quantitative and detailed temporal expression profiles. In leaf and young seeds at free-nuclear endosperm stages (12 and 19 DAP), FAH is expressed at negligible levels (Fig. 2i). FAH is expressed at a high level when seeds progress into cellular endosperm development and storage TAG/ricinoleate biosynthesis during 26–54 DAP (Fig. 1b), showing an immediate 5,000-fold induction at the beginning (26 DAP, Fig. 2i) and continuous sharp increases during the rest of the stages. As a consequence, the FAH expression showed a linear-rise pattern and a maximum induction of 43,083-fold at the end of the seed development (54 DAP, Fig. 2i; Table 2). These results would indicate that the FAH gene is transcriptionally expressed at similar high rates throughout the cellular endosperm development and, at the same time, the FAH transcripts are probably also very stable in cellular endosperm, resulting in its high accumulation. Furthermore, such high accumulation of the FAH gene transcript would suggest transcriptional regulation is the mechanism for maintaining high ricinoleate synthesis to fulfill the growing demand of TAG synthesis in developing cellular endosperm.
The function of the FASI-like gene remains unknown due to its incomplete sequence information. When searching the Genbank protein database with the truncate FASI-like protein sequence as a query, it retrieved a plant sequence encoding phenylpropenal double-bond reductase from Pinus taeda (Genbank accession: DQ829775.1) among five top hits (alignment score from 60 to 62). Although this enzyme is involved in phenylpropanoid metabolism for plant defense [28], it also shows sequence similarity to enzymes involved in plant lipid metabolism [28]: the enoyl acyl carrier protein reductase (Genbank accession: CAC41367) from Brassica napus and the 2-alkenal reductase (Genbank accession: At5g16970) from Arabidopsis. The castor FASI-like gene was highly expressed during seed development and oil synthesis, showing a maximum induction of 940-fold and a similar linear-rise pattern as that of FAH (Fig. 3e; Table 2). The results reveal intriguing information on the FASI-like gene, which should lead to future investigations on its enzymatic and cellular function during castor seed development.
Expression Profile of Castor DGAT1
Diacylglycerol acyltransferase catalyzes the final step in TAG synthesis that acylates diacylglycerol (DAG) with acyl-CoA to form TAG [29]. Castor DGAT1 has been shown to preferentially incorporate ricinoleate into diricinolein to form triricinolein [30]. Among the lipid genes examined, the DGAT1 is the only member in cluster 1 displaying a declining pattern. During seed development, the DGAT1 mRNA accumulated to highest level at early stages (19–26 DAP, Fig. 2j) and then decreased to lower levels over time (33–54 DAP), showing a maximum of 2.4-fold reduction from 26 to 54 DAP. The temporal pattern of DGAT1 (Fig. 3a) is in agreement with previous Northern data [16] and quantitative RT-PCR data [31]. However the DGAT1 protein level and enzymatic activity during seed development increased at 26 DAP, reached a maximum at 40 DAP, and both protein and activity were at least 50% maximum level from 26 to 54 DAP [16], consistent with the pattern of TAG synthesis (Fig. 1a). The shifted pattern of DGAT1 protein/activity in comparison to its mRNAs was interpreted as post-transcriptional regulation contributing in part to the mechanism of TAG synthesis [16]. Since we observed the onset of cellular endosperm development that also started at 26 DAP (Fig. 1d), it is possible that the cellular endosperm plays a role in regulating the DGAT1 protein/activity by providing a micro-environment for its cellular function. The DGAT1-involved TAG synthesis takes place mainly in developing seed and pollen as storage reserves [29], but it also occurs in various vegetative tissues of Arabidopsis, such as root, stem, senescing leaf, flower and silique [32, 33]. We found that the DGAT1 mRNA in leaf tissue was higher than that in developing seeds (Fig. 2j; Table 2). Also, in cotyledons and true leaves of germinating castor seedlings, the DGAT1 mRNA, protein and enzyme activity were found to be expressed and temporally associated with increased levels of newly synthesized TAG [34]. These results suggest that DGAT1 may also play a regulatory role in balancing DAG and acyl-CoA molecules in membrane lipid metabolism to maintain membrane function in vegetative tissue. Besides DGAT1, there are at least other two independent reactions contributing to the final step of TAG synthesis in castor. A castor DGAT2 gene which has little amino sequence homology to DGAT1 but can also use diricinolein as acceptor and ricinoleoyl-CoA as the donor in TAG synthesis has been identified recently ([31], our unpublished data). Studies show that the DGAT2 displays a bell-shaped temporal pattern during seed development and is more highly expressed in seeds than in leaves ([31], our unpublished data). A castor enzyme, phospholipid:diacylglycerol acyltransferase (PDAT), has also been shown to contribute to TAG synthesis by acyl-CoA-independent acylation using ricinoleoyl-PC as acyl donors and ricinoleoyl-DAG as acceptors [35]. More independent gene families, such as bifunctional wax ester synthase/DGAT [36] and cytosolic DGAT [37] have been identified to catalyze the formation of TAG. It is possible that these gene families also participate in the final step in TAG synthesis in castor. Taken together, the final step in TAG synthesis in castor and other oilseeds is complicated and the question remains as to which of these genes and at what developmental stage do they play more or less important roles in TAG synthesis.
Overall, our series of seed studies, including endosperm development, storage TAG accumulation and gene expression, are all in parallel with the same time-course, which provides integrative information for understanding their relationships during castor seed development. It is evident that synthesis of TAG requires coordinated regulation controlled by seed developmental programs at multiple levels, such as endosperm cellularization, gene transcription and post-transcriptional regulation. Obviously, gene transcription exerts a primary control in castor oil biosynthesis. The clustering analysis on temporal expression of lipid genes reveals five distinct patterns, indicative of different transcriptional regulatory mechanisms among these lipid genes. Our results of analysis of gene expression represent snapshots of a global transcriptional activities taking place during castor seed development and oil biosynthesis.
Abbreviations
- ACP:
-
Acyl-carrier protein
- ACBP:
-
Acyl-CoA binding protein
- BC:
-
Biotin carboxylase
- DAP:
-
Day after pollination
- DGAT1:
-
Diacylglycerol acyltransferase 1
- ER:
-
Endoplasmic reticulum
- EAR:
-
Enoyl-ACP reductase
- FAH:
-
Fatty acid hydroxylase
- FAME:
-
Fatty acid methyl ester
- FAS:
-
Fatty acid synthase
- KASA:
-
46 kDa β-ketoacyl-ACP synthase
- KASB:
-
50 kDa ß-ketoacyl-ACP synthase
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- RT-PCR:
-
Reverse transcription-polymerase chain reaction
- SAD:
-
Stearoyl-acyl-carrier protein desaturase
- TAG:
-
Triacylglycerol
References
Caupin HJ (1997) Products from castor oil: past, present, and future. In: Gunstone FD, Padley FB (eds) Lipid technologies and applications. Marcel Dekker, New York, pp 787–795
Goodrum JW, Geller DP (2005) Influence of fatty acid methyl esters from hydroxylated vegetable oils on diesel fuel lubricity. Bioresour Technol 96:851–855
Van De Loo F, Broun P, Turner S, Somerville C (1995) An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc Natl Acad Sci USA 92:6743–6747
Broun P, Somerville C (1997) Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol 113:933–942
Thelen JJ, Ohlrogge JB (2002) Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng 4:12–21
Lu C, Fulda M, Wallis JG, Browse J (2006) A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J 45:847–856
Cernac A, Benning C (2004) WRINKLED 1 encodes and AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40:575–585
Bafor M, Smith M, Jonsson L, Stobart K, Stymne S (1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280:507–514
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007
Eastmond PJ (2004) Cloning and characterization of the acid lipase from castor beans. J Biol Chem 279:45540–45545
Herwig R, Poustka A, Muller C, Bull C, Lehrach H, O’Brien J (1999) Large scale clustering of cDNA-fingerprinting data, Genome Res 9:1093–1105
Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R (2005) EXPANDER—an integrative program suite for microarray data analysis. BMC Bioinformatics 6:232
Christie WW (1989) The preparation of derivatives of fatty acids. In: Christie WW (ed) Gas chromatography and lipids: a practical guide. The Oily Press Ltd, Dundee, pp 66–84
Roberts LM, Lord JM (1981) Protein biosynthetic capacity in the endosperm tissue of ripening castor bean seeds. Planta 152:420–427
Chen GQ, He X, Liao LP, McKeon TA (2004) 2S albumin gene expression in castor plant (Ricinus communis). J Am Oil Chem Soc 81:867–872
He X, Chen GC, Lin JT, McKeon TA (2004) Regulation of diacylglycerol acyltransferase in developing seeds in castor. Lipids 39:865–871
Chen GQ, He X, McKeon TA (2005) A simple and sensitive assay for distinguishing the expression of Ricin and Ricinus communis agglutinin genes, in developing castor seed (Ricinus communis L.) J Agric Food Chem 53:2358–2361
Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, Mhaske VB, Cho Y, Ohlrogge JB (2003) Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and web-based database. Plant Physiol 132:681–697
Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HRH (2005) Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn 5:209–219
Somerville C, Browse J, Jaworski JG, Ohlrogge JB (2000) Lipids. In: Buchanan B, Gruissem W, Jones R (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 456–527
Roughan PG, Ohlrogge JB (1996) Evidence that isolated chloroplasts contain an integrated lipid-synthesizing assembly that channels acetate into long-chain fatty acids. Plant Physiol 110:1239–1247
Ke J, Wen TN, Nikolau BJ, Wurtele ES (2000) Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetlyl-coenzyme a carboxylase, Plant Physiol 122:1057–1071
O’Hara P, Slabas AR, Fawcett T (2002) Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol 129:310–320
Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14:1191–1206
Shanklin J, Somerville C (1991) Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA 88:2510–2514
Murphy DJ (2005) Lipid-associated proteins. In: Murphy DJ (ed) Plant lipids: biology, utilisation and manipulation. CRC Press, Florida, pp 226–269
Sadeghipour HR, Bhatla SC (2002) Differential sensitivity of oleosins to proteolysis during oil body mobilization in sunflower seedlings. Plant Cell Physiol 43:1117–1126
Kasahara H, Jiao Y, Bedgar DL, Kim SJ, Patten AM, Xia ZQ, Davin LB, Lewis NG (2006) Pinus taeda phenylpropenal double-bond reductase: purification, cDNA cloning, heterologous expression in Escherichia coli, and subcellular localization in P. taeda. Phytochemistry 67:1765–1780
Weselake RJ (2005) Storage lipids. In: Murphy DJ (ed) Plant lipids: biology, utilisation and manipulation. CRC Press, Florida, pp 162–225
He X, Turner C, Chen GQ, Lin JT, McKeon TA (2004) Cloning and characterization of a cDNA encoding diacylglycerol acyltransferase from castor bean. Lipids 39:311–318
Kroon JTM, Wei W, Simon WJ, Slabas AR (2006) Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67:2541–2549
Kaup MT, Froese CD, Thompson JE (2002) A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol 129:1616–1626
Lu CL, Bayon de Noyer S, Hobbs DH, Kang J, Wen Y, Krachtus D, Hills MJ (2003) Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol Biol 52:31–41
He X, Chen GQ, Lin JT, McKeon TA (2006) Diacylglycerol acyltransferase activity and triacylglycerol synthesis in germinating castor seed cotyledons. Lipids 41:281–285
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyses the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA 97:6487–6492
Kalscheuer R, Steinbuchel A (2003) A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem 278:8075–8082
Saha S, Enugutti B, Rajakumari S, Rajasekharan R (2006) Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol 141:1533–1543
Acknowledgments
The authors thank Susan Altenbach, Glenn Bartley and Paul Roessler for critical reading of the manuscript. This work was supported in part by the USDA-ARS Current Research Information System (CRIS) Project 5325-2100-012-00D, USDA Initiative for Future Agriculture and Food System Grant No. 2000-04820 and a cooperative agreement (No. 58-3K95-2-918) with Dow Chemical Company (Midland, MI).
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Chen, G.Q., Turner, C., He, X. et al. Expression Profiles of Genes Involved in Fatty Acid and Triacylglycerol Synthesis in Castor Bean (Ricinus communis L.). Lipids 42, 263–274 (2007). https://doi.org/10.1007/s11745-007-3022-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11745-007-3022-z