Abstract
Salt overly sensitive (SOS) pathway genes, SOS1 (plasma membrane Na+/H+ antiporter), SOS2 (CBL interacting protein kinase 24), and SOS3 (calcineurin B like protein 4) are associated with active efflux of toxic sodium ions (Na+) from cytosol and thus confer salinity tolerance in glycophytic plants such as Arabidopsis. The role of SOS pathway genes SOS2 and SOS3 in salinity tolerance of wheat is rarely studied. One-month-old seedlings of three bread wheat genotypes namely, HD 2009, HD2687 and Kharchia 65 were imposed with two levels of salinity stress (100 and 200 mM NaCl) for 30 days duration. Based on the physiological parameters, genotype Kharchia 65 was highly tolerant, HD 2009 was moderately tolerant and HD 2687 was sensitive to salinity stress. Tolerant genotypes accumulated lesser amount of Na+ in root, stem and leaf tissues. Transcript abundance of SOS1, SOS2 and SOS3 genes was significantly higher in salt tolerant genotypes under long-term salinity and correlated with improved sodium exclusion, and higher potassium/sodium (K+/Na+) ratio. Expression levels of genes involved in vacuolar partitioning of Na+, NHX1 (vacuolar Na+/H+ antiporter) and VP1 (Vacuolar pyrophosphatase) were also higher in salt tolerant wheat genotypes under 200 mM NaCl stress. Partial coding sequences of SOS1, SOS2, SOS3, NHX1 and VP1 genes were cloned and sequenced from the above mentioned three wheat genotypes. The results in the present study demonstrated that SOS pathway of ion homeostasis under salinity stress is conserved across species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity is a problem affecting approximately 20 % of the irrigated agricultural land and about 10 million ha arable land is abandoned yearly due to excessive salt accumulation (Szabolcs 1989). Development of salinity tolerant, high yielding genotypes requires thorough understanding of traits associated with salinity tolerance (Munns and Tester 2008). Most potent mechanism underlying the salinity tolerance is restriction of the entry of sodium ions (Na+) into the cytosol by selective regulation of Na+ uptake and efflux systems (Zhu 2003; Ji et al. 2013). Due to similarities in ionic features, Na+ compete with and are taken up through potassium ions (K+) uptake systems (Plett and Moller 2010). Due to the negative effect of Na+ on K+ nutrition, salinity stress leads to inhibition of photosynthesis, enzyme activities and growth of plants (Zhu 2003). To reduce cytosolic Na+ levels, plants activate Na+ efflux from roots and sequester Na+ into vacuoles (Plett and Moller 2010). Bread wheat exhibits lower net Na+ uptake possibly by counter-balancing the high Na+ influx rate by an efficient Na+ efflux mechanism (Davenport et al. 2005). The Salt Overly Sensitive 1 (SOS1), a plasma membrane Na+/H+ antiporter is the first well-characterized Na+ efflux protein in plants (Shi et al. 2000). SOS1 has been characterized in different plant species including Arabidopsis thaliana (Shi et al. 2000), Thellungiella salsuginea (Oh et al. 2009), Solanum lycopersicum (Olías et al. 2009) and Oryza sativa (Martinez-Atienza et al. 2007). SOS1 mediates Na+ efflux at the root surface and regulates Na+ transport from root to shoot (Tester and Davenport 2003) thereby maintaining favorable K+/Na+ ratio in leaves, where the major metabolic activities of plants take place. SOS1 is activated through protein phosphorylation by the SOS2/SOS3 kinase complex (Pardo et al. 2006). SOS2 is a serine–threonine protein kinase, belonging to the sucrose non-fermenting 1-related kinase 3 (SnRK3) family (Liu et al. 2000). SOS3 is a Ca2+ sensor belonging to calcineurin B-like (CBL) protein family, highly similar to neuronal calcium sensor of animals (Kolukisaoglu et al. 2004). SOS3 upon sensing the salinity stress-induced calcium oscillations binds to FISL motif of SOS2 and activates SOS2. The SOS3/SOS2 complex controls the expression of SOS1 gene and also activates the Na+/H+ antiport activity of SOS1 (Sanchez-Barrena et al. 2005). Mutants deficient in both SOS2 and SOS3 exhibits salt-sensitive phenotype similar to sos1 plants (Zhu 2001).
Na+ sequestration into the vacuole depends on expression and activity of Na+/H+ antiporters (NHX), vacuolar type H+-ATPases and H+-pyrophosphatases (VP). The proton pumps generate the necessary proton motive force required for activity of NHX (Gaxiola et al. 2001). Transgenic Arabidopsis plants over-expressing a vacuolar H+-pyrophosphatase exhibited increased partitioning of Na+ into the vacuole and maintained higher relative water content in leaves and improved salinity stress tolerance (Gaxiola et al. 1999). The NHX1 gene is known to be induced, both by salinity and ABA in Arabidopsis (Shi and Zhu 2002) and rice (Fukuda et al. 2004).
Even though active efflux of Na+ is one of the important mechanisms of salinity tolerance, SOS pathway has not yet been experimentally established in wheat. Although SOS1 homologues have been identified and functionally characterized in durum (Xu et al. 2008) and bread wheat (Cuin et al. 2011), none of these studies examined the whole SOS pathway including SOS2 and SOS3 genes. Ramezani et al. (2013) studied the expression of SOS1 and SOS4 genes in cultivated and wild wheat plants and concluded that all the three alleles of SOS1 gene are active in their function. In present study we aimed to examine the expression of all the genes of SOS pathway in bread wheat genotypes with contrasting salinity tolerance and to analyze their association with salinity tolerance.
Materials and methods
Plant growth conditions and stress treatments
Three bread wheat (Triticum aestivum L.) genotypes viz., Kharchia 65, HD 2009 and HD 2687 were sown in 30 cm earthen pots filled with clay loam soil and farmyard manure in 3:1 ratio, pots were lined with 2-layers of 400 gauge polythene sheets to avoid leakage of the salt solution. Pots were fertilized with recommended dose of N, P, and K. Two different levels of salinity treatments i.e. 100 and 200 mM NaCl (2.5 liter of NaCl solution per pot containing 10 kg soil)were given to the plants at 25 days after sowing(DAS), while control plants were supplied with 2.5 liter of water. To avoid sudden osmotic shock to plants, salt solutions were supplied as batches of 0.5 liter per pot per day, finally reaching the desired concentration at 30 DAS. Scheduled routine of irrigation was practiced for both control and treated pots throughout the crop growth period. Each treatment was replicated 20 times. Soil samples were collected at three stages viz. 30DAS, 45 DAS and 60 DAS, and mean electrical conductivity of extract (ECe) of three stages were taken as the actual salinity level, viz, 1.21, 8.43 and 13.04 dS m−1 for control (C), 100 mM NaCl (S1) and 200 mM NaCl (S2), respectively.
Physiological and growth parameters
Physiological observations were taken at 60DAS. Upper most expanded leaf (third leaf from top) sampled in duplicate from 3 pots were used for recording observations. Relative water content (RWC) and membrane stability index (MSI), were estimated using standard protocols (Sairam et al. 1997; Weatherley 1950). For estimating leaf area, all the green leaves were sampled from two plants each from three pots and area was determined using leaf area meter, LICOR-3000 (Li-COR Ltd., Lincoln, Nebraska, USA). Roots were thoroughly washed and used for 2D root image analysis using Win Rhizo (Regent Instruments Inc., Canada).
Determination of potassium and sodium contents
For ion estimations, two plants each were harvested from 3 different pots and separated into leaves, stem and roots. The plant samples were dried and a representative ground plant sample (0.5 g) was digested in a di-acid mixture (20 ml) containing HNO3 and HClO4 acid (9:4) on digestion unit (Turbotherm, Gerhardt analytical systems, Germany) following the established procedure (Tandon 1995). The digested material was cooled, diluted with distilled water and filtered through Whatman No. 42 filter paper. The volume was made up to 25 ml and stored in a polypropylene container. The K+ and Na+ content in the standard solutions, leaf, stem, and root samples were estimated using flame photometer (Systronics FF128).
RNA extraction and semi quantitative RT-PCR
For gene expression analysis, leaf and root samples were collected 24 h after the desired level of salinity concentration was achieved, i.e., on 31DAS. Third leaf from top was collected from two plants each of 4 different pots. Similarly roots were collected after thoroughly washing the roots in running tap water. Total RNA was extracted from leaf and root tissues using RNAeasy plant minikit (Qiagen Inc., Chatsworth CA 91311, USA, Cat No: 749040). DNA contamination from the RNA samples was removed using on-column digestion with DNase I (Qiagen Science, Maryland, USA). For RT-PCR expression analysis and cloning of cDNAs, wheat homologues of the SOS pathway genes identified based on the sequence similarity to O. sativa SOS genes. Homologs in wheat genome database were identified by tBLASTn (wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST) with default parameters, and further verified by updated expressed sequence tags (ESTs) or/and full-length cDNAs (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and tentative consensus sequences (TC) at DFCI gene index (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=wheat). The chromosomal localization and physical position in bacterial artificial chromosome (BAC) were analyzed on the basis of wheat high throughput genomic sequences (urgi.versailles.inra.fr/Seq-Repository/BLAST). Due to the hexaploid genome of wheat all of the genes are having copies in A, B and D genomes. Primers were designed from highly conserved regions of the sequence to amplify all the possible alleles of the genes. Primer sequences of the genes are given in Online Resource 1. One µg of total RNA was reverse transcribed using Qiagen one-step RT-PCR kit on QN 96 Thermal cycler (Quanta biotech, England). Linear amplification for RT-PCR was obtained with 27 cycles and products were electrophoresed on 1.2 % agarose gel at 120 volts in TBE buffer (0.4 M Tris-borate, 0.001 M EDTA, pH 8.0) along with known concentration DNA ladders. Gels were stained with ethidium bromide and visualized on AlphaImager® imaging system (Protien simple, USA). Actin was used as internal standard. The electrophoretic bands were quantified using AlphaView® Software of AlphaImager® imaging system, and the gene/actin ratio was calculated by dividing the value of candidate gene bands by that of actin in each sample.
Sequence analysis of SOS pathway genes
To confirm the specificity of amplification, RT-PCR products were fractionated on agarose gel and purified. The purified cDNAs for each gene were cloned into pDrive vector and transformed into Escherichia coli (strain DH5α) cells. Colonies with recombinant plasmids were identified by blue-white screening and the presence of recombinant plasmids were confirmed by restriction analysis. Recombinant plasmids were isolated, purified and sequenced (Chromous Biotech Pvt. Ltd., Bengaluru, India). All the partial ESTs were submitted to GenBank. To study the conservation of genes across genera, ClustalW2.1 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) multiple sequence alignment of wheat ESTs were done along with corresponding sequences from model organisms. Deduced protein sequences were analyzed for the presence of conserved domains using protein blast (http://blast.ncbi.nlm.nih.gov/) with default parameters.
Statistical analyses
The experiment was conducted following factorial CRD design. Data presented are means of 6 observations (n = 6) for physiological estimations. F test was carried out to test the significance of the treatment differences, and the least significant differences (LSD) were computed to test the significance of the different treatments at 5 % level of probability by using SPSS 16.0. Mean separation by Duncan’s multiple range tests (DMRT) at P < 0.01 was done using SAS® vers. 9.3 (SAS Institute, Cary, NC, USA).
Results
Physiological parameters
Wheat genotypes utilized in the present study were selected from a set of 100 genotypes (data not shown) based on germination and seedling growth assay under salinity stress (100 mM NaCl). The contrasting genotypes, tolerant (Kharchia 65), moderately tolerant (HD 2009) and sensitive (HD 2687) were selected and further evaluated for their response to salinity stress in pot culture growth conditions during rabi season in New Delhi, India. RWC of leaves were measured after 30 days of stress imposition. Salinity significantly reduced RWC of plants (Table 1). Kharchia 65 (salinity tolerant genotype) showed lowest decline in RWC as compared with control. It maintained significantly higher RWC as compared to HD 2687 (salinity sensitive genotype). MSI values of all the three genotypes also showed significant decrease in response to salinity stress (Table 1). The mean percentage reduction over control was 19 and 23.8 % under S1 and S2 salinity levels, respectively. Salinity tolerant and moderately tolerant genotypes exhibited significantly higher MSI values than the sensitive genotype (Table 1).
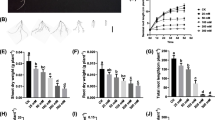
Salinity stress drastically reduced leaf area of all the genotypes (Table 2) as indicated by the mean reduction (58 %) in leaf area of all the genotypes under S2 treatment. Salinity tolerant genotype exhibited lowest reduction in leaf area (36 %), whereas sensitive genotype showed severe reduction in total leaf area (74 %) under S2 level. Severe reduction in root surface area was observed under S2 stress level in all the genotypes (Table 2). Salinity tolerant genotype maintained significantly higher root surface area under both S1 and S2 level of stresses as compared to other genotypes. Similarly maximum root length was also observed in salinity tolerant genotype followed by moderately tolerant genotype (Table 2).
Sodium and potassium contents and K+/Na+ ratio in different plant parts
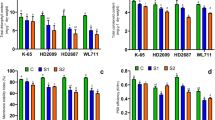
Salinity stress resulted in excessive accumulation of Na+ in different plant parts such as leaves, stem and roots of all the wheat genotypes (Fig. 1a). In stem and roots, Na+ content increased under both S1 and S2 salinity treatments. In stem, highest level of Na+ content was found in sensitive genotype both under S1 and S2 treatments. Among the plant parts, Na+ accumulation was highest in roots in all the genotypes. In roots, 0.7- to 2-fold increase in Na+ content was observed in tolerant genotype, but other genotypes showed 1.0- to 3.4-fold increase in Na+ content.
Effect of salinity stress on sodium (a) and potassium (b) contents and potassium sodium ratio (c) of leaf, stem, and root tissues of wheat genotypes. Plants were subjected to three treatments: normal irrigation (control); treatment with 100 mM NaCl solution; treatment with 200 mM NaCl solution. Vertical bars represent standard error. Within each tissue type, different letters indicate significant differences between the means (P ≤ 0.01)
Salinity stress lead to decrease in K+ content, irrespective of type of tissues and genotypes (Fig. 1b). The average decrease in salinity tolerant genotype was much lesser than other genotypes. While moderately tolerant genotype showed medium levels of K+ content, sensitive genotype showed very low levels of K+ under salinity stress. In stem and roots also K+ content decreased under both S1 and S2 salinity treatments. Salinity tolerant genotype maintained the highest level of K+ content in stem and roots under both S1 and S2 salinity treatments.
Salinity stress decreased potassium/sodium (K+/Na+) ratio in leaves, stem and roots in all the genotypes (Fig. 1c). Salinity tolerant genotype maintained highest K+/Na+ ratio in leaves under salinity treatment. Similar trend was observed in stem and root tissues also. Salinity tolerant genotype was able to reduce net Na+ uptake and sustain K+ uptake, and thus exhibited favorable K+/Na+ ratio in all the tissues.
Effect of salinity stress on expression of SOS pathway genes
To elucidate the role of SOS pathway, expression of SOS1, SOS2 and SOS3 genes were studied in leaves and roots of salinity tolerant and sensitive genotypes. Both constitutive and salinity induced expression of SOS1 gene was approximately 2 fold higher in leaf tissues of salinity tolerant genotype (Fig. 2a). Both moderately tolerant and sensitive genotypes also showed salinity induced expression SOS1 gene in leaf tissue, but the level of expression was significantly lower than tolerant genotype. Up-regulation of SOS pathway genes in root tissue was very high, 100- to 750-fold higher in tolerant genotype as compared to 10- to 50-fold increase in leaf tissue. In root tissues, salinity induced up regulation of SOS1 was observed only in tolerant genotype and expression increased with the increase in salinity levels (Fig. 2b), while other genotypes showed little or undetectable expression of SOS1 under control and salinity stress conditions.
Effect of salinity stress on transcript abundance of SOS1 (a) SOS2 (b) and SOS3 (c) genes in leaf and root tissues of wheat genotypes. Plants were subjected to three treatments: normal irrigation (control); treatment with 100 mM NaCl solution; treatment with 200 mM NaCl solution. Vertical bars represent standard error. Within each tissue type, different letters indicate significant differences between the means (P ≤ 0.01)
In leaves, stress-induced SOS2 gene expression was observed in tolerant and moderately tolerant genotypes, while significantly lower expression was detected in sensitive genotype (Fig. 2c). In root tissues, tolerant genotype showed significant salinity induced up regulation of SOS2 gene at both salinity levels, while moderately tolerant genotype showed expression only at S2 level. Similar to SOS1 gene expression, transcript abundance of the SOS2 gene increased with increase in salinity levels. Sensitive genotype showed minimal expression under control and stress conditions (Fig. 2c).
Constitutive as well as salinity stress-induced expression of SOS3 gene in leaf tissue was highest in tolerant genotype. Moderately tolerant and sensitive genotypes also exhibited stress induced over expression of SOS3 gene in leaves (Fig. 2c). Similar to SOS1 gene expression, only tolerant genotype showed expression of SOS3 gene in of root tissues (Fig. 2c).
VP1 gene expression in leaf tissue was down-regulated by salinity stress in all the three genotypes; however, the reduction in gene expression was lowest in salinity tolerant and moderately tolerant wheat genotypes (Fig. 3). NHX1gene expression was induced by salinity stress in all the wheat genotypes. Salinity tolerant and moderately tolerant wheat genotypes showed very high salinity induced expression of NHX1, while comparatively low gene expression was observed in sensitive genotype.
Effect of salinity stress on transcript abundance of VP1 and NHX1 genes in leaf tissue of wheat genotypes. Plants were subjected to three treatments: normal irrigation (control); treatment with 100 mM NaCl solution; treatment with 200 mM NaCl solution. Vertical bars represent standard error. Within each tissue type, different letters indicate significant differences between the means (P ≤ 0.01)
Partial coding sequences of SOS1, SOS2, SOS3, VP1 and NHX1 were cloned and sequenced from all the 3 genotypes and were submitted to GenBank (Online Resource 2). Comparison of deduced partial protein sequences of these genes revealed high levels of similarity to their homologs in rice and A. thaliana (Online Resource 3). The partial protein sequence showed a conserved domain: sodium/hydrogen exchanger 3 (amino acid residues 1–126), Monovalent Cation: Proton Antiporter-1 (CPA1) Family (TC 2.A.36). Transmembrane domain prediction revealed the presence of 2 transmembrane (TM) spans in the partial protein sequences (Online Resource 2).The partial sequence showed two conserved domains: (i) cd05123 (amino acid residues 1–78), catalytic domain of serine/threonine kinases (STKs) (ii) pfam03822 (amino acid residues 120–178)—NAF domain-a conserved protein–protein interaction module found in Ca2+-regulated kinases. Blast analysis indicated the presence of a conserved domain: cd00051: amino acid residues 19–89: EFh, the calcium binding EF-hand. The VP1 partial protein sequence showed a conserved domain: (amino acid residues 1–218) H+-translocating inorganic pyrophosphatase. Topology prediction using TMHMM program predicted the presence of 3 TM spans in the partial protein sequence. The NHX1 deduced partial protein sequences of wheat genotypes showed a conserved domain: (amino acid residues 1–173) TIGR00840: b_cpa1 sodium/hydrogen exchanger 3, the Monovalent Cation: Proton Antiporter-1 (CPA1) Family (TC 2.A.36). Topology prediction using TMHMM program predicted presence of 4 TM spans in the partial protein sequences (Online Resource 3).
Discussion
High genetic variability exists for salinity tolerance in wheat gene pool. Kharchia 65 is well-known salinity tolerant bread wheat genotype developed in India (Hollington 2000). However, the role of SOS pathway in salinity tolerance of Kharchia 65 has not been examined before. Further, the relationship between SOS pathway expression and Na+ exclusion also has not been established in wheat. Hence in this study, expression levels of SOS pathway genes and salinity tolerance were examined in wheat genotypes with contrasting salinity tolerance.
The presence of salt in the root zone reduces soil water potential, which leads to osmotic stress, affects water uptake and retards growth of crop plants (Chinnusamy et al. 2005). Salinity induced decline in RWC was significantly lesser in tolerant genotype as compared to moderately tolerant and sensitive genotypes (Table 1). Maintenance of higher RWC (%) by tolerant genotype indicates either sustenance of water uptake and/or minimization of transpirational water keeping optimum leaf turgor. It is deduced from the present results that, better root growth of salinity stressed plants of tolerant genotype (Table 2) might have resulted in better water relations.
Membrane stability index (MSI) is one of the important physiological traits suitable for screening of drought tolerance in various crops including wheat (Sairam et al. 2001, Iseki et al. 2014). Sairam et al. (2002) have used MSI and RWC as key parameters to differentiate wheat genotypes differing in salinity tolerance. MSI exhibited a positive correlation with osmotic potential, K+ concentration, osmotic adjustment, and/or RWC under salinity stress (Munns 2002). Salinity induced decline in MSI was significantly less in tolerant genotype followed by moderately tolerant genotypes under salinity stress as compared to sensitive genotypes (Table 1). Comparatively better maintenance of RWC and MSI in salinity tolerant genotype might be due to the higher tissue tolerance, resulting in lesser salinity induced decline of leaf area (Tables 1, 2).
Maintenance of K+ uptake and/or reduction in Na+ entry, efflux of Na+ from the cytosol, and sequestration of Na+ for osmotic adjustment are strategies commonly used by plants to maintain the desirable K+/Na+ ratios in the cytosol (Zhu 2003). In this study, tolerant genotype accumulated lower amount of Na+ and higher amount of K+ and thus maintained higher K+/Na+ ratio in different plant parts as compared with sensitive genotype under both salinity stress levels (Fig. 1). Though salinity stress increased the Na+ content in all the genotypes irrespective of the tissue, leaf Na+ content was significantly higher in sensitive genotypes as compared with tolerant genotype of wheat. Interestingly, the tolerant genotype Kharchia 65 accumulated greater proportion of Na+ in the root and stem tissues, and lesser amount in the leaf. Thus, tolerant genotypes excluded Na+ ions selectively from leaves, the source tissue that supplies photosynthates for grain development.
Molecular basis of ion homeostasis under salinity stress was elucidated by the discovery of Arabidopsis salt overly sensitive (sos) mutants (Zhu 2003). SOS pathway regulates Na+ and K+ homeostasis at the root–soil interface and also regulates long-distance Na+ transport from roots to shoots. Salinity stress invokes cytosolic calcium oscillations that are sensed by a specialized calcium binding protein SOS3, which further transduces the signal by binding to and activating SOS2, a serine/threonine protein kinase (Qiu et al. 2002). The SOS2 kinase phosphorylates and activates SOS1, the plasma membrane Na+/H+ antiporter (Quintero et al. 2002). Activated SOS1 pumps out Na+ from cytosol to apoplast of root tissues. SOS1 also helps in exporting Na+ from the leaves into the phloem and then unloading it into the roots for efflux (Munns and Tester 2008). Physiological analysis clearly showed that salinity tolerant Kharchia 65 effectively excluded Na+ away from leaves and also maintained lower levels of Na+ in all the plant parts. This suggests that Kharchia 65 may have low net uptake of Na+ at the root level and able to exclude the absorbed Na+ from entering xylem stream and reaching leaves. Both of these processes are mediated by plasma membrane Na+/H+ exchanger encoded by the SOS1gene. Hence, we analyzed the expression of SOS1. The expression of SOS1 was up-regulated under salinity in tolerant genotype in both root and leaf tissues, which had low tissue Na+ levels and maintained better leaf K+/Na+ ratio (Figs. 1, 2a). Higher expression of SOS1 in tolerant genotypes as compared with sensitive genotype has probably facilitated exclusion of toxic Na+ into root apoplast and thus resulted in higher K+/Na+ ratio of leaves. A putative Na+/H+ antiporter from bread wheat enhanced Na+/H+ exchange activity and salinity tolerance of yeast (Xu et al. 2008). Similarly, durum wheat SOS1 protein effectively arbitrated Na+/H+ antiport in yeast and Arabidopsis (Feki et al. 2011, 2014).
The SOS2 gene expression was induced in roots of tolerant genotype under salinity stress as compared with control. However, in roots of sensitive genotypes, SOS2 was not induced and expression levels were lowest as compared with tolerant genotype. In leaf tissue also, SOS2 expression was highest in tolerant genotype. Greater expression of SOS2 in tolerant genotype (Kharchia 65) thus might have helped sustained activity of SOS1 under salinity stress (Fig. 2b). The SOS3 is required for the activation of SOS2 (Halfter et al. 2000).The SOS3 gene expression was up-regulated in the roots of tolerant genotype under salinity stress, while no induction was observed in sensitive genotypes. Similar to SOS1, constitutive expression of SOS3 gene was high in both leaf and root tissues of Kharchia 65 (Fig. 2c). Comparatively less expression of SOS3 in sensitive genotype HD 2687 under salinity stress was probably responsible for susceptibility to salinity stress. Thus, transcript abundance of SOS pathway genes is correlated with salinity tolerance in wheat. Constitutive as well as stress induced expression of SOS pathway genes correlated with salinity tolerance of Brassica genotypes (Kumar et al. 2009).
The expression levels of genes involved in Na+ sequestration namely VP1 and NHX1 were also higher in tolerant genotype at S2 level of salinity as compared with sensitive genotypes (Fig. 3). Results showed that salinity treatment sharply increased gene expression of VP1 in tolerant and moderately tolerant genotypes, which might have created favorable proton gradient leading to increased activity of NHX1, which sequesters Na+ into vacuole (Apse et al. 1999). In case of sensitive genotypes, the VP1and NHX1 gene expressions were impaired under severe salinity stress.
ClustalW (2.1) multiple sequence alignment and comparison of deduced partial protein sequences of wheat SOS pathway genes with corresponding genes of Arabidopsis thaliana and Oryza sativa revealed the inter-generic similarity among the sequences (Online Resource 3). Apart from sequence homology, presence of unique conserved domains and TM spans (in SOS1, VP1 and NHX1) unequivocally prove that the genes studied here, actually belong to wheat SOS pathway. Transgenic over expression of SOS pathway genes imparted favorable Na+ and K+ contents in Arabidopsis plants (Yang et al. 2009; Nie et al. 2015). Martinez-Atienza et al. (2007) concluded that in rice, higher expression of SOS1, SOS2 and SOS3, correlated with better sodium exclusion from shoot. Cuin et al. (2011) reported that Na + efflux mediated probably by a SOS1 like protein was highest in Kharchia 65 genotype, which also exhibited better Na+ sequestration into the vacuoles of root cells as revealed by confocal microscopy using sodium binding fluorescent dye. Similarly, Ramezani et al. (2013) studied the expression of SOS1 and SOS4 genes in cultivated and wild wheat genotypes and concluded that all the three alleles of SOS1 gene are active in salinity tolerance function.
It can thus be concluded that significantly higher transcript abundance of SOS1, SOS2 and SOS3 in tolerant genotype, Kharchia 65 as compared with sensitive genotype HD 2687 in roots and leaves might have restricted the entry of Na+ into the leaves and resulted in higher K+/Na+ ratio. Expression of VP1 and NHX1 genes were also high in leaves of tolerant genotypes, resulting in efficient sequestration of Na+ into vacuoles. The results showed that the salinity tolerant varieties maintained proper growth by restricting the absorption of toxic Na+ and lesser reduction in tissue K+ content. Calcium ions which are the major signaling molecules in SOS pathway was maintained at significantly higher level in salinity tolerant genotypes exposed to long term salinity stress (Lekshmy et al. 2013). Our results provide evidence of the importance SOS pathway genes (SOS1, SOS2, and SOS3), VP1 and NHX1 in maintaining ion homeostasis in the well-known salinity tolerant wheat cv. Kharchia 65.
Author contribution statement
Lekshmy Sathee conducted the experiments and contributed to the manuscript writing. R. K. Sairam and Chinnusamy V designed the experiment and wrote the manuscript. S. K. Jha did the cloning, bioinformatics and statistical analyses.
References
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by over expression of a vacuolar Na+/H+ antiporter in Arabidopsis. Science 285:1256–1258
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437–448
Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S (2011) Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: In planta quantification methods. Plant Cell Environ 34:947–996
Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R (2005) Control of sodium transport in durum wheat. Plant Physiol 137:807–818
Feki K, Quintero FJ, Pardo JM, Masmoudi K (2011) Regulation of durum wheat Na+/H+ exchanger TdSOS1 by phosphorylation. Plant Mol Biol 76:545–556
Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, Pardo JM, Brini F (2014) A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep 33:277–288
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45:146–159
Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton tranporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96:1480–1485
Gaxiola RA, Li J, Undurraga S, Dang V, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt- tolerant plants result from over expression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98:11444–11449
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97:3735–3740
Hollington PA (2000) Technological breakthroughs in screening/breeding wheat varieties for salt tolerance. In: Gupta SK, Sharma SK, Tyagi NK (eds) National conference on salinity management in agriculture. Central Soil Salinity Research Institute, Karnal, pp 273–289
Iseki K, Homma K, Shiraiwa T, Jongdee B, Mekwatanakarn P (2014) The effects of cross-tolerance to oxidative stress and drought stress on rice dry matter production under aerobic conditions. Field Crops Res 163:18–23
Ji H, Pardo JM, Batelli G, Van Oosten MJ, Bressan RA, Li X (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Mol Plant 6:275–286
Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Kumar G, Purty RS, Sharma MP, Singla-Pareek SL, Pareek A (2009) Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J Plant Physiol 166:507–520
Lekshmy S, Sairam RK, Kushwaha SR (2013) Effect of long-term salinity stress on growth and nutrient uptake in contrasting wheat genotypes. Ind J Plant Physiol 18(4):344–353
Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci USA 97:3730–3734
Martinez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Nie WX, Xu L, Yu BJ (2015) A putative soybean GmsSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma 252:127–134
Oh DH, Leidi E, Zhang Q, Hwang SM, Li YZ, Quintero FJ, Jiang XY, D’Urzo MP, Lee SY, Zhao YX (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151:210–222
Olías R, Eljakaoui Z, Li J, Marín- Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ 32:904–916
Pardo JM, Cubero B, Leidi EO, Quintero FJ (2006) Alkali cation exchangers: roles on cellular homeostasis and stress tolerance. J Exp Bot 57:1181–1199
Plett DC, Moller IS (2010) Na+ transport in glycophytic plants: what we know and would like to know. Plant Cell Environ 33:612–626
Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441
Quintero FJ, Ohta M, Shi HZ, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99:9061–9066
Ramezani A, Ali N, Ali AA, Mahboobeh ZB, Tahereh D, Mahmod E, Hosein A, Esmail E (2013) Quantitative expression analysis of TaSOS1 and TaSOS4 genes in cultivated and wild wheat plants under salt stress. Mol Biotechnol 53:189–197
Sairam RK, Deshmukh PS, Shukla DS (1997) Tolerance to drought and temperature stress in relation to increased antioxidant enzyme activity in wheat. J Agron Crop Sci 178:171–177
Sairam RK, Chandrasekhar V, Srivastava GC (2001) Comparison of hexaploid and tetraploid wheat cultivars in their responses to water stress. Biol Plant 44:89–94
Sairam RK, Rao KV, Srivastava GC (2002) Differential response of wheat genotypes to long-term salinity stress in relation to oxidative stress, antioxidant activity and osmolyte concentration. Plant Sci 163:1037–1046
Sanchez-Barrena MJ, Martinez-Ripoll M, Zhu JK, Albert A (2005) The structure of the Arabidopsis thaliana SOS3: molecular mechanism of sensing calcium for salt stress response. J Mol Biol 345:1253–1264
Shi H, Zhu JK (2002) Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and ABA. Plant Mol Biol 50:543–550
Shi H, Ishitani M, Kim CS, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901
Szabolcs I (1989) Salt-Affected Soils. CRC Press, Boca Raton
Tandon HLS (1995) Estimation of sodium and potassium. Methods of analysis of soils, plants, water and fertilizers. FDCO, New Delhi, pp 62–63
Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot 91:503–527
Weatherley PE (1950) Studies in water relations of cotton plants. I. The field measurement of water deficit in leaves. New Phytol 49:81–87
Xu H, Jiang X, Zhan K, Cheng X, Chen X, Pardo JM, Cui D (2008) Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch Biochem Biophys 473:8–15
Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z (2009) Over expression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Acknowledgments
Authors are thankful to the ICAR-Indian Agricultural Research Institute for funding and providing the necessary facilities. Lekshmy Sathee also gratefully acknowledges the Junior and Senior Research Fellowship provided to her by the Council of Scientific and Industrial Research, New Delhi during the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Sowinski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lekshmy Sathee, Sairam, R.K., Chinnusamy, V. et al. Differential transcript abundance of salt overly sensitive (SOS) pathway genes is a determinant of salinity stress tolerance of wheat. Acta Physiol Plant 37, 169 (2015). https://doi.org/10.1007/s11738-015-1910-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1910-z