Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that negatively regulate special target mRNAs at the post-transcriptional level by directing target mRNA cleavage or translational inhibition. Plant miRNAs regulate gene expression mainly by guiding cleavage of target mRNAs and subsequently play important roles in diverse developmental processes, nutrient homeostasis and responses to biotic and abiotic stresses. MiRNA393 plays important and diverse roles in defense against bacterial pathogens by negatively targeting transport inhibitor response 1 (TIR1) in plant development. It will be essential for understanding complex feedback regulations in the development pathway by unraveling the miR393 network in a temporal and spatial manner. Here, we report that Zma-miR393b down-regulates its putative target TIR1-like (F-box) gene by guiding the cleavage of their mRNAs in development of leaf sheaths in response to R. Solani infection, Zma-miR393b and its putative target gene TIR1 were confirmed through Q-PCR and the spatial expression of Zma-miR393b was further analyzed by in situ hybridization. These findings suggested that, as a negative feedback regulation of TIR1-like (F-box) gene, Zma-miR393b plays an important role in defense against R. Solani infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants respond to variable and severe environmental stress via a series of physiological, cellular, and molecular processes (Sunkar et al. 2007). An adaptive mechanism has developed to cope with various types of environmental stresses, such as starvation, oxidative stress, drought stress and invasion by phytopathogens during their differentiation, development, and aging processes. Recent studies indicate that post-transcriptional regulations of gene expression play an important role in how plants respond to abiotic stresses (Mirouze and Paszkowski 2011). MicroRNAs (miRNAs) act as ubiquitous post-transcriptional gene regulatory molecules in plants. Plant miRNAs are approximately 21-nt-long small regulatory RNAs that recognize their mRNA targets based on imperfect sequence complementarity, thereby suppress expression of the target gene by guiding degradation and/or translational repression of the cognate mRNA target, which are involved in the regulation of plants growth and development (Schwab et al. 2005; Garcia 2008; Zhang et al. 2011). Increasing evidence demonstrates that miRNAs play an important role in many biological and metabolic processes including regulation of plant growth, development and response to biotic and abiotic stresses via interactions with their specific target mRNAs (Shukla et al. 2008; Liu et al. 2008; Jones-Rhoades and Bartel 2004). A multitude of small RNAs accumulate in plant tissue, among these miRNAs, miR393 has been first reported to enhance innate immunity in response to bacterial infection (Zhang et al. 2011; Jones-Rhoades and Bartel 2004; Sunkar et al. 2007), which was firstly discovered in a small RNA library of Arabidopsis seedlings exposed to dehydration, salinity, cold stress and to the plant stress hormone abscisic acid. In addition, miR393 family was also identified in Arabidopsis thaliana and rice by comparative genomic approaches. Moreover, it is showed that plant miR393 family shared different spatial and temporal expression models in response to various environments, which was involved in regulation network in response to stresses by targeting TIR1 (transport inhibitor response 1, TIR1), such as auxin signaling transduction pathway (Navarro et al. 2006). Though the regulatory mechanism of miR393 in response to pathogen infection has been comprehensively studied in rice and Arabidopsis thaliana, how Zma-miR393 is responsive to banded leaf and sheath blight (BLSB) in maize and how Zma-miR393 is expressed in a temporal and spatial manner in the development pathway still remain unknown.
Maize is one of the most important cereal crops as agricultural feed and bio-ethanol production in the world. However, it has been increasingly affected by BLSB caused by Rhizoctonia solani Kuhn, causing severe losses in several Asian countries (Chung et al. 2005). Currently, many genes involved in maize under BLSB stress have not been isolated and the molecular mechanism of R. solani tolerance in maize still remains limited, the knowledge about role of miRNAs responsive to BLSB stress in maize is still scared as well. In this study, temporal and spatial manner of miRNA393b was detected and located by ISH technology and then the specific expression of BLSB-responsive miR393b and its putative target TIR1 were identified by quantitative real-time PCR (Q-PCR).
Materials and methods
Plants material and pathogen infection
High-resistance maize inbred line seedlings of “R15” and high-sensitive maize inbred line seedlings of “478” were treated with 7 % hypochlorite solution for 30 min, respectively, followed by three washes with sterilized water before being sowed in pots with autoclaved soil. Control plants were maintained under the same conditions. R. solani AG1-IA (kindly provided by the Rice Institution of Sichuan Agricultural University, Sichuan, China) was cultured on potato dextrose agar (PDA) and grown at room temperature (22–24 °C) under continuous light; agar blocks (0.5-cm squares) were cut and prepared from the outer edge of a 3-day-old culture. Soak barley grains were prepared in water for 24 h and dispensed 40 g in 250-mL conical flask. Two- to three-day-old pure culture is suspended in distilled sterile water, to make suspension and seed 5 mL of the suspension in each flask, and then incubated at 27 °C for 10 days. The impregnated grains can later on be used for inoculation. Impregnated barley grains were placed at the junction of sheath and leaf during the rainy days when moist condition is prevailed and crop is 30–40 days old. Two to four grains should be inserted between stalk and sheath on second or third internode level from soil for better inoculation. Subsequently, the leave sheaths were covered with plastic bags to ensure high humidity. The inoculated plants and mock-inoculated plants grew in the same growth chambers in Maize Research Institute of Sichuan Agricultural University. The day/night temperature during seedling development ranged from 25 to 33 °C, which was similar to field temperatures during maize seedling stage development (Liu et al. 2012; Qiu et al. 2007). The leaf sheaths without any leaves were harvested from each of the three maize plants, and the three leaf sheaths were combined to represent one replicate. Three independent replicates were collected for each sample. Control samples (CK) were harvested from water-treated leaf sheaths incubated under the same conditions. Partially, inoculated (Treat) and water-treated leaf sheaths incubated (CK) bract tissues were collected at 0, 6, 12, 24 h after inoculation for four stages, respectively.
Maize sample collection and RNA isolation
All samples from R15 and Ye478 were cleaned and immediately frozen in liquid nitrogen for further biochemical and molecular studies. According to the manufacturer’s instructions, small RNA was isolated from each sample using the mirVana™miRNA Isolation Kit (Ambion) and total RNA was isolated from each sample using Trizol Reagent (Invitrogen, Nottingham, UK).
Bioinformatics analyses
After downloading the precursor sequences of miR393 in plants from miRBase (http://www.mirbase.org/) (Griffiths-Jones et al. 2008). We conducted their conservation and evolutional relationship analysis using Geneious software, and target genes of miR393 were predicted using WMD3 website (http://wmd3.weigelworld.org/) using the Zea mays ZmB73 v4a.53 (MGC) database (Ossowski et al. 2008).
Expression analysis of Zma-miRNA393b and association target gene by Q-PCR
To monitor the expression patterns of miRNA393b (MIMAT0013999c), the corresponding sequences for miRNA393b (5′-CTCCAAAGGGATCGCATTGATA-3′) and 5 s RNA (5′-TAAGGTAGCGGCGAGACGAGC-3′) was used as the forward primers, respectively, and the 3′-corresponding primer (5′-GCTGTCAACGATACGCTACGTAACG-3′, Qiagen), as the common reverse primer. MiR393b were validated by relative real-time quantitative RT-PCR according to the manufacturer’s protocol, mitochondrial 5S rRNA was used as an internal control to normalize all data. Briefly, 2 μg of miRNA was reverse transcribed using the One Step PrimeScript® miRNA cDNA Synthesis Kit (TaKaRa Biotechnology Co., Ltd., Chengdu, China). The reverse transcription reaction system included 10 μL of 2 × miRNA reaction buffer, 2 μL of 0.1 % BSA, 2 μL of miRNA PrimeScript® RT Enzyme Mix, 2 μL of total RNA (10 pg/μL–1 μg/μL), and RNase-free dH2O to a final volume of 20 μL. The reaction mixtures were incubated in a 96-well plate at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s and 72 °C for 30 s.
To monitor the expression target gene TIR1-like (F-box) (GRMZM2G135978) of candidate miRNA393b, the sequences 5′-AGAACTCGGAACAGGAAGA-3′ and 5′-AAAGGAGCAGAAGGGAAA-3′ were used as forward and reverse primers, respectively, for amplification of TIR1-like gene, the sequences 5′-CTGAGAAACGGCTACCACA-3′ and 5′-CCCAAGGTCCAACTACGAG-3′ were used as forward and reverse primers, respectively, for amplification of mitochondrial 18S rRNA. cDNA synthesis was carried out using 1 μg total RNA with Prime Script RT reagent kit (TaKaRa). The reaction mixture (20 μL) contained 0.5 μL of each primer and the appropriate amounts of enzymes, cDNA and fluorescent dyes. All runs used a negative control without adding target cDNA, resulting in no detectable fluorescence signal from the reaction. A range of five dilutions of the total cDNA was tested under the same conditions as the samples. Amplification reactions were initiated with a pre-denaturing step at 95 °C for 10 s and followed by denaturing (95 °C for 5 s), annealing (60 °C for 10 s) and extension (72 °C for 15 s) steps for 49 cycles during the second stage, and a final stage of 55–95 °C to determine dissociation curves of the amplified products. All samples were performed in three biological replicates with three technical replicates. The mean and standard deviation (SD) are determined from the triplicate samples. We used the 2−∆∆CT method to calculate the absolute amount of miRNA393b according to the standard curve. Each sample was replicated for three times. The each miRNA level was expressed as 2−∆∆CT mean ± SEM (Schefe et al. 2006). The statistical significances of the results were compared and analyzed with two-way analysis of variance (ANOVA) and multiple comparisons using uncorrected Fisher’s LSD test. Differences were scored as statistically significant at P < 0.05.
Zma-miR393 in situ hybridization (ISH) assays
For in situ hybridization, we utilized miRCURY 5′-DIG- and 3′-DIG-labeled LNA-miRNAs (Zma-miR393b; Exiqon, Woburn, MA, USA) detection probes. The probe of Zma-miR393b is 5′-DIG-UCCAAAGGGAUCGCAUUGAUCC-DIG-3′. Sections of 30-μm thickness were performed using locked nucleic acid (LNA) probes, and all steps were carried out as described previously (Wibrand et al. 2010; Pena et al. 2009). TBS (Tris-buffered saline) and proteinase K are used to rinse and incubate all sections for 5 min at 37 °C, respectively, and then post-fixed for 5 min in 4 % PFA after washing once in 0.2 % glycine/TBS and twice in TBS. Subsequently, sections immersed in 1-ethyl-3- (3-dimethylaminopropyl) carbodiimide (EDC) fixative for 60 min at room temperature, followed by incubation in freshly prepared 1-methylimidazole solution. To inactivate endogenous alkaline phosphates and peroxidases, we washed sections again followed by acetylation with triethanolamine and acetic anhydride. 4 pmol of LNA probe diluted in 200 μL hybridization buffer was selected to incubate sections overnight after 10 min of prehybridization. In addition, a hybridization temperature of 20 °C below Tm was used to determine miRNA-LNA probe duplex. Before being blocked and incubated with anti-POD-AP for 1 h at RT, 3 % hydrogen peroxide was used to wash the sections, and then stained using the TSA Plus Cy3 System.
Results
Bioinformatics analysis of miRNA393 and its target gene TIR1
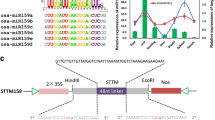
Plant miRNAs are highly conserved among distant-related plant species, both in terms of primary and mature miRNAs, the conservation of mature miRNAs and pre-miRNAs provides the chance to investigate their evolutionary relationships. Comparison of the precursor sequences of the predicted miRNAs with other members in the same family showed that most members could be found to have a high degree of sequence similarity with others, to investigate the conservative of mi393 among plant species, the pre-miRNA sequences of miR393 in Arabidopsis thaliana (ath), Oryza sativa (Osa), Zea mays (Zma), Sorghum bicolor (Sbi), Populus trichocarpa (Ptc) and other plants whose genomes were partially or completely sequenced were downloaded from miRBase (http://www.mirbase.org/), and subsequently the evolutionary relationships of Zma-miRNA393 with other members from the same families were analyzed using the Geneious (Kearse et al. 2012), which represents an ideal platform for the bioinformatics community to leverage existing components and to integrate their own specific requirements for the discovery, analysis and visualization of biological data (Fig. 1). It showed that the precursor sequence similarity between Zma-miR393b and other Zma-miR393 members, that between sbi-MIR393b and osa-MIR393b, Zma-miR393b was similar to each other (Fig. 1). Only one or two bases are different at the end of the mature sequences of miR393 in different plant species, even some sequences of miR393 are identical in the same plants. It could be seen from the phylogenetic trees that the evolutionary relationships of Zma-miRNA393 with other species were similar in same families; the conservatism of miR393 family in different plant species suggests that miR393 probably plays the similar vital role in stress responses.
Conservation analysis of precursor sequences of miR393 among plant species. Identical and conserved nucleotides of miR393 family showed higher conservation among plant species, only few changes has found in nucleotide sequences in some family members. Four colors represent the A, T, C and G, which show the relationship significance among the precursor sequence (color figure online)
The high level of complementarity between plant miRNAs and their target genes allows an effective prediction system for predict their target genes (Alves-Junior et al. 2009; Fattash et al. 2007). In our research, target genes of zma-miR393 were predicted by WMD3 (http://wmd3.weigelworld.org/) using the Zea mays ZmB73 v4a.53 (MGC) database. Protein transport inhibitor response 1 (TIR1) (GRMZM2G135978) was predicted as Zma-miR393′ target gene, which was consistent with the result reported in Zhang’s research (Zhang et al. 2009).
In the previous study, in Arabidopsis, miR393 targets mRNAs that code for the auxin receptors (TIR1, AFB2 and AFB3) (Jones-Rhoades and Bartel 2004). Previous studies have shown that miR393 appears to regulate TIR1/AFB expression by complementary to the auxin receptors. TIR1, AFB1, AFB2, and AFB3 are broadly transcribed throughout the plant by acting as auxin receptors. MiR393 acts as an upstream regular of these receptors, responses to adverse conditions by auxin signal pathway. Moreover, many studies have reported that miR393 could regulate transport inhibitor response 1 (TIR1) and auxin signaling F-box proteins (AFBs), which play key roles in signaling transduction as the other auxin receptors. However, the targeting genes of Zma-miR393 have not been determined in maize. The effects of Zma-miR393 on leaf growth and development responsive to BLSB are still unclear. In our previous study, we validated miRNA393 by Solexa deep sequencing (Data unpublished) and predicted the targets of miRNA393, it is showed that Zma-miRNA393b is down-regulated in treatment compared with the expression in CK, however, Zma-miR393a and Zma-miR393c are up-regulated. In the current study, Zma-miR393b and its putative target gene TIR1-like were confirmed through Q-PCR and the spatial expression of Zma-miR393b was further analyzed by in situ hybridization technology.
Expression analysis of Zma-miR393b and its target gene
If miR393b degrades target mRNA transcripts, expression levels should be negatively correlated with each other. In this study, we compared the expression patterns of predicted miRNA targets and with miRNA accumulation levels in Q-PCR assay. The expression levels of Zma-miR393b targets are inversely correlated with the accumulation levels of corresponding Zma-miR393b (Figs. 2, 3). Zma-miR393b was especially down-expressed in different periods after inoculation with BLSB in the leaf sheath of R15 and reached to minimum in 24 h (10.4-fold) compared with control group (0 h), while there were not obvious changes among the corresponding expression in control group (CK); conversely, Zma-miR393b was especially up-regulated before 24 h (5.8-fold at 6 h and 3.1-fold at 12 h) infection in Ye478 and decreased in 24 h (9.6 fold) compare with CK, while there was obviously uptrend among the corresponding expression in CK. However, TIR1-like (F-box) gene was especially up-expressed in different periods after inoculation with R. Solani in the leaf sheath of R15, reached to maximum in 24 h (8.3-fold), but only up-expressed in the leaf sheath of Ye478 after 6 h (0.5-fold at 6 h, 1.2-fold at 12 h and 7.8-fold at 24 h) compare with CK. The results suggested that Zma-miR393b down-regulates TIR1 genes in high-resistance maize inbred line R15 could respond to R. Solani infection earlier by accumulating more TIR1 genes than high-sensitive maize inbred line Ye478.
Q-PCR analysis of Zma-miR393b responsive to inoculation. R. Solani CK samples correspond to mock-inoculated plants and treat samples correspond to inoculated plants. The color nodes represent the expression of fold change. Expression of BLSB-responsive Zma-miR393b was mapped left in Ye478, right in R15. Error bars show the standard error calculated from three biological replicates. Stars indicate the significant (P < 0.05) difference identified by uncorrected Fisher’s LSD test in multiple comparisons after two-way ANOVA (color figure online)
Q-PCR analysis of putative target gene TIR1-like (F-box). Mock-inoculated plants correspond to CK sample and inoculated plants correspond to treat sample. The color nodes represent the expression of fold change. Expression of BLSB-responsive TIR1-like (F-box) gene was mapped left in Ye478, right in R15. Error bars show the standard error calculated from three biological replicates. Stars indicate the significant (P < 0.05) difference identified by uncorrected Fisher’s LSD test in multiple comparisons after two-way ANOVA (color figure online)
The special expression of Zma-miR393b was further analyzed by in situ hybridization technology (Fig. 4). The existence and location of Zma-miR393b were observed by the blue-black in positive area, and the microscopic results were quantified by the blue-black (in situ hybridization) for positive signal in per unit area. It is showed that the blue/black-positive hybrid signals were detected throughout the four stressed stages (0, 6, 12, 24 h) in the leaf sheath of R15 and Ye478 (Fig. 5), and Zma-miR393b was especially down-expressed in different periods after inoculation with BLSB in the leaf sheath of R15, reached to minimum in 24 h (6.2 ± 1.0 %) compare with control groups (0 h) (100 ± 9.0 %), conversely, Zma-miR393b was especially up-regulated in 6 h (130 ± 17.0 %) infection in Ye478 and decreased in 24 h (10 ± 2.0 %) compare with control groups (0 h) (100 ± 8.7 %). Similar results were revealed that Zma-miR393b was up-regulated in 6 h in Ye478 and R15 by Q-PCR, so we speculated that R15 could be more earlier to accumulate more TIR1 genes responsive to R. Solani infection than Ye478 (Figs. 2, 3, 4, 5). In addition, according to expression levers of the BLSB-responsive Zma-miR393b, it is demonstrated that leaf sheaths were infected by R. Solani spreading from outside to inside with the prolong of infection time and especially expressed in surrounding tissues of the primary xylem and metaxylem after 24 h.
In situ hybridization analysis of the special expression of Zma-miR393b. ISH was performed on fixed tissue with oligoribonucleotide (LNA) DIG-labeled probe as described in the text. a–d Correspond to Ye478-0h ck; Ye478-6h ck; Ye478-12 h ck; Ye478-24 h ck; definition of corresponding e–h correspond to Ye478-0h treat; Ye478-6h treat; Ye478-12h treat; Ye478-24h treat, respectively; and i–l correspond to R15-0h ck; R15-6h ck; R15-12h ck; R15-24h ck; corresponding m–p correspond to R15-0h treat; R15-6h treat; R15-12h treat; R15-24h treat, respectively. Ck samples correspond to mock-inoculated plants and treat samples correspond to inoculated plants
Change of relative quantity of Zma-miR393b in R15 and Ye478 ISH-quantized by image analysis. The microscopic results were quantified by the blue–black (in situ hybridization) for positive signal in per unit area (n = 9). Error bars show the standard error calculated from three biological replicates. Stars indicate the significant (P < 0.05) difference identified by uncorrected Fisher’s LSD test in multiple comparisons after two-way ANOVA (color figure online)
Discussion
Zma-miR393 targeted TIR1-like (F-box) gene responsive to BLSB stress
Previous researches showed that the expression of different sets of TIR1-like (F-box) gene was regulated by miR393 following pathogen infection or nitrate treatment, which plays a key role in critical receptors in the auxin signaling. Here, we report that miR393b helps regulate auxin-related development of leaf sheaths infection with BLSB. We found that Zma-miR393b is the predominant source for zma-miR393 family in leaf sheaths by Solexa deep sequencing in our previous study (unpublished results), and we demonstrated the expression of the TIR1-like (F-box) gene (GRMZM2G135978) was negatively regulated by Zma-miR393b in response to BLSB stress in maize leaf sheaths (Figs. 2, 3). An integrated putative Zma-miR393b-TIR1 module has been proposed for interaction between Zma-miR393b and its target TIR1-like (F-box) gene, which is required for auxin responses in plant development and stress responses.
It will provide useful information about their physiological functions by analyzing the spatial and temporal expression patterns of miRNAs. In addition, it is showed that many miRNAs have differential accumulation in specific developmental stages and tissues of plants. In our LNA-ISH assay, it is demonstrated that Zma-miR393b significantly expressed in the basic tissue and primary phloem of leaf sheath infected with BLSB at the early stage, and then penetrated deeply in metaphloem, bundle sheath and primary xylem tissue at the medium stage, and last completely invaded in surrounding tissues of the primary xylem and metaxylem.
Molecular mechanism of Zma-miR393 in response to pathogen infection
Zea mays has developed various mechanisms to cope with and adapt to different types of abiotic stress (such as pathogen infection) by regulating the expression of specific genes at the transcriptional or post-transcriptional level, which are directly involved in metabolism, physiological and regulatory or antibacterial processes (Cordoba et al. 2009). Emerging data suggest that the biogenesis of miRNAs, the expression of mRNA targets, and the activities of miRNA-protein complexes can be altered by stress conditions. In addition, the level of target gene repression is also dependent on the concentration of mRNA targets relative to the miRNA by mRNA cleavage and translational repression, indirectly involved in the metabolism and physiological process, thus to resist stresses. Several studies have focused on miRNAs related to the stress response (Kruszka et al. 2012). MiR393 acts as a representative of miRNAs’ response to stresses and has been studied comparatively and incisively. The function study of miR393 and targets under plant biotic stresses can deepen our understanding of the expression mechanism and response mechanism of genes regulated by miR393 at the post-transcriptional level (Vidal et al. 2010).
Recent studies indicate that Aux/IAA and auxin-regulated transcription are mediated by TIR1 that acted as an auxin receptor in plant auxin signal pathway (Gray et al. 1999) and their activity is regulated by auxin (Kepinski and Leyser 2005). TIR1 acts as a big part of the ubiquitin ligases complex SCFTIR1 in the auxin regulating pathway, ubiquitin-mediated and 26S proteasome-dependent protein degradation pathway and performs a very useful role in the most hormone signal-transmitting pathways (Vierstra 2009), thus it is a crucial step of degradation of auxin-dependent negative regulators in auxin signaling pathway (Dharmasiri et al. 2005). It is proposed that TIR1 acts as a main target gene of Zma-miR393. Furthermore, TIR1 expression was regulated by miR393 that expressed and acted as the auxin receptors in plant. In addition, miR393 was the first miRNA identified in plants responsive to bacterial pathogens, especially by repressing auxin signaling, and responds to resistance against virulent Pseudomonas syringae pv. tomato strain DC3000 (Pto DC3000) in Arabidopsis, implying that miR393 is a key component of plant basal defense (Parry et al. 2009; Tan et al. 2007).
Based on our studies, we proposed a model of the regulation mechanism of miR393b-targeted TIR1-like (F-box) gene in maize leaf sheaths under BLSB treatment, we demonstrate that Zma-miR393b down-regulated TIR1-like (F-box) gene in response to BLSB infection, and its induction was due to enhanced miR393b transcription. Target genes TIR1 was up-regulated by Zma-miR393b, enhancing auxin sensitivity and taking effects on maize leaf sheaths development; Zma-miR393b was down-expressed under BLSB stress, thereby increasing the expression of TIR1-like (F-box) gene and thus the putative target gene TIR1 being consequently accumulated in response to the pathogen attack by inducing auxin signals, at last, signal transduction pathways were activated to strengthen the morphological and metabolic adaptation in maize.
Author contribution
GPT and ZMZ conceived and designed the research. JG and ML collected samples, generated experimental data, performed the whole data analysis, and drafted the earlier versions of the manuscript. JG and HP involved the sample collection and partially revised the manuscript. ZMZ and ML designed the study, partially analyzed the data, and revised the manuscript. All authors read, reviewed and approved the final manuscript.
References
Alves-Junior L, Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T (2009) Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res 37(12):4010–4021
Chung W, Huang J, Huang H (2005) Formulation of a soil biofungicide for control of damping-off of Chinese cabbage (Brassica chinensis) caused by Rhizoctonia solani. Biol Control 32(2):287–294
Cordoba E, Salmi M, León P (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J Exp Bot 60(10):2933–2943
Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9(1):109–119
Fattash I, Voß B, Reski R, Hess WR, Frank W (2007) Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol 7(1):13
Garcia D (2008) A miRacle in plant development: role of microRNAs in cell differentiation and patterning. In: Seminars in cell and developmental biology, vol 6. Elsevier, pp 586–595
Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M (1999) Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13(13):1678–1691
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(suppl 1):D154–D158
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435(7041):446–451
Kruszka K, Pieczynski M, Windels D, Bielewicz D, Jarmolowski A, Szweykowska-Kulinska Z, Vazquez F (2012) Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol 169(16):1664–1672
Liu H-H, Tian X, Li Y-J, Wu C-A, Zheng C-C (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14(5):836–843
Liu Z, Kumari S, Zhang L, Zheng Y, Ware D (2012) Characterization of miRNAs in response to short-term waterlogging in three inbred lines of Zea mays. PLoS One 7(6):e39786
Mirouze M, Paszkowski J (2011) Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 14(3):267–274
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312(5772):436–439
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53(4):674–690
Parry G, Calderon-Villalobos L, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray W, Bennett M, Estelle M (2009) Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci 106(52):22540–22545
Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M (2009) miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods 6(2):139–141
Qiu F, Zheng Y, Zhang Z, Xu S (2007) Mapping of QTL associated with waterlogging tolerance during the seedling stage in maize. Ann Bot 99(6):1067–1081
Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s C T difference” formula. J Mol Med 84(11):901–910
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8(4):517–527
Shukla LI, Chinnusamy V, Sunkar R (2008) The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regul Mech 1779(11):743–748
Sunkar R, Chinnusamy V, Zhu J, Zhu J-K (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12(7):301–309
Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446(7136):640–645
Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci 107(9):4477–4482
Vierstra RD (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10(6):385–397
Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR (2010) Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci 31(4):636–645
Zhang L, Chia J-M, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5(11):e1000716
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75(1–2):93–105
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (30900901), the Research Project on Rice Functional Genes related to the Maize Large-scale Gene Discovery and Functional Genomics Resource, Technology, Information Platform Construction Program (863) of China (SS2012AA100107, 2012AA10A300), the Ph.D. Programs Foundation of Ministry of Education of China (20095103120002) and the Major Project of China on New varieties of GMO Cultivation (2011ZX08003-003) and the Youth Natural Science Foundation of Department of Education of Sichuan Province of China (11ZB123).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by B. Barna.
M. Luo and J. Gao contributed equally to this work.
Rights and permissions
About this article
Cite this article
Luo, M., Gao, J., Peng, H. et al. MiR393-targeted TIR1-like (F-box) gene in response to inoculation to R. Solani in Zea mays . Acta Physiol Plant 36, 1283–1291 (2014). https://doi.org/10.1007/s11738-014-1509-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11738-014-1509-9