Abstract
One of the oldest crops, foxtail millet (Setaria italica), is regarded as a model plant for studying drought tolerance owing to its short life-cycle and small genome size. The miRNA member miR394 plays multifaceted roles in plants. In this study, we identified the Sit-miR394 target gene SiFBP6 using 5′ RLM-RACE (RNA ligase mediated rapid amplification of 5′ cDNA ends). An expression analysis in different tissues showed that Sit-miR394 was highly expressed in leaves and had a lower relative expression in roots, while the target gene SiFBP6 exhibited the opposite expression pattern, being strongly expressed in roots and relatively lowly expressed in leaves. Furthermore, Sit-miR394 was upregulated in foxtail millet under drought-stress conditions and by exogenous methyl jasmonate, ethephon, salicylic acid and abscisic acid treatments. The overexpression of Sit-miR394 in transgenic Arabidopsis thaliana increased its tolerance to drought stress. The germination rates and root lengths were significantly greater compared with the wild-type line, which suggested that Sit-miR394 plays a positive role in responding to drought conditions. Moreover, a transcription analysis indicated that the overexpression of Sit-miR394 affected the expression of a set of stress-related genes, thereby conferring an increased abiotic or biotic stress tolerance to transgenic Arabidopsis. Thus, Sit-miR394 functions in the positive modulation of abiotic or biotic stress tolerance and has potential applications in molecular breeding to enhance stress tolerance in crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought is a major abiotic factor limiting plant growth and negatively affecting crop productivity. To cope with drought, plants have evolved several mechanisms at the physiological and molecular levels. Plant microRNAs are a class of short endogenous non-coding small RNAs that play important roles in biological and metabolic processes by cleaving target mRNAs or repressing translation. Increasing evidence suggests that miRNAs play key roles in plant responses to abiotic stress (Liu et al. 2008; Song et al. 2010; Cai et al. 2009). For example, the overexpression of Sly-miR169c in transgenic tomato (Solanum lycopersicum) plants results in a lowered leaf water loss and enhanced drought tolerance (Zhang et al. 2011). In barley, Hv-miR827 (Hordeum vulgare L.) overexpressing transgenic plants show an improved water-use efficiency with no effects on growth or reproductive timing compared with the wild-type (WT) plants (Ferdous et al. 2017). miR394 is a highly conserved family of miRNAs in plants, and 132 miR394s from 32 plant species have been deposited in the miRBase database (http://www.mirbase.org/). miR394 was first identified in Arabidopsis thaliana and rice been found in other crops, such as soybean (Joshi et al. 2010; Zhai et al. 2011), rapeseed (Zhao et al. 2012), cotton (Pang et al. 2009) and maize (Zhang et al. 2009; Tobias Dezulian 2005). The target genes of miR394s were found to encode F-box proteins in most species (Jones-Rhoades and Bartel 2004). In Arabidopsis, miR394 and its target gene leaf curling responsiveness (LCR) are involved in the regulation of leaf curling-related morphology (Song et al. 2012). Rice miR394 and its target gene LEAF INCLINCATION 4 (LC4) regulate leaf lamina joint development and rice architecture by modulating the expansion and elongation of adaxial parenchymal cells (Qu et al. 2018). Additionally, miR394s may play important roles in responses to abiotic and biotic stresses. For example, the overexpression of miR394 in Arabidopsis improves plant drought and cold tolerance, whereas miR394 overexpression increases the susceptibility of plants to salinity (Song et al. 2016; Yang et al. 2016). Similarly, the overexpression of soybean miR394 enhances drought tolerance in transgenic A. thaliana (Ni et al. 2012). In tomato, miR394 is downregulated in response to Botrytis cinerea infection (Jin and Wu 2015). Chand et al. found that garlic miR394 is significantly induced in response to Fusarium oxysporum f. sp. cepae (FOC) infection, suggesting that miR394 plays critical roles in plant microbe interactions (Chand et al. 2016). miR394 also plays important roles in several other biological processes. In rapeseed (Brassica napus), the overexpression of miR394 not only delays flowering time and enlarges the sizes of leaf blades, pods and seed bodies, but it also alters their fatty acid compositions (Song et al. 2015). Furthermore, miR394 is induced by iron deficiency-related stress in Malus xiaojinensis (Yu et al. 2012). To date, miR394s have been found to be involved in the modulation of various developmental processes and stress responses by negatively mediating their targets.
Foxtail millet (Setaria italica) is one of the oldest crops, and is characterized by a short life-cycle, small genome size and prominent drought resistance (Jia et al. 2013). Foxtail millet is a model plant for studying drought tolerance owing to these characteristics (Li et al. 2018; Doust et al. 2009; Lata et al. 2012). In a previous study, we found that Sit-miR394 was induced by drought in foxtail millet, and we predicted the target gene of Sit-miR394 using bioinformatics (Wang et al. 2016). Here, to further understand the mechanisms of Sit-miR394a, we characterized Sit-miR394 expression patterns after hormone treatments and in various foxtail millet tissues. The target gene of Sit-miR394 was validated using RLM-RACE (Leng et al. 2016). Furthermore, we generated transgenic A. thaliana plants overexpressing Sit-miR394 and found that the plants showed enhanced drought-stress tolerance. Thus, Sit-miR394 appears to be a positive regulator of drought-stress tolerance in foxtail millet.
Results
Validation of the Sit-miR394 Target
In a previous study, Sit-miR394 was predicted to target SiFBP6 mRNA (Si001465m) encoding an F-box protein. To identify the cleavage sites within SiFBP6 mRNA, 5′ RLM-RACE was performed. Sequencing of 10 5′-RLM-RACE clones indicated that the cleavage site was between 10 and 11 bp from the 5′ end of the miRNA394-SiFBP6 complementary region (Fig. 1a). The tissue expression patterns of Sit-miR394 and the target gene SiFBP6 in leaves, roots and stem were determined using real-time PCR. Sit-miR394 was highly expressed in leaves and had a lower relative expression in roots (Fig. 1b), while the target gene SiFBP6 exhibited the opposite expression pattern, being strongly expressed in roots and relatively lowly expressed in roots and leaves. These results suggested that SiFBP6 mRNA is an in vivo miR394-cleavage target gene in foxtail millet (Fig. 1c).
Alignments of miR394 with target gene validated and expression in difference Setaria italica tissue. a Arrows indicate the cleavage sites of target mRNAs, as detected with the 5′-RACE assay and the factions above the red arrows show the numbers of clones with an identified 5′end detected in the total sequenced clones. b Sit-miR394 relative expression in difference tissue. c The target gene sitFBP6 relative expression in difference tissue. The expression level in the root was set as 1. Setaria italica U6 and actin7 sequences were used as controls (p < 0.05*, p < 0.01**)
Expression Patterns of Sit-miR394 in Response to Stress and Phytohormone Treatments
To investigate the responses of the Sit-miR394 to drought stress, foxtail millet seedlings were subjected to natural soil drying and water recovery processes. The relative expression levels of Sit-miR394 were evaluated at 2, 4, 6 and 10 days after initiating the drought treatment and after a 2-day water recovery period. After 4, 6 and 10 days, the RWC of the drought-treated leaves decreased to 81.67%, 80.07% and 54.90%, respectively, while the RWC of the soil decreased to 90.12%, 58.30% and 13.30%, respectively. We observed that the expression of Sit-miR394 increased more than twofold after 4 days of drought treatment and then decreased until day 10. After re-watering for 2 days, the leaves of drought-treated plants recovered, and the expression of Sit-miR394 returned to normal. The SiFBP6 expression shows the opposite trend (Fig. 2a, b). We also analyzed the expression of the Sit-miR394 after plants were subjected to four phytohormones (ETH, ABA, MeJA and SA). Sit-miR394 showed an initial slight downregulation and then upregulation in response to high levels of ABA, SA and ETH. After the MeJA treatment, the expression of Sit-miR394 increased gradually and then decreased, followed by a sharp rise at 24 h (Fig. 2c). While the SiFBP6 expression showed opposite trend, SiFBP6 initial upregulation and then downregulation in response to four phytohormones. Notably, the expression level of Sit-miR394 increased significantly after 24 h of treatment with each of the four phytohormones, implying that Sit-miR394 may play a large role in foxtail millet’s tolerance to various environmental stresses.
miR394 and SiFBP6 relative expression in drought and phytohormone treatments. a Dynamic changes of leaf relative water content and soil water content under drought treatment. b miR394 and SiFBP6 relative expression in drought treatment. c miR394 and SiFBP6 relative expression in four phytohormone treatments. The expression level of the miRNAs in the control sampled at 0 d and 0 h was set as 1. Setaria italica U6 and actin7 sequences were used as controls (p < 0.05*, p < 0.01**)
Overexpression of Sit-miR394 Confers Drought Tolerance to Arabidopsis
To identify the role of Sit-miR394 in regulating plant responses to drought, transgenic Arabidopsis plants overexpressing Sit-miR394 were generated. The expression of miR394 in two 35S:Sit-miR394a transgenic lines was upregulated about threefold compared to WT plants (Fig. 3c). Two transgenic lines were used to analyze seed germination and seedling growth under drought-stress conditions. The seeds of the transgenic lines and the WT line germinated normally and no significant differences were observed on 1/2 MS medium without PEG (Fig. 3a, b). However, on 1/2 MS medium supplemented with 20% PEG, the germination rates of the two transgenic lines were 88.75% and 93.33%, respectively, while that of the WT line was 67.05% (Fig. 3d). The seed germination rates of the transgenic lines were significantly greater compared with that of the WT line, and the transgenic lines showed longer roots than WT seedlings after growing on the 1/2 MS medium containing 20% PEG for 12 days (Fig. 3e, g). To evaluate the effects of Sit-miR394 expression on plant drought tolerance, 30-day-old transgenic and WT Arabidopsis plants were treated with natural drought stress. After 15 days of the drought treatment, WT plants displayed severe wilting and eventually died, while most of the transgenic plants appeared to be healthy (Fig. 3f, h). Thus, the overexpressing Sit-miR394 plants showed increased drought tolerance, and Sit-miR394 appears to play an important role in plant drought tolerance.
The effect of sit-miR394 expression in transgenic Arabidopsis plants. a The transgenic Arabidopsis and WT seed germination and seedling growth seed germination and seedling growth in 1/2MS medium without PEG. b The transgenic Arabidopsis and WT seed germination and seedling growth in 1/2MS medium with PEG. c Verification of Sit-miR394 overexpression in Sit-miR394-overexpressing transgenic Arabidopsis lines by q-PCR. d The transgenic Arabidopsis and WT germinate rate in 1/2MS medium with PEG. f, h The transgenic Arabidopsis and WT growth and survial rate in natural drought. e, g The transgenic Arabidopsis and WT root length in 1/2MS medium with PEG (p < 0.05*, p < 0.01**)
Differential Expression Analysis of Transcriptomes between WT and Transgenic Lines
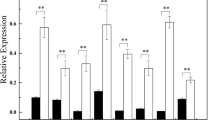
To gain further insights into the mechanisms of Sit-miR394, the gene expression profiles between 30-day-old transgenic and WT plants grown under normal conditions were compared. On the basis of the high-throughput sequencing results, 187 (184 upregulated and 3 downregulated) genes were considered as DEGs (Supplementary Table 2), and we found that the expression level of SiFBP6 homolog gene (At1g27340) decreased (log2 fold-change = − 0.55, corrected p-value = 0.03) but did not reach the standard of significantly different genes. Among these DEGs, 38 transcription factors belonging to 10 families were identified. The three major transcription factor families, ERF, MYB and C2H2, contained 19, 5 and 5 genes, respectively. A number of stress-responsive genes, such as DREB1D, ZAT12, JAZ5 and CML24, were significantly upregulated. Moreover, we select 13 genes for real-time fluorescence quantitative verification. The differential expression was consistent with the trend of the sequencing results, with both being upregulated (Fig. 4b), indicating that the sequencing results were reliable.
Differential gene GO enrichment and part of DEGs qPCR analysis. a Enrichment analysis of GO term of all DEGs analysis. Red, blue and green font denote biological process, cellular component, molecular function. The size of the dot represents the number of DEGs. b Part of DEGs qPCR analysis. The genes expression level of WT in the control sampled was set as 1. Arabidopsis EF1α sequences were used as controls (p < 0.05*, p < 0.01**)
We also performed GO enrichment analyses of the DEGs (Fig. 4a), and the top ten enriched GO terms based on false discovery rates were identified (Supplementary Table 2). The analysis of molecular function revealed that the DEGs were significantly enriched in response to chitin (GO:0010200), carbohydrate stimulus (GO:0009743), organic substance (GO:0010033), chemical stimulus (GO:0042221), stimulus (GO:0050896) and stress (GO:0006950). For the biological process and cellular component categories, DEGs were significantly enriched in transcription factor activity (GO:0003700) and nucleus (GO:0005634), respectively. The results of the GO enrichment analyses suggested that DEGs were involved in responding to multiple hormones, such as ABA, JA, SA an ETH, which was consistent with the qPCR results. Furthermore, some GO terms related to stress response were significantly enriched, such as cold, salt, water and wound responses. These results indicated that the miR394-F-box regulatory pathway plays a key role in plant stress resistance.
Discussion
F-box Genes are Highly Conserved Targets of miR394s
miR394 is a highly conserved miRNA family that exists in many plant species. Although miR394 precursors vary in different species, the lengths and sequences of mature miR394s are conserved, as are the target genes. Kumar et al. predicted 43 target genes of miR394 across 40 plant species using bioinformatics and found 32 target genes belonging to the F-box gene family and 11 target genes belonging to other families, such as GRAS and GDSL. To date, most of the miR394 target genes that have been experimentally verified in different species belong to the F-box gene family (Kumar et al. 2019). Only in garlic was a miR394 target gene encoding CYP450 verified using RLM-RACE. In foxtail millet, five putative target genes of miR394 were identified using bioinformatics. However, only SiFBP6, encoding an F-box, was verified using RLM-RACE in this study. These results suggested that F-box genes are highly conserved targets of miR394s. F-box proteins are essential for protein post-translational regulation, and may be involved in various plant biological processes, including phytohormone signaling, plant development, cell signaling, circadian clock regulation, biotic and abiotic stress responses and self-incompatibility (Gupta et al. 2015; Abd-Hamid et al. 2020; Xu et al. 2009). The functional diversity of F-box proteins may result in miR394 having multiple functions, unlike some other miRNAs, such as miR528, that regulate different target genes, including AAO, LAC, CBP and PPO, resulting in a variety of functions (Chen et al. 2019; Zhu et al. 2020).
miR394 is Regulated by Multiple Hormones
Plant hormones are important growth regulators (Pandey et al. 2003). ABA, SA, JA and ETH play major roles in mediating plant development and in responses against pathogens and abiotic stresses (Tiwari et al. 2017; Bari and Jones 2009; Neller et al. 2018). In Arabidopsis, miR394 was slightly induced by the ABA treatment, and the over-expression of miR394 led to ABA hypersensitivity and ABA-associated phenotypes. The expression of miR394 produces a similar pattern in soybean after an ABA treatment, showing an initial decrease and then a significant increase. In garlic, an exogenous JA treatment of JA also regulates miR394 signaling responses. Typically, abiotic stresses, such as drought, salinity and temperature, as well as wounding, trigger increases in ABA levels. SA, JA and ETH play major roles in responses to biotic stress conditions, with their levels increasing after pathogen infection. In this study, the qRT-PCR showed that the transcript levels of Sit-miR394 significantly increased in foxtail millet plants after ABA, ETH, JA and SA treatments. In particular, at 24 h after the ETH treatment, the expression level of Sit-miR394 increased by more than eightfold (Fig. 2c). Furthermore, we observed that large numbers of the hormone-responsive genes, such as ERF5 (Son et al. 2012; Moffat et al. 2012), JAZ5 (Torres et al. 2016; Pauwels and Goossens 2011) and CML24 (Delk et al. 2005; Tsai et al. 2007), were differentially expressed in Sit-miR394-overexpressing plants. Thus, miR394 may play a central coordinating role in the regulation of multiple hormone-response pathways.
miR394 Plays Multifaceted Roles in Plants
Previous studies suggested that miR394 and its target gene LCR are involved in regulating leaf curling-related morphology in Arabidopsis. Subsequently, miR394 was found to be induced by salt, drought and cold stresses. Additionally, the overexpression of miR394 results in enhanced drought and cold tolerance levels, but it also results in a hypersensitivity to salt stress. Moreover, miR394 may have negative effects on F. oxysporum f. sp. cepae and B. cinerea resistance, which suggested that miR394 participates in biotic stress responses (Jin and Wu 2015; Tian et al. 2018). Furthermore, miR394 is also involved in metabolism. Research shows that miR393, miR394 and miR395 act on target genes that affect flavonoid accumulation. In this study, Sit-miR394 was also induced by drought in foxtail millet, and the overexpression of Sit-miR394 confers tolerance to drought in transgenic A. thaliana. A GO analysis of DEGs between Sit-miR394-overexpressing and WT plants showed that some stress-related GO terms were enriched, such as response to cold (GO:0009409), salt stress (GO:0009651), water deprivation (GO:0009414), wounding (GO:0009611) and fungus (GO:0009620). Compared with WT plants, some stress-related genes, such as DREB1B/C/D/F, ZAT10 and ERF13, are significantly upregulated in Sit-miR394-overexpressing plants (Supplementary Table 2). Previous studies demonstrated that DREB1A/B promotes a higher freezing tolerance, and DREB1D/E/F is related to plant drought tolerance (Yang et al. 2016; Guttikonda et al. 2014). ZAT10 elevates the expression of reactive oxygen defense transcripts, which enhances plant tolerance to salinity, heat and osmotic stresses (Gupta et al. 2015; Nguyen et al. 2016). Additionally, ERF13 negatively regulates defenses against Pseudomonas syringae, which is consistent with the negative effects of mir394 on plant diseases (Miyamoto et al. 2019). These results suggested that miR394 has highly diverse functions in growth and development, as well as biotic stress responses.
miR394 participates in the regulation of gene expression by regulating its target LCR F-box. An F-box protein is the core component of the SCF complex (a well-characterized E3 ligase) and confers specificity for target substrate degradation (McGinnis et al. 2003; Pierce et al. 2013). Litholdo et al. (2016) identified a member of the major latex protein (MLP) family as a potential LCR F-box target using a proteomic analysis (Litholdo et al. 2016). The MLP genes are involved in plant development and stress resistance (Iwabuchi et al. 2020). For instance, cotton MLP28 might act as a positive regulator of ERF6 in defense against Verticillium dahliae (Yang et al. 2015). MLP423 negatively regulates defenses against fungal infections in apple (He et al. 2020). MLP43 is involved in ABA signal transduction and acts upstream of SnRK2s by directly interacting with SnRK2.6 and ABF1. The overexpression of MLP43 confers drought tolerance to A. thaliana (Wang et al. 2015). On the basis of these results, we proposed a model in which the miR394-F-box-MLP pathway regulates abiotic or biotic plant stress responses and plant development (Fig. 5). This increases our understanding of the molecular mechanisms underlying miR394-mediated plant abiotic/biotic stress tolerance.
Materials and Methods
Plant Materials and Treatments
Foxtail millet cultivar Yugu-1 was used to isolate the miRNA of Sit-miR394 and to examine its expression patterns in different tissue and after hormone treatments. The Yugu-1 seedlings were grown in a greenhouse (16-h day/8-h night at 28 °C day/20 °C night) with nutrient soil, and 15-day-old seedlings were used in the treatments. For the drought treatment, deficit irrigation was applied by withholding watering, while control plants were well watered. The soil water content and relative leaf water content (RWC) were measured using instrument HengMei HM-S and a formula RWC = (fresh weight − dry weight)/(saturated fresh weight − dry weight), respectively, as indicators of stress levels. The plants were harvested at 2, 4, 6 and 10 days after the drought treatment, and then plants were re-watered to permit recovery. For hormone treatments, the seedlings were sprayed independently with 100 µM methyl jasmonate (MeJA), 100 µM ethephon (ETH), 100 µM salicylic acid (SA) and 100 µM abscisic acid (ABA). The plants were collected at 0, 0.5, 2, 6, 12 and 24 h after hormone treatments. The roots, stems and leaves from 15-day-old seedling were also harvested for tissue-specific expression analyses. All the samples were frozen in liquid nitrogen immediately and stored at − 80℃ until use.
Target Validation
RLM-RACE was carried out for target gene and cleaved site validation using a GeneRacer Kit (Invitrogen, Carlsbad, California, US) in accordance with the manufacturer’s instructions. Total RNA was isolated from seedling plants using a Plant RNA reagent (Invitrogen). Then, total RNA was directly ligated to the RNA adapters before being reverse transcribed. The cDNA samples were amplified using nested PCR. The initial PCR was carried out using the GeneRacer 5′-primer (provided in the kit) and gene-specific primers (Supplementary Table 1). Nested PCR was carried out using 2 µL of the initial PCR reaction, the GeneRacer nested primer and gene-specific primer 001465-R. After amplification, the products were gel purified and cloned into the pCAMBIA1305.1-EYFP vector, and at least 20 independent clones were sequenced.
Quantitative Real-Time PCR (qPCR)
Total RNA was isolated from each sample as described above. The first-strand cDNA synthesis of miRNA was performed using miRcute Plus miRNA First-strand cDNA kit and, and the qPCR was carried out using a miRcute Plus qPCR Kit, which contained antisense adaptor primers and the sense primer, as shown in Supplementary Table 1. The U6 and Actin7 gene was used as an internal control, and the primers are listed in Supplementary Table 1. The transgenic Arabidopsis lines and WT RNA were isolated applying RNAprep pure plant plus kit. The first-strand cDNA synthesis of RNA was performed using RNA First-strand cDNA kit and the qPCR was implement using superreal premix plus(SYBR Green). The internal reference was used AtEF1α in Arabidopsis. The above described kits are purchased from Tiangen (Tiangen, Beijing, China).The reaction conditions were set as follows: 95 ℃ for 10 min, followed by 40 cycles of 95 ℃ for 15 s and 58 ℃ for 1 min, followed by a final dissociation curve analysis. All the reactions were performed with three biological replicates per sample. All primer sequences Supplementary Table 1. The relative expressions were calculated using the comparative CT (2−ΔΔCt) method.
Transgenic Arabidopsis Generation and Drought-resistance Evaluation
To generate an overexpression construct, the pre-miR394 with 128-bp flanking the mature miR394 sequence was amplified from foxtail millet genomic DNA. The primers sequence are shown in Supplementary Table 1. The amplified fragment was introduced into the pCAMBIA1305.1-EYFP vector for sequencing confirmation and was then subcloned into the pCAMBIA1305.1-EYFP vector having the 35S promoter (Sangon Biotech Bioengineering Shanghai Co., Ltd.). The construct was transferred into Agrobacterium tumefaciens GV3101 and was transformed into Arabidopsis plants using the vacuum infiltration method. Homozygous plants were used for further analyses.
To assay the germination rate and root growth, transgenic and WT seeds were placed on MS-containing agar plates supplemented with 50% polyethylene glycol (PEG) under a 16-h light/8-h dark photoperiod at 22 ℃. After 5 and 10 days, the germination rates and the root lengths were measured. Drought tolerance assays were carried out on seedlings. Transgenic and WT seeds were planted in pots containing mixed soil (1:1 nutrient soil:vermiculite) and well watered. The seedlings were cultured in a greenhouse under a 16-h light/8-h dark photoperiod at 22 ℃ with 60% humidity. After 12 days, transgenic and WT seedlings were subjected to natural drought until phenotypic differences were evident between the plant types.
Transgenic and WT Arabidopsis Transcriptome Sequencing and Analysis
Transgenic and WT plants were planted under well-watered conditions as described above. After 30 days of growth, total RNAs were extracted form transgenic and WT leaves. The qualified RNAs were sent to Beijing Biomics Technology Co., Ltd (Beijing, China) to prepare and sequence the RNA sequencing libraries using an Illumina HiSeq 2000 sequencer.
The differential expression between drought and control conditions was calculated using DESeq2 software (Love et al. 2014). mRNAs with an absolute log2 fold-change (transgenic/WT) value ≥ 1 and a significance threshold ≤ 0.01 were considered to be significantly differentially expressed mRNAs. A gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was performed (corrected p value < 0.05) using the online tool AgriGO (http://bioinfo.cau.edu.cn/agriGO/).
Conclusion
In this study, we identified the target of sit-miR394. RLM-RACE experiments demonstrated that Sit-miR394 directs the cleavage of the SiFBP6 mRNA. Sit-miR394 was induced by drought stress, and the overexpression of Sit-miR394 in Arabidopsis increased the plants tolerance to drought stress, which suggested that sit-miR394 plays a positive role in responding to drought. Furthermore, Sit-miR394 was also upregulated in foxtail millet after independent exogenous applications of JA, ETH, SA and ABA. A transcriptional analysis indicated that the overexpression of Sit-miR394 affects the expression of a set of stress-related genes, thereby conferring an increased abiotic or biotic stress tolerance to transgenic Arabidopsis.
References
Abd-Hamid NA, Ahmad-Fauzi MI, Zainal Z, Ismail I (2020) Diverse and dynamic roles of F-box proteins in plant biology. Planta 251(3):1–31
Bari R, Jones JDG (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69(4):473–488
Cai Y, Yu X, Hu S, Yu J (2009) A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinform 7(4):147–154
Chand SK, Nanda S, Joshi RK (2016) Regulation of miR394 in response to Fusarium oxysporum f. sp. cepae (FOC) infection in garlic (Allium sativum L.). Front Plant Sci 7:1–12
Chen C, Liu Y, Xia R (2019) Jack of many trades: the multifaceted role of miR528 in monocots. Mol Plant 12(8):1044–1046
de Torres ZM, Zhai B, Jayaraman S, Eleftheriadou G, Winsbury R, Yang R, Truman W, Tang S, Smirnoff N, Grant M (2016) Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol 209(3):1120–1134
Delk NA, Johnson KA, Chowdhury NI, Braam J (2005) CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol 139(1):240–253
Doust AN, Kellogg EA, Devos KM, Bennetzen JL (2009) Foxtail millet: a sequence-driven grass model system. Plant Physiol (Bethesda) 149(1):137–141
Ferdous J, Whitford R, Nguyen M, Brien C, Langridge P, Tricker PJ (2017) Drought-inducible expression of Hv-miR827 enhances drought tolerance in transgenic barley. Funct Integr Genomic 17(2–3):279–292
Gupta S, Garg V, Kant C, Bhatia S (2015a) Genome-wide survey and expression analysis of F-box genes in chickpea. BMC Genomics 16(1):67. https://doi.org/10.1186/s12864-015-1293-y
Guttikonda SK, Valliyodan B, Neelakandan AK, Tran LP, Kumar R, Quach TN, Voothuluru P, Gutierrez-Gonzalez JJ, Aldrich DL, Pallardy SG, Sharp RE, Ho TD, Nguyen HT (2014) Overexpression of AtDREB1D transcription factor improves drought tolerance in soybean. Mol Biol Rep 41(12):7995–8008
He S, Yuan G, Bian S, Han X, Liu K, Cong P, Zhang C (2020) Major latex protein MdMLP423 negatively regulates defense against fungal infections in apple. Int J Mol Sci 21(5):1879. https://doi.org/10.3390/ijms21051879
Iwabuchi A, Katte N, Suwa M, Goto J, Inui H (2020) Factors regulating the differential uptake of persistent organic pollutants in cucurbits and non-cucurbits. J Plant Physiol 245:153094. https://doi.org/10.1016/j.jplph.2019.153094
Jia G, Huang X, Zhi H, Zhao Y, Zhao Q, Li W, Chai Y, Yang L, Liu K, Lu H, Zhu C, Lu Y, Zhou C, Fan D, Weng Q, Guo Y, Huang T, Zhang L, Lu T, Feng Q, Hao H, Liu H, Lu P, Zhang N, Li Y, Guo E, Wang S, Wang S, Liu J, Zhang W, Chen G, Zhang B, Li W, Wang Y, Li H, Zhao B, Li J, Diao X, Han B (2013) A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet 45(8):957–961
Jin W, Wu F (2015a) Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol 15(1):1. https://doi.org/10.1186/s12870-014-0410-4
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799
Joshi T, Yan Z, Libault M, Jeong D, Park S, Green PJ, Sherrier DJ, Farmer A, May G, Meyers BC, Xu D, Stacey G (2010) Prediction of novel miRNAs and associated target genes in Glycine max. BMC Bioinform 11(Suppl 1):S14
Kumar A, Gautam V, Kumar P, Mukherjee S, Verma S, Sarkar AK (2019) Identification and co-evolution pattern of stem cell regulator miR394s and their targets among diverse plant species. BMC Evol Biol 19(1):1–16. https://doi.org/10.1186/s12862-019-1382-7
Lata C, Gupta S, Prasad M (2012) Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 33(3):328–343
Leng X, Song C, Han J, Shangguan L, Fang J, Wang C (2016) Determination of the precise sequences of computationally predicted miRNAs in Citrus reticulata by miR-RACE and characterization of the related target genes using RLM-RACE. Gene 575(2):498–505
Li HY, Cheng HY, Guo Y, Duan M, Ma FF (2018) Progress in the mechanisms of drought tolerance in foxtail millet. J Shanxi Agric Univ (Nat Sci Ed) 38(01):6–10
Litholdo CG, Parker BL, Eamens AL, Larsen MR, Cordwell SJ, Waterhouse PM (2016) Proteomic identification of putative MicroRNA394 target genes in Arabidopsisthaliana identifies major latex protein family members critical for normal development. Mol Cell Proteomics 15(6):2033–2047
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsisthaliana. RNA 14(5):836–843
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. https://doi.org/10.1186/s13059-014-0550-8
McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T, Steber CM (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-Box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15(5):1120–1130
Miyamoto T, Uemura T, Nemoto K, Daito M, Nozawa A, Sawasaki T, Arimura G (2019) Tyrosine kinase-dependent defense responses against herbivory in Arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2019.00776
Moffat CS, Ingle RA, Wathugala DL, Saunders NJ, Knight H, Knight MR (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytiscinerea in Arabidopsis. PLoS ONE 7(4):e35995
Neller K, Klenov A, Guzman JC, Hudak KA (2018) Integration of the pokeweed miRNA and mRNA transcriptomes reveals targeting of jasmonic acid-responsive genes. Front Plant Sci 9:589. https://doi.org/10.3389/fpls.2018.00589
Nguyen XC, Kim SH, Hussain S, An J, Yoo Y, Han HJ, Yoo JS, Lim CO, Yun D, Chung WS (2016) A positive transcription factor in osmotic stress tolerance, ZAT10, is regulated by MAP kinases in Arabidopsis. J Plant Biol 59(1):55–61
Ni Z, Hu Z, Jiang Q, Zhang H (2012) Overexpression of gma-MIR394a confers tolerance to drought in transgenic Arabidopsisthaliana. Biochem Biophys Res Commun 427(2):330–335
Pandey DM, Goswami CL, Kumar B (2003) Physiological effects of plant hormones in cotton under drought. Biol Plant 47(4):535–540
Pang M, Woodward AW, Agarwal V, Guan X, Ha M, Ramachandran V, Chen X, Triplett BA, Stelly DM, Chen ZJ (2009) Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol 10(11):R122
Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23(9):3089–3100
Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RLJ, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan S, Deshaies RJ (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F Box proteins. Cell 153(1):206–215
Qu L, Lin LB, Xue HW (2018) Rice miR394 suppresses leaf inclination through targeting an F-box gene, leaf inclination 4. J Integr Plant Biol 61(4):406–416
Son GH, Wan J, Kim HJ, Nguyen XC, Chung WS, Hong JC, Stacey G (2012) Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol Plant Microbe Interact 25(1):48–60
Song C, Wang C, Zhang C, Korir N, Yu H, Ma Z, Fang J (2010) Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrustrifoliata). BMC Genomics 11(1):431–431
Song JB, Huang SQ, Dalmay T, Yang ZM (2012) Regulation of leaf morphology by microRNA394 and its target leaf curling responsiveness. Plant Cell Physiol 53(9):1669–1669
Song JB, Shu XX, Shen Q, Li BW, Song J, Yang ZM (2015) Altered fruit and seed development of transgenic rapeseed (Brassicanapus) Over-Expressing MicroRNA394. PLoS ONE 10(5):e0125427
Song JB, Gao S, Wang Y, Li BW, Zhang YL, Yang ZM (2016) miR394 and its target gene LCR are involved in cold stress response in Arabidopsis. Plant Gene 5:56–64
Tian X, Song L, Wang Y, Jin W, Tong F, Wu F (2018) miR394 acts as a negative regulator of Arabidopsis resistance to B. cinerea infection by targeting LCR. Front Plant Sci 9:903. https://doi.org/10.3389/fpls.2018.00903
Tiwari S, Lata C, Chauhan PS, Prasad V, Prasad M (2017) A functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Curr Genomics 18(6):469–482
Tobias Dezulian JFPD (2005) Conservation and divergence of microRNA families in plants. Genome Biol 11(6):1–14
Tsai YC, Delk NA, Chowdhury NI, Braam J (2007) Arabidopsis potential calcium sensors regulate nitric oxide levels and the transition to flowering. Plant Signal Behav 2(6):446–454
Wang Y, Yang L, Chen X, Ye T, Zhong B, Liu R, Wu Y, Chan Z (2015) Major latex protein-like protein 43 (MLP4) functions as a positive regulator during abscisic acid responses and confers drought tolerance in Arabidopsis thaliana. J Exp Bot 67(1):421–434
Wang Y, Li L, Tang S, Liu J, Zhang H, Zhi H, Jia G, Diao X (2016) Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genet 17(1):57. https://doi.org/10.1186/s12863-016-0364-7
Xu G, Ma H, Nei M, Kong H (2009) Evolution of F-Box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA 106(3):835–840
Yang CL, Liang S, Wang HY, Han LB, Wang FX, Cheng HQ, Wu XM, Qu ZL, Wu JH, Xia GX (2015) Cotton major latex protein 28 functions as a positive regulator of the ethylene responsive factor 6 in defense against Verticilliumdahliae. Mol Plant 8(3):399–411
Yang Y, Liao W, Yu X, Wang B, Peng M, Ruan M (2016a) Overexpression of MeDREB1D confers tolerance to both drought and cold stresses in transgenic Arabidopsis. Acta Physiol Plant 38(10):243. https://doi.org/10.1007/s11738-016-2258-8
Yu CJ, Sun R, Wang Y, Zhang XZ, Han ZH (2012) Cloning and expression analysis of miR394a under iron deficiency in malus xiaojinensis. Chin Agric Sci Bull 28(28):158–162
Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Gene Dev 25(23):2540–2553
Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D (2009) A genome-wide characterization of microRNA genes in maize. PLoS Genet 5(11):e1000716
Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z (2011) Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett 33(2):403–409
Zhao Y, Wang M, Fu S, Yang W, Qi C, Wang X (2012) Small RNA profiling in two Brassicanapus cultivars identifies MicroRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiol 158(2):813–823
Zhu H, Chen C, Zeng J, Yun Z, Liu Y, Qu H, Jiang Y, Duan X, Xia R (2020) MicroRNA528, a hub regulator modulating ROS homeostasis via targeting of a diverse set of genes encoding copper-containing proteins in monocots. New Phytol 225(1):385–399
Acknowledgements
We thank Lesley Benyon, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This research was funded by Youth Science Foundation of Hebei Province (C2018301088).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Geng, Z., Liu, J., Li, D. et al. A Conserved miR394-Targeted F-Box Gene Positively Regulates Drought Resistance in Foxtail Millet. J. Plant Biol. 64, 243–252 (2021). https://doi.org/10.1007/s12374-021-09303-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-021-09303-8