Abstract
Background
Microsporidia may cause infection in both immunocompromised and immunocompetent populations. The best strategy to control microsporidiosis is obtaining thorough knowledge of its outbreak and pathogenicity.
Purpose
Because of the lack of precise estimation of microsporidia prevalence among Iranian children with cancer, the current study aimed at evaluating the rate of intestinal microsporidia in children undergoing chemotherapy.
Methods
Patients with cancer undergoing chemotherapy in a children’s hospital in Northwestern Iran were studied; 132 stool samples were collected and stained by the Weber and Ryan-blue modified trichrome staining techniques. The extracted DNA samples were evaluated by the nested polymerase chain reaction (PCR) method. All positive isolates were sequenced for genotyping and phylogenetic analysis.
Results
A total of 17 (12.8%) samples were microscopically positive for microsporidia infection, whereas only 14 (10.6%) cases were positive based on nested PCR results. In the positive samples detected with nested PCR, the frequency of Enterocytozoon bieneusi and Encephalitozoon intestinalis infections was 71.4% (n = 10) and 28.6% (n = 4), respectively. After sequencing and phylogenetic analysis, the genotype of E. bieneusi was type D and the sequences of the isolated species were similar to those of the registered ones.
Conclusion
E. bieneusi is a major contributor to microsporidiosis in young immunocompromised patients in Iran. Microsporidia species are well-detected when confirmatory techniques such as molecular methods are in agreement with staining. So, to ensure this, a suggestion has been made to introduce a certain diagnostic test for microsporidiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A large array of data regarding obligate intracellular eukaryotes indicated that they are fungal-related parasites infecting both animals and humans, which belong to the phylum microsporidia [8], and can be quite life-threatening in patients with immunodeficiency [36]. Initially, microsporidia were assumed as protozoa; however, molecular studies put forward the hypothesis that the so-called microorganisms belong to the kingdom of fungi [34]. Over 1300 species of microsporidia are detected, of which 15 species were isolated from humans [4, 21]. Further studies revealed that Enterocytozoon bieneusi (E. bieneusi) and Encephalitozoon intestinalis (E. intestinalis) were the most prevalent human infectious agents; E. bieneusi was directly related to chronic diarrhea and weight loss in immunocompromised individuals [16, 29, 33]. The lifecycle of microsporidia consists of proliferative and spore formation stages, spores presenting as infective agents. Spore formation is the only stage in the lifecycle of the microorganism, which let it even survive outside the host cell [8, 19].
The disease has several transmission methods including fecal–oral or oral–oral route, inhalation or ingestion of spores, and human to human (anthroponotic) or animal to human (zoonotic) transmissions [6, 9]. The clinical symptoms of microsporidiosis are fever, weight loss, and chronic diarrhea, although in rare cases disseminated infections are also reported [7, 20]. Microsporidia may cause infection both in immunocompromised (i.e., patients with AIDS, the ones with cancer receiving high-dose chemotherapy, and organ transplant recipients) and immunocompetent populations [9, 17, 22]). The immune status determines the clinical signs of microsporidiosis [34].
The clinical samples suspected of microsporidiosis are usually traced by various techniques such as light microscopy, transmission electron microscopy (TEM), immunofluorescence assays (IFAs), cell culture, and molecular methods [25, 4], among which the light microscopy using modified trichrome staining technique is the most routinely utilized method [28, 35]. It is noteworthy that due to the limitations of microscopic method to detect the minute spores of these microorganisms, today, the application of sensitive molecular methods for diagnostic purposes is common [26, 33]. Besides, thanks to molecular analysis, identification of microsporidia species, which are otherwise undetectable, is feasible, contributing to the diagnosis of infection [16, 33]. The estimated prevalence of microsporidiosis in immunocompromised patients, the ones with gastroenteritis, and those without diarrhea were 8.18%, 0.5%, and 4.1%, respectively [13].
Current studies are indicative of the fact that children with immunodeficiency are susceptible to microsporidia infections, thus, the best strategy to control this disease is gaining thorough knowledge of its outbreak and pathogenicity [7, 24]. Due to the lack of precise estimations of microsporidia prevalence in Iranian children with cancer, the current study aimed at evaluating the rate of intestinal microsporidia in pediatric patients undergoing chemotherapy.
Materials and Methods
Study Design and Sample Collection
Children with cancer undergoing chemotherapy in a children’s hospital affiliated to Tabriz University of Medical Sciences, Tabriz, Iran, were enrolled in the current cross-sectional study from May 2015 to June 2016. The protocol of the study was approved by the Ethics Committee of the university (TBZMED.REC.1394.61); parents’ consent was also obtained before undertaking the study. Next, demographic and clinical data such as age, gender, chemotherapy rounds, place of residence, contact with animals, and cancer type were accurately recorded (Table 1). Stool samples were collected from 132 children (77 males and 55 females) within the age range of 3–15 years (mean age: 9 years; majority 3–5 years) with either leukemia (n = 79) or other malignancies (n = 53). The majority of the patients were urban dwellers (n = 95), whereas the rest came from rural areas. Merely, seven cases had contact with animals. All of the patients underwent chemotherapy from 1 month to over 1 year before the onset of the study. Initially, a 2 mL blood sample was collected from each patient for neutrophil count, as a laboratory indicator of immune status. Then, stool samples with no preservatives were stored at − 20 °C.
Light Microscopy
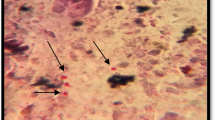
All macroscopic features of stool samples including odor, color, consistency, and presence of mucus or blood were thoroughly examined. Then, stool smears were stained with the Weber and Ryan-blue modified trichrome staining techniques [28, 35].
DNA Extraction
DNA was extracted from frozen samples using the stool DNA extraction mini kit (Yekta Tajhiz Azma, Iran). Briefly, 500 μL of stool suspension was washed three times with phosphate-buffered saline (pH = 7.2) followed by centrifugation at 10,000×g for 5 min, and then the supernatant was discarded. The final pellets were lysed by the addition of 600 μL of lysis buffer, 40 μL of proteinase K, and 200 mg of glass beads and vortexed for 5 min, followed by incubation at 60 °C for 2 h. Samples were then vortexed for further 30 s every 30 min. Finally, the supernatant was transferred to the stool DNA extraction mini kit and the procedure continued according to the manufacturer’s instructions. Eventually, the purified DNA was stored at − 20 °C.
PCR Amplification
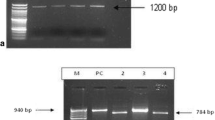
In the current study, SSU-ITS-LSU (rRNA) fragment was amplified by nested polymerase chain reaction (PCR) technique using the specific primers previously described by Katzwinkel-Wladarsch [15, 27]. Forward primers MSP-1 (TGAATG [G/T] GT CCC TGT) and MSP-3 (GGA ATT CAC ACC GCC CGTC [A/G] [C/T] TAT) targeted the 3′region of the SSU and identified the microsporidia species. Reverse primers MSP-2A (TCA CTC GCC GCT ACT) and MSP-4A (CCA AGC TTA TGC TTA AGT [C/T] [A/C] AA [A/G] G GGT) identified E. intestinalis and MSP-2B (GTT CAT TCG CAC TAC T) and MSP-4B (CCA AGC TTA TGC TTA AGT CCA GGG AG) targeted the 5′ region of the LSU of E. bieneusi. PCR was performed with a final volume of 25 μL, containing 20 pmol of each primer, 10X PCR buffer, 10 mM of dNTP, 50 mM of MgCl2, and 5 U/µL of Taq DNA polymerase. The first-round PCR mixture included 1 µL of each primer, MSP-1, MSP-2A (for E. intestinalis), as well as MSP-2B (for E. bieneusi) and 1 µL of the template DNA. The second-round PCR reaction contained 1 µL of each primer, MSP-3, MSP-4A (for E. intestinalis), MSP-4B (for E. bieneusi), and 1 µL of the first amplification reaction mixture. Amplicon size for the MSP-3–MSP-4B (E. bieneusi) product was about 500 bp, and for the MSP-3–MSP-4A (E. intestinalis), the product was nearly 300 bp. The amplification process was conducted under the following conditions: initial denaturation at 95 °C for 5 min, followed by 46 cycles including 45 s at 95 °C as denaturation, 45 s of annealing at 55 °C, and extension at 72 °C for 1 min. A 10-min final extension at 72 °C was added after 46 cycles. All the products were electrophoresed on 2% agarose gel and stained with safe staining for the amplified PCR products to be visualized under ultraviolet (UV) light.
Genotyping and Phylogenetic Analysis
ABI PrismTM 3130 Genetic Analyzer (Applied Biosystems, USA) was used for sequencing of all PCR-positive isolates. After comparing the obtained sequences with data available on GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi), multiple alignments were conducted by ClustalW using BioEdit version 7.2.5 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Then, to determine the similarity and difference rates of sequences, the MegAlign program (DNASTAR, Madison, WI, USA) was employed. Finally, maximum parsimony algorithm with kimura 2-parameter model in MEGA 5.05 software [32] by 1000 bootstrap re-sampling was considered for phylogenetic analysis.
Statistical Analysis
The obtained data are expressed as percentage. STATA version 11 (STATA Corp., USA) was used for statistical analysis. The rate of microsporidiosis and the associated risk factors were analyzed by Chi square and the Fisher exact tests. Logistic regression analysis was used to determine the relationship between the rate of microsporidiosis and the identified risk factors. P value < 0.05 was considered statistically significant.
Results
In the current study, 17 (12.8%) out of 132 samples were microsporidia positive based on microscopic evaluations, whereas 14 (10.6%) cases were positive based on nested PCR results, of which 12 (85.7%) belonged to urban citizens and 2 (14.3%) to rural residents. Moreover, 2 out of 17 positive cases (11.7%) were coinfected with microsporidia and Cryptosporidium spp. Among the samples found positive by the nested PCR, E. bieneusi and E. intestinalis depicted 71.4% (n = 10) and 28.6% (n = 4) of total cases, respectively. In addition, the number of male patients with microsporidiosis was higher than female ones, although there was no significant difference between the genders. Moreover, the age group of 3–5 years, patients with leukemia, and a clear history of taking corticosteroid drugs demonstrated the highest susceptibility to microsporidia (11.79%). Other disease variables did not depict any significant difference between age groups, according to the results (P = 0.59). Although 62 patients with neutrophil count of 1000–1500 cell/mm3 were considered positive, microsporidiosis was not a contributing factor [95% confidence interval (CI) 6.69–23.45; P = 0.56]. Merely, 9 out of 66 (13.6%) patients with diarrhea were positive for microsporidia and only 8 (19.51%) cases were reported with soft diarrhea. Thus, there was an insignificant difference between the cases with and without diarrhea (P = 0.15). Finally, there were no significant relationships between microsporidiosis and other variables such as contact with domestic animals, stage of chemotherapy, and history of immunodeficiency based on the statistical analyses. Additional results are summarized in Table 1. Other enteric protozoa revealed under the light microscopy examinations were Giardia lamblia cysts in four (3%) cases, Chilomastix mesnili trophozoites in one (0.8%) case, and Entamoeba coli cysts in two (1.5%) cases. In the present study, sequencing results of all positive isolates showed a similarity of 98–100% compared with GenBank sequences. SSU-ITS-LSU rRNA multiple sequence alignment compared the nucleotide sequences with specify similarities and differences between sequences. Sequence divergence and percent identity are shown in Figs. 1 and 2. E. bieneusi isolates in this study were genotype D and based on phylogenetic analysis, all E. bieneusi and E. intestinalis isolates were similar to those of other registered sequences (Figs. 3, 4). KX685189 and KX685190 are the accession numbers of E. bieneusi and E. intestinalis, respectively, registered in GenBank based on the current investigation.
Discussion
Humans and an expansive range of animals are prone to infection with microsporidia worldwide [27, 29]. A wide variety of diagnostic methods and investigations in terms of gender, age, and immunodeficiency types are considered as barriers to determine and compare the precise outbreak of the infection globally, which necessitates further comprehensive molecular and phylogenic studies with reliable results. Immunocompromised patients are in urgent need for the accurate diagnosis of such infectious diseases, since grave problems as well as interference with disease control procedures may arise which can eventually exacerbate their quality of life [3, 5, 18]. There is no thorough investigation of microsporidia species in stool samples of Iranian children with cancer, especially the ones undergoing chemotherapy, and that is why the current study solely focused on intestinal microsporidia.
In the current study, the staining method revealed 17 (12.8%) positive cases, whereas nested PCR uncovered 14 (10.6%) E. bieneusi and E. intestinalis-positive patients out of 132, with E. bieneusi being the main culprit affecting immunocompromised patients. Askari et al. [2], reported the infection prevalence of 10.56% among all cases, confirming the relatively high prevalence of E. bieneusi compared with E. cuniculi, which was quite parallel to the findings of the current study. Furthermore, Tavalla et al. [33] reported E. bieneusi as the dominant species isolated from immunocompromised individuals, followed by E. intestinalis. In a study conducted by Eligio-Garcia et al. [11], PCR was more sensitive than staining in pathogen detection, and they did not report any relationships between microsporidiosis and diarrhea. Results of the current study were consistence with those of their study. These results affirmed a high prevalence of microsporidiosis among patients with diarrhea (13.63%); however, the current study did not confirm any significant relationships between microsporidiosis and diarrhea.
A number of factors can contribute to the differences between the results reported from microscopic and molecular investigations. Lack of experienced microscopists can be the first reason; second, as indicated previously, the miniscule spores of this microorganism can lead to false-negative results. Despite the high sensitivity of the staining method, false-positive results might be attained due to the striking resemblance of microsporidia spores to other fungal spores [28]. However, the probability of infection with species other than E. bieneusi and E. intestinalis cannot be ruled out due to the absence of proper primers in the deployed molecular techniques. It should also be noted that the likelihood of false-positive results for microsporidiosis obtained from staining methods and false-negative results from PCR can be attributed to the presence of inhibitors during DNA extraction. To further confirm the higher sensitivity of the molecular techniques compared with routine microscopic methods, El Sobky et al., asserted that PCR should be deployed along with staining methods as a confirmatory technique to enhance the differentiation of Microsporidia species [10]. Likewise, the enhanced sensitivity of PCR method was further proved by a study conducted on HIV+ patients in Iran by Ghorbanzadeh et al. [12], as well as the study by Hamamci et al. [14], that reported the prevalence of microsporidia infection as 69.9% in 93 patients with cancer undergoing chemotherapy, with fifty-one patients complaining of diarrhea. Unlike the present study, a significant correlation was observed between microsporidia infection and diarrhea in the mentioned studies, but in line with the current findings, the highest prevalence was observed in male patients, a finding well-substantiated by other studies [1, 23, 31]. Various cultural, social, and environmental factors trigger such gender-dependent microsporidiosis distribution.
Molecular techniques can also be very useful for identifying the genotypes and phylogenetic analysis of microsporidia. Khanduja et al. [17] reported eight genotypes of E. bieneusi in renal transplant recipients. In the current study, the genotype of E. bieneusi was type D and based on phylogenetic analysis, all isolates were similar to those of other registered sequences.
Despite a report by Sokolova et al., regarding the co-existence of more than one species in human and animal stool samples [30], the present findings reported no mixed infections. In this study, patients with leukemia receiving corticosteroid drugs and the ones with neutrophil count of < 1500 cells/mm3 showed positive results, lending support to the fact that opportunistic pathogen hazards are commonly posed to immunocompromised patients as a result of the disease itself, cytotoxic treatments, or both. Furthermore, 7 positive cases out of 55 young patients with cancer (12.7%) had a rather long-term exposure to chemotherapy (1 year or above). Thus, an urgent need is felt to incorporate epidemiological investigations to detect microsporidia in stool samples of children with cancer, particularly when the patient complains of diarrhea and no other pathogens are isolated.
Conclusion
Enterocytozoon bieneusi is known as a major contributor to microsporidiosis in young immunocompromised patients in Northwestern Iran, with immunosuppressivity and childhood as the main risk factors. Microsporidia species are well-detected when confirmatory techniques such as molecular methods (nested PCR) are consistent with staining. It is recommended that oncology ward of children’s hospitals should introduce a diagnostic method for microsporidiosis.
References
Agholi M., Hatam G.R., Motazedian M.H. 2013. Microsporidia and coccidia as causes of persistence diarrhea among liver transplant children: incidence rate and species/genotypes. The Pediatric Infectious Disease Journal, 32, 185–187. https://doi.org/10.1097/inf.0b013e318273d95f
Askari Z., Mirjalali H., Mohebali M., Zarei Z., Shojaei S., Rezaeian T., Rezaeian M. 2015. Molecular detection and identification of zoonotic microsporidia spore in fecal samples of some animals with close-contact to human. Iranian Journal of Parasitology, 10, 381–388
Berahmat R., Mahami-Oskouei M., Rezamand A., Spotin A., Aminisani N., Ghoyounchi R., Madadi S. 2017. Cryptosporidium infection in children with cancer undergoing chemotherapy: how important is the prevention of opportunistic parasitic infections in patients with malignancies? Parasitology Research, 116, 2507–2515. https://doi.org/10.1007/s00436-017-5560-5
Cali A., Takvorian N.R.P.M. 2011. Microsporidiosis, p 61–76. In Meyers WM, Firpo A, Wear DJ (ed), Topics on the pathology of protozoan and invasive arthropod diseases. Armed Forces Institute of Pathology (AFIP), Washington, DC. http://oai.dtic.mil/oai/oai?verb_getRecord&metadataPrefix_html&identifier_ADA545141.
Chandramathi S., Suresh K., Anita Z.B., Kuppusamy U.R. 2012. Infections of Blastocystis hominis and microsporidia in cancer patients: are they opportunistic? Transactions of the Royal Society of Tropical Medicine and Hygiene, 106, 267–269. https://doi.org/10.1016/j.trstmh.2011.12.008
Dengjel B., Zahler M., Hermanns W., Heinritzi K., Spillmann T., Thomschke A., Löscher T., Gothe R., Rinder H. 2001. Zoonotic potential of Enterocytozoon bieneusi. Journal of Clinical Microbiology, 39, 4495–4499. https://doi.org/10.1128/jcm.39.12.4495-4499.2001
Desportes I., Charpentier YL., Galian A., Bernard F., Cochand‐Priollet B., Lavergne A., Ravisse P., Modigliani R. 1985. Occurrence of a new microsporidan: Enterocytozoon bieneusi ng, n. sp., in the enterocytes of a human patient with AIDS. The Journal of Protozoology, 32, 250–254.
Didier E.S., Vossbrinck C.R., Stovall M.E., Green L.C., Bowers L., Fredenburg A., Didier P.J. 2004. Diagnosis and epidemiology of microsporidian infections in humans. The Southeast Asian Journal of Tropical Medicine and Public Health, 35, 65–81.
Didier E.S., Weiss L.M. 2006. Microsporidiosis: current status. Current Opinion in Infectious Diseases, 19, 485–492. https://doi.org/10.1097/01.qco.0000244055.46382.23
El Sobky M., El Nahas N. 2012. Detection and differentiation between Enterocytozoon bieneusi and Encephalitozoon intestinalis species in cancer patient’s stools using PCR compared with different staining methods. Parasitology United Journal, 5, 20–26.
Eligio-García L., Cano-Estrada A., Cortés-Campos A., Medina-Sansón A., Jiménez-Cardoso E. 2013. Identification of Microsporidium spp. in patients with acute lymphoblastic leukemia. Boletín Médico del Hospital Infantil de México, 70, 26–30.
Ghorbanzadeh B., Sadraei J., Emadi H. 2012. Diagnosis of Cryptosporidium and intestinal Microsporidia in HIV/AIDS patients with staining and PCR methods on 16srRNA gen. Arak Medical University Journal, 15, 37–47.
Ghoyounchi R., Ahmadpour E., Spotin A., Mahami-Oskouei M., Rezamand A., Aminisani N., Ghojazadeh M., Berahmat R., Mikaeili-Galeh T. 2017. Microsporidiosis in Iran: a systematic review and meta-analysis. Asian Pacific Journal of Tropical Medicine, 10, 341–350. https://doi.org/10.1016/j.apjtm.2017.03.017
Hamamcı B., Çetinkaya Ü., Berk V., Kaynar L., Kuk S., Yazar S. 2015. Prevalence of Encephalitozoon intestinalis and Enterocytozoon bieneusi in cancer patients under chemotherapy. Mikrobiyoloji Bülteni, 49, 105–113.
Katzwinkel-Wladarsch S., Lieb M., Helse W., Loscher T., Rinder H. 1996. Direct amplification and species determination of microsporidian DNA from stool specimens. Tropical Medicine & International Health, 1, 373–378.
Khanduja S., Ghoshal U., Agarwal V., Pant P., Ghoshal U.C. 2017a. Identification and genotyping of Enterocytozoon bieneusi among human immunodeficiency virus infected patients. Journal of Infection and Public Health, 10, 31–40. https://doi.org/10.1016/j.jiph.2016.01.005
Khanduja S., Ghoshal U., Ghoshal U.C. 2017b. Phylogenetic analysis of genetically distinct Enterocytozoon bieneusi infecting renal transplant recipients. Acta Parasitologica, 62, 63–68. https://doi.org/10.1515/ap-2017-0007
Lobo M.L., Xiao L., Antunes F., Matos O. 2012. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and-negative patients. International Journal for Parasitology, 42, 197–205. https://doi.org/10.1016/j.ijpara.2011.12.002
Martín-Hernández R., Higes M., Sagastume S., Juarranz Á., Dias-Almeida J., Budge G.E., Meana A., Boonham N. 2017. Microsporidia infection impacts the host cell’s cycle and reduces host cell apoptosis. PLoS One, 12, e0170183. https://doi.org/10.1371/journal.pone.0170183
Mathews A., Hotard A., Hale-Donze H. 2009. Innate immune responses to Encephalitozoon species infections. Microbes and Infection, 11, 905–911. https://doi.org/10.1016/j.micinf.2009.06.004
Mathis A., Weber R., Deplazes P. 2005. Zoonotic potential of the microsporidia. Clinical Microbiology Reviews, 18, 423–445. https://doi.org/10.1128/cmr.18.3.423-445.2005
Mirjalali H., Mirhendi H., Meamar A.R., Mohebali M., Askari Z., Mirsamadi E.S., Rezaeian M. 2015. Genotyping and molecular analysis of Enterocytozoon bieneusi isolated from immunocompromised patients in Iran. Infection, Genetics and Evolution, 36, 244–249. https://doi.org/10.1016/j.meegid.2015.09.022
Mirjalali H., Mohebali M., Mirhendi H., Gholami R., Keshavarz H., Meamar A.R., Rezaeian M. 2014. Emerging intestinal microsporidia infection in HIV +/AIDS patients in Iran: microscopic and molecular detection. Iranian Journal of Parasitology, 9, 149–154.
Nadham Kadham M., Maysloon A.A.S. 2012. Microsporidiosis among children with malignant diseases in Basrah, Iraq. Pakistan Journal of Medical Sciences, 28, 621–624.
Nasarudin S.N., Zainudin N.S., Bernadus M., Nawi A.M., Hanafiah A., Osman E. 2015. Loop-mediated isothermal amplification for rapid molecular detection of Enterocytozoon bieneusi in faecal specimens. Journal of Medical Microbiology, 64, 1329–1334. https://doi.org/10.1099/jmm.0.000156
Nooshadokht M., Sharifi I., Mohammadi M.A., Pirestani M., Afgar A., Mahootchi A., Salari S. 2017. Intestinal microsporidiosis in Iran: infection in immune-compromised and immunocompetent patients. Current Medical Mycology, 3, 30–36. https://doi.org/10.18869/acadpub.cmm.3.1.30
Pirestani M., Sadraei J., Forouzandeh M. 2013. Molecular characterization and genotyping of human related microsporidia in free-ranging and captive pigeons of Tehran, Iran. Infection, Genetics and Evolution, 20, 495–499. https://doi.org/10.1016/j.meegid.2013.10.007
Ryan N.J., Sutherland G., Coughlan K., Globan M., Doultree J., Marshall J., Baird R., Pedersen J., Dwyer B. 1993. A new trichrome-blue stain for detection of microsporidial species in urine, stool, and nasopharyngeal specimens. Journal of Clinical Microbiology, 31, 3264–3269.
Sak B., Brady D., Pelikánová M., Květoňová D., Rost M., Kostka M., Tolarová V., Hůzová Z., Kváč M. 2011. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. Journal of Clinical Microbiology, 49, 1064–1070. https://doi.org/10.1128/jcm.01147-10
Sokolova O.I., Demyanov A.V., Bowers L.C., Didier E.S., Yakovlev A.V., Skarlato S.O., Sokolova Y.Y. 2011. Emerging microsporidian infections in Russian HIV-infected patients. Journal of Clinical Microbiology, 49, 2102–2108. https://doi.org/10.1128/jcm.02624-10
Tabatabaie F., Tafreshi Z.A., Shahmohammad N., Pirestani M. 2015. Molecular detection of microsporidiosis in various samples of Iranian immunocompromised patients. Journal of Parasitic Diseases, 39, 634–638. https://doi.org/10.1007/s12639-014-0432-8
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 2731–2739. https://doi.org/10.1093/molbev/msr121
Tavalla M., Mardani-Kateki M., Abdizadeh R., Nashibi R., Rafie A., Khademvatan S. 2017. Molecular identification of Enterocytozoon bieneusi and Encephalitozoon spp. in immunodeficient patients in Ahvaz, Southwest of Iran. Acta Tropica, 172, 107–112. https://doi.org/10.1016/j.actatropica.2017.04.015
Texier C., Vidau C., Viguès B., El Alaoui H., Delbac F. 2010. Microsporidia: a model for minimal parasite–host interactions. Current Opinion in Microbiology, 13, 443–449. https://doi.org/10.1016/j.mib.2010.05.005
Weber R., Bryan R.T., Owen R.L., Wilcox CM., Gorelkin L., Visvesvara G.S. 1992. Improved light-microscopical detection of microsporidia spores in stool and duodenal aspirates. The enteric opportunistic infections working group. The New England Journal of Medicine, 326, 161–166. https://doi.org/10.1056/nejm199201163260304
Zainudin N.S., Nasarudin S.N.S., Periyasamy P., Moktar N., Noordin R., Osman E. 2016. Diagnosis of disseminated microsporidiosis: detection of circulating Enterocytozoon bieneusi DNA in blood of HIV/AIDS patients. Tropical Biomedicine, 33, 761–770.
Acknowledgements
This study was financially supported by Pediatric Health Research Center, Tabriz University of Medical Sciences, Iran. This is a report of a database from the master’s thesis of the first author registered in Tabriz University of Medical Sciences (Thesis number: 93/2–4/12).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ghoyounchi, R., Mahami-Oskouei, M., Rezamand, A. et al. Molecular Phylodiagnosis of Enterocytozoon bieneusi and Encephalitozoon intestinalis in Children with Cancer: Microsporidia in Malignancies as an Emerging Opportunistic Infection. Acta Parasit. 64, 103–111 (2019). https://doi.org/10.2478/s11686-018-00012-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11686-018-00012-w