Abstract
The relative expression of developmentally regulated genes was analyzed during zygotic embryo development in Pinus taeda and somatic embryo development/maturation in P. taeda and Pinus oocarpa. The following four embryo samples were studied: zygotic pro-embryos and somatic embryogenic suspensor masses; round/globular, early cotyledonary, and late cotyledonary. The relative transcript levels of six genes of interest: legumin-/vicilin-like, group 4 late embryogenesis abundant, homeodomain-leucine zipper I, 26S proteasome regulatory subunit S2, and clavata-like, associated with different aspects of embryo development, were analyzed by real-time PCR. In both pine species, the relative transcript levels for legumin-/vicilin-like storage proteins and the late embryogenesis abundant protein accumulated gradually through somatic embryo maturation, in contrast to zygotic embryos, where transcripts increased significantly to their highest levels at the late cotyledonary stage. The homeodomain-leucine zipper I relative transcript accumulation pattern differed between somatic and zygotic embryos. This difference was attributed to differences in cell-type compositions between the embryogenic suspensor masses and pro-embryos. Additionally, in P. oocarpa, the transcript levels of homeodomain-leucine zipper I remained high after the transfer of somatic embryos to maturation conditions, but declined during maturation in P. taeda. The highest 26S proteasome regulatory subunit S2 relative transcript levels in P. taeda were in round/globular somatic and zygotic embryos and in embryogenic suspensor masses and round/globular P. oocarpa somatic embryos. Finally, the relative transcripts levels for the clavata-like gene were more predominant in round/globular, early cotyledonary, and embryogenic suspensor masses in P. taeda and P. oocarpa, respectively. Somatic embryos exhibited relative gene expression patterns similar to their zygotic counterparts, although some differences were noted between zygotic and somatic embryos, as well as between the different pine somatic embryo systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In higher plants, zygotic embryo formation is divided into the following 3 phases: morphogenesis, maturation and desiccation (West and Harada 1993). These phases correspond to the definition of the shoot–root plant body pattern, the formation of storage organs (such as cotyledons), and the conversion to dormancy (Goldberg et al. 1994). Somatic embryogenesis is the process in which cells within the plant are induced to form somatic embryos through the manipulation of culture medium components and plant growth regulators (PGRs; von Arnold et al. 2002) without the involvement of gamete fusion. Somatic embryos pass through similar stages of morphological development as they are exposed to culture conditions that attempt to mimic the growth conditions of their zygotic embryo counterparts (Ikeda and Kamada 2006). Thus, somatic embryogenesis has been used as a model system to study physiological, biochemical, and morphological pathways during zygotic embryogenesis (Zimmerman 1993) and to reveal novel pathways and gene interactions during embryo development (Stasolla et al. 2002; Quiroz-Figueroa et al. 2006).
Somatic embryogenesis in loblolly pine (Pinus taeda) was first reported more than 20 years ago (Gupta and Durzan 1987). Various improvements have been made to increase the number of “normal” plants produced, with a major emphasis placed on the improvement of initiation rates, especially in P. taeda (Pullman and Johnson 2002; Pullman et al. 2003a; Pullman et al. 2005; Pullman et al. 2006). However, many of the current protocols for somatic embryogenesis report low numbers of mature embryos per gram of fresh weight. In some cases, somatic embryos do not fully mature, resulting in slow germination and initial growth (Pullman et al. 2003a). With respect to Oocarpa pine (Pinus oocarpa), the most common pine species in the southern half of Mexico and Central America (Dvorak et al. 2009), somatic embryogenesis has only recently been reported (Lara-Chavez et al. 2011). Full seedling development was obtained from somatic embryos initiated from immature zygotic embryos; however, improvements are still needed to achieve a more reliable protocol across additional cell lines.
Currently, it is possible to distinguish embryogenic tissue from non-embryogenic tissue through visual observations and the use of histochemical stains and molecular markers. Genes associated with embryogenesis (SERK, LEC1, FUS3, and ABI3) are differentially expressed between embryogenic and non-embryogenic cultures (Yang and Zhang 2010). Following the induction and multiplication of embryogenic tissue, maturation of somatic embryos has been monitored using markers for storage protein gene expression and storage protein accumulation (Sterk and de Vries 1993). In addition, late embryogenesis abundant or LEA proteins are associated with the accumulation of abscisic acid (ABA) during maturation and embryo protection during desiccation (Stasolla et al. 2002). A better understanding of the underlying physiological differences between zygotic and somatic embryo development through the analysis of gene expression patterns has the potential to improve in vitro protocols (Bonga et al. 2010). Gene expression analyses can be a useful tool for monitoring embryo development from the acquisition of embryogenic competence through to maturation (Feher et al. 2003). Ideal “marker” genes should be universal, sensitive, detectable in small amounts of tissue, and reveal specific processes characteristic of each of the developmental stages or transitions.
Of the various PGRs, ABA regulates several essential processes during embryo maturation. More than 150 genes from a range of species were ABA-inducible (Giraudat et al. 1994). ABA is supplied to the pine (conifer) zygotic embryo through the megagametophyte (MG); a lack of this tissue during in vitro developmental conditions has made the application of ABA essential for the normal maturation of somatic embryos. The concentration of ABA supplied in vitro for the culture of somatic embryos in Pinus species varies from 15 μM in Pinus elliottii (Newton et al. 2005) to 120 μM in Pinus pinaster (Lelu-Walter et al. 2006). The aim of this study was to assess the relative expression of selected genes associated with embryogenesis in 4 samples of embryo development for both P. taeda and P. oocarpa. Analysis of gene expression could demonstrate the fidelity of the somatic embryo induction and development, identify changes that could be made to the culture medium and conditions to enhance the quality of the somatic embryos, and provide a developmental comparison between somatic embryos of two pine species. The following genes were analyzed for expression: legumin-like, vicilin-like (storage proteins), a group 4 LEA (late embryo development and desiccation tolerance), homeodomain-leucine zipper I (HD-Zip I) protein transcription factor, 26S proteasome regulatory subunit S2 (RPN1; ubiquitin-mediated proteolysis), and clavata-like protein (meristem development). This is the first report describing gene expression in somatic embryos of P. oocarpa and a comparison with a similar developmental series for somatic embryos of P. taeda.
Materials and Methods
Plant material.
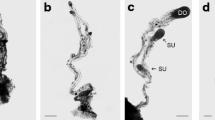
Collection of zygotic embryo samples. Cones of P. taeda were collected weekly from the Reynolds Homestead (Critz, VA), from June 10 to September 15, 2009. Cones were opened and seeds were collected for isolation of zygotic embryos. MGs were excised from the seeds and zygotic embryos were dissected. Zygotic embryos were classified in the following four stages (Fig. 1a–d ): (1) pro-embryo (PE), round/globular (RG), early cotyledonary (EC), and late cotyledonary (LC). Cones were collected from June 10, but discernable PE were not identified until July 14 and were then collected during the week of July 28. RG, EC, and LC samples were observed and collected from August 5, 12, and 25, respectively. Tissue was extracted from the cone collections until September 9, when the majority of the embryos were at the LC stage. Observations were made with a zoom stereomicroscope SZX7 (Olympus America Inc. Center Valley, PA), and collected samples were frozen in liquid nitrogen and kept at −80°C until RNA was extracted.
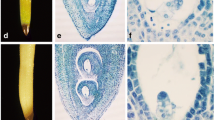
Developmental stages of zygotic embryos of P. taeda (a–d) and somatic embryos for P. taeda (e–h) and P. oocarpa (i–l): (a) pro-embryo; (e, i) embryogenic suspensor mass (ESM) after double staining; (b, f, j) round/globular (RG) stage; (c, g, k) early cotyledonary (EC) stage; and (d, h, l) late cotyledonary (LC) stage. (Bar in e, h, i, l = 1 mm; Bar in a, b, c, d, f, g, j, k = 0.1 mm). Circle denotes zygotic embryo within megagametophyte (b–d).
Collection of somatic embryo samples. Embryogenic P. taeda (courtesy of Dr. Scott Merkle, University of Georgia, Athens, GA) and P. oocarpa cell cultures (Lara-Chavez et al. 2011) were used to produce the somatic embryos. Initial embryogenic suspensor masses (Fig. 1e, i ) were confirmed by histological methods using double staining with acetocarmine and Evan’s blue (Lara-Chavez et al. 2011). These cultures produced morphologically correct embryos through all stages, eventually yielding LC embryos capable of germination (Fig. 1e–l ). Classification of somatic embryo samples in both pine species was based on morphological similarities with their counterpart zygotic embryos of P. taeda, following a similar characterization for somatic embryos of Picea glauca (Dong et al. 1997). Briefly, the samples were characterized as follows: (1) ESM (similar to zygotic PEs) collected from cultures on maintenance medium consisting of small cytoplasmic cells and long vacuolated suspensor-like cells; (2) RG, in which the heads of the embryos were opaque and round-shaped; (3) EC embryos, characterized by small cotyledon primordia below the circumference of a prominent central meristem; and (4) LC embryos, fully mature in appearance with well-formed, elongated cotyledons and still hydrated. The last three samples were collected after cultures were transferred to maturation medium. For maturation, ESM cultures of both species were placed on pre-maturation medium (multiplication medium lacking PGRs) for 1 wk. For P. taeda, the cultures were resuspended with PGR-free maintenance medium at concentration of 20 % (w/v); 2 ml was spread per plate containing maturation medium, and the remaining liquid medium was removed from each plate. For P. oocarpa, small clumps of ESM were placed on the maturation medium. For both pine species, the maturation medium consisted of 927 basal medium (Pullman et al. 2003b) with 6 % (w/v) maltose, 40 μM ABA, and 0.3 % (w/v) Phytagel (Phytotechnology Labs, Shawnee Mission, KS) instead of Gelrite. Maturation duration was 8 and 15 wk for P. taeda and P. oocarpa, respectively. Collected samples were frozen in liquid nitrogen and kept at −80°C until RNA extraction.
RNA extraction and complementary deoxyribonucleic acid (cDNA) synthesis.
Total RNA was extracted from zygotic and somatic embryo samples using the Concert™ Plant RNA Reagent (Invitrogen, Carlsbad, CA,). To eliminate any residual genomic DNA present in the samples, P. taeda RNA samples were treated with RQ1 RNase-Free DNase (Promega, Madison, WI) and P. oocarpa RNA samples with DNA-free kit (Ambion, Austin, TX) according to the manufacturer’s protocols. The quantity of isolated RNA both before and after DNase treatment was measured using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). Total RNA was quantified and the integrity verified by agarose gel electrophoresis. Because of the small amount of tissue collected from PE of P. taeda zygotic embryos, the amount of RNA was too limiting to proceed through the entire range of experiments. Thus, all of the P. taeda zygotic embryo RNA samples were subjected to in vitro amplification using the T7 RNA Polymerase amplification method (van Gelder et al. 1990). Therefore, total RNA from each sample underwent one round of amplification using the MessageAmp II aRNA kit (#AM1751, Ambion), according to the manufacturer’s protocol, using an in vitro transcription time of 14 h. Subsequently, RNA of each sample was used for cDNA synthesis using the iScript™ cDNA synthesis kit (Bio-Rad, Hercules, CA) for P. taeda somatic embryo samples; and SuperScript III, First-Strand Synthesis System for RT-PCR (Invitrogen) for P. taeda zygotic embryo samples and all P. oocarpa samples, using random hexamer and oligo (dT) primers, respectively. Subsequently, cDNA was verified via PCR using the two endogenous control primers (Table 1).
Relative real-time PCR.
The relative transcript levels of the six genes of interest (Table 1) were analyzed by real-time PCR. Publicly accessible databases were searched to identify Pinus nucleotide sequences for primer design. If Pinus sequences were not available, closely related Picea sequences were used for primer design. Primers were initially tested for their ability to generate the correct-sized PCR products in both Pinus species (Supplemental Fig. 1), and no other PCR products. The reactions were carried out using a Bio-Rad iQ5 Multicolor Real Time Detection system under the same conditions used by Ratnaparkhe et al. (2009). Briefly, reactions were carried out in 20 μl, containing 10 μl iQ SYBR green Supermix (Bio-Rad), 1 μl each of forward and reverse primer (each primer a 10-μM stock), 2 μl cDNA template, and 6 μl nuclease-free water. Tubulin and ubiquitin-conjugating enzyme 1 (based on Picea abies UBC1 [Palovaara and Hakman 2008]) genes (Table 1) was used for normalization. To analyze gene expression within each embryo developmental series in P. taeda and P. oocarpa, the relative transcript levels were normalized using UBC1, and the transcript levels were compared relative to LC for somatic and zygotic samples; LC samples showed more similarities between the LC somatic embryos with the LC zygotic embryos. To perform the analysis of P. taeda gene expression comparison between individual somatic and zygotic embryo samples, the relative transcript levels were normalized using tubulin and compared relative to the zygotic samples. Each sample was subjected to three technical replicates, and the relative transcript level expression was analyzed by the 2−ΔΔCt method (Livak and Schmittgen 2001). Mean values are shown with standard errors (SE). Statistical comparisons were evaluated by the t test using the ΔCt values (Yuan et al. 2006). Differences of P < 0.05 were regarded as significant. The JMP 8.0 (SAS, Inc., Cary, NC) was used for statistical analysis.
Results
Gene expression in P. taeda—relative transcript level changes during zygotic and somatic embryo development.
Two endogenous controls were tested in this study, and UBC1 showed less variation than tubulin within each developmental series for zygotic and somatic embryos. Thus, UBC1 was used for the normalization of the data.
Storage proteins (legumin-like and vicilin-like). During both somatic and zygotic P. taeda embryo development, legumin-like relative transcript levels followed the same pattern in which the relative transcript levels were minimal in the PE/ESM, increased during the RG stage, decreased slightly in the EC stage and significantly increased to a maximum level during the LC stage (Fig. 2a ). A similar vicilin-like expression pattern was observed during both zygotic and somatic embryo development (Fig. 2b ), except in the somatic embryos, the relative transcript levels gradually increased from ESM to LC. The maximum vicilin-like relative transcript levels were also observed at the LC stage, as noted for the other storage protein gene transcript.
Relative transcript levels for target genes at four stages of P. taeda somatic and zygotic embryos, using ubiquitin-conjugating enzyme 1 (UBC1) as endogenous control. Genes under study: (a) legumin-like, (b) vicilin-like, (c) LEA, (d) HD-Zip I, (e) 26S proteasome subunit S2 (RPN1), and (f) clavata-like. ESM embryogenic suspensor masses, PE pro-embryo, RG round/globular, EC early cotyledonary, LC late cotyledonary. Values were normalized to the value of LC set at 1. Each value represents a mean+SE of 3 technical replicates. Bars with different letters are statistically different (P < 0.05), with lower-case letters used to represent the comparison among somatic samples and upper-case letters for zygotic samples.
Late embryogenesis abundant (LEA). Relative transcript levels for the group 4 LEA gene were detected at 1/3 maximum level in the ESM, gradually increased during P. taeda somatic embryo maturation and peaked at the LC stage (Fig. 2c ). In contrast, the relative transcript levels in P. taeda zygotic embryos accumulated later at the RG stage (about 1/6 the maximum level), showed little difference at the EC stage and then rapidly increased to maximum levels at the LC stage (Fig. 2c ). The temporal accumulation of LEA transcripts in somatic embryos was somewhat different from that observed in P. taeda zygotic embryos, with a more gradual accumulation representing a 2.5-fold change in abundance over the somatic embryo development processes, and a more controlled accumulation in zygotic embryos, representing an approximate 100-fold change over the zygotic embryo developmental series. The highest levels of expression were observed in LC embryos in both somatic and zygotic systems.
Homeodomain-leucine zipper I (HD-Zip I). The expression of HD-Zip I in both P. taeda somatic and zygotic embryos decreased during overall embryo development and maturation from peak levels at the RG stage (somatic embryos) or in PEs (zygotic embryos; Fig. 2d ) through to LC stage. There was some difference between the earliest somatic and zygotic embryo stages, in which the highest relative expression level was found at the zygotic embryo PE stage, while the somatic embryo ESM showed the lowest relative expression level for somatic embryo development, before increasing to peak levels at the RG stage and subsequently declining.
26S Proteasome subunit S2 (RPN1). The highest relative expression for RPN1 was detected at the RG stage in both zygotic and somatic embryo developmental samples. In the somatic embryo system, RPN1 transcripts maintained a high level with a slight decline at the LC stage (Fig. 2e ). In zygotic embryos, the relative expression declined more drastically at the subsequent stage (EC) and then slightly increased at the LC stage.
Clavata-like. The relative expression profiles of the clavata-like gene exhibited differences between P. taeda somatic and zygotic embryos when comparing the level of transcript change during embryo maturation (Fig. 2f ). The overall variation in transcript abundance was similar in both cases when comparing lowest and highest abundance (approximately 31-fold and 32-fold in somatic and zygotic embryos, respectively). During somatic embryogenesis, the relative expression levels went from their lowest levels at the ESM stage to peak levels at the RG stage but did not vary much during the developmental period between the RG and LC stages. There was a slight decline from the maximum abundance at the RG stage to a similar level in both EC and LC stages. Zygotic embryos also exhibited their lowest clavata-like transcript levels at the PE stage, followed by a significant increase in transcript level at the RG stage, which increased slightly by the EC stage and then declined significantly at the LC stage. The decline between the EC and LC stages represented an approximate 19-fold change, much greater than observed for the somatic embryo developmental series.
Gene expression in P. taeda—comparison of relative transcript levels between specific somatic and zygotic embryo developmental stages.
The previously described work assessed the relative transcript level changes within somatic or zygotic embryo samples during the developmental series relative to the level in their respective LC stage embryos. These changes did not provide an assessment of the relative transcript level differences between the somatic and zygotic embryos themselves at different stages of development. To provide this assessment, relative transcript levels were compared at each individual embryo stage between zygotic and somatic embryo samples of P. taeda. Samples were normalized using the housekeeping gene tubulin chosen because it exhibited less variation between the individual somatic and zygotic embryo samples than UBC1.
The comparison between P. taeda embryo samples at the PE and ESM stages showed that with most of the genes, with the exception of legumin-like and RPN1, the zygotic PE had higher expression levels compared to the somatic ESM (Fig. 3a ). The greatest variation in expression level was observed for the vicilin-like transcript (ESM was 333-fold lower than PE). The least variation was observed for the clavata-like transcript (ESM was 5.3-fold lower than PE). The relative expression level of legumin-like and RPN1 in ESM was 1.3- and 2.2-fold higher than in the PE, respectively.
For the remaining three embryo samples (RG, EC, and LC), the somatic embryo samples exhibited higher relative expression levels than the similar-stage zygotic embryo samples for all genes except for LEA (Fig. 3b–g ). The expression levels of vicilin-like, clavata-like, HD-Zip I, legumin-like, and RPN1 genes were approximately 5.7-, 2.0-, 1.4-, 30.0-, and 12.0-fold higher in somatic RG sample than at the same zygotic stage (Fig. 3b, c ). During the EC stage, the levels of vicilin-like, clavata-like, HD-Zip I, legumin-like, and RPN1 transcripts were approximately 30.0-, 2.0-, 9.0-, 49.0-, and 90.0-fold greater, respectively, in the somatic embryos compared with the zygotic embryos (Fig. 3d, e ). This trend continued through to the LC sample, in which the relative transcript levels for vicilin-like, clavata-like, HD-Zip I, legumin-like, and RPN1 were 38-, 55-, 40-, 100-, and 45-fold greater, respectively, in the somatic LC sample (Fig. 3f, g ).
Comparison of relative target gene transcript levels between somatic and zygotic embryo stages of P. taeda: (a) somatic embryogenic suspensor masses (ESM) and zygotic pro-embryos (PE); (b, c) round/globular (Som-RG) and zygotic round/globular (Zyg-RG); (d, e) early cotyledonary (Som-EC) and zygotic early cotyledonary (Zyg-EC); (f, g) late cotyledonary (Som-LC) and zygotic late cotyledonary (Zyg-LC). Tubulin was used as endogenous control, and values were normalized to the value of their respective zygotic sample set at 1. Each value represents a mean+SE of 3 technical replicates. All means are statistically different (P < 0.05) except between ESM and PE for legumin-like.
Thus, for almost all of the marker genes used in this study, the differences in relative expression level between somatic and zygotic embryos increased during maturation. As an example, for the legumin-like transcripts, the somatic embryo RG relative transcript level was 30-fold higher compared to the zygotic embryo RG, and this difference increased to 50- and 100-fold at the EC and LC samples, respectively. These higher transcript levels in the somatic embryo samples compared to their counterpart zygotic samples meant that the transcripts could be detected more readily in the somatic embryos.
Gene expression in P. taeda and P. oocarpa—comparison of relative transcript level changes during somatic embryo development.
Storage proteins (legumin-like and vicilin-like). During P. oocarpa somatic embryo development, both legumin-like (Fig. 4a ) and vicilin-like (Fig. 4b ) relative transcript levels were low in the ESM and RG stages, peaked at the EC stage and declined at the LC stage. Furthermore, the levels of legumin-like transcripts declined to near minimal levels at the LC stage. These results were in distinct contrast to those observed during P. taeda somatic embryogenesis, in which both storage protein transcripts increased from minimal levels at the ESM stage to peak levels at the LC stage (Fig. 2a, b ). Hence, the decline in relative transcript abundance during the LC stage, especially for the legumin-like transcripts, was a very distinct difference during P. oocarpa somatic embryogenesis.
Relative transcript levels for target genes at four stages of P. oocarpa somatic embryo development using ubiquitin-conjugating enzyme 1 (UBC1) as endogenous control. Genes under study: (a) legumin-like, (b) vicilin-like, (c) LEA, (d) HD-Zip I, (e) 26S proteasome subunit S2 (RPN1), and (f) clavata-like. ESM embryogenic suspensor masses, RG round/globular, EC early cotyledonary, LC late cotyledonary. Values were normalized to the value of LC set at 1. Each value represents a mean+SE of 3 technical replicates. Bars with different letters are statistically different (P < 0.05).
Late embryogenesis abundant (LEA). During P. oocarpa somatic embryo development, relative transcript levels for LEA increased from minimal levels at the ESM stage to the RG stage, remained relatively steady at the EC stage and increased to peak levels at the LC stage (Fig. 4c ). This developmental expression pattern was very similar to that observed during P. taeda somatic embryogenesis (Fig. 2c ). Similar to P. taeda, the P. oocarpa ESM stage also showed 1/3 the maximum levels of LEA, which occurred at the LC stage.
Homeodomain-leucine zipper I (HD-Zip I). The lowest relative transcript level for HD-Zip I occurred at the ESM stage, increased by the RG stage and remained relatively constant during the subsequent somatic embryo EC and LC stages of maturation (Fig. 4d ). This general trend was similar to that observed during P. taeda somatic embryogenesis (Fig. 2d ), where the lowest relative transcript levels were observed at the ESM stage and increased once the embryos were transferred to maturation medium. However, a gradual decline in HD-Zip I transcripts during the P. oocarpa cotyledonary stage progression was not observed as it was for P. taeda.
26S Proteasome subunit S2 (RPN1). Similar high relative transcript levels for RPN1 were detected at both the ESM and EC stages (Fig. 4e ) during P. oocarpa somatic embryo development. There was a significant decline in these levels between these two stages, with the RG stage exhibiting the lowest relative levels of RPN1 during the stages assessed. The expression levels also decreased drastically at the LC stage, such that the overall pattern of RPN1 transcript accumulation appeared biphasic. These results were in major contrast to those observed for P. taeda somatic embryos, where the overall variation in transcript levels during the developmental period was not as great, and with a single peak of accumulation at the RG stage (Fig. 2e ), followed by a gradual decline in transcript during continued maturation.
Meristem development (clavata-like). The relative expression of the clavata-like gene in P. oocarpa somatic embryos peaked at the early ESM stage, decreased drastically to minimal levels at the RG stage, slightly increased at the EC stage and declined slightly at the LC stage (Fig. 4f ). Again, this pattern of transcript accumulation was very different from that observed during P. taeda somatic embryo development, where there was little variation in transcript levels between the RG, EC, and LC stages, with the highest transcript levels at the RG stage and the lowest levels at the ESM stage (Fig. 2f ).
Discussion
To assess the developmental similarity of somatic embryos of two pine species matured under the same conditions, as well as the similarity with zygotic embryos, we characterized the relative expression of six developmentally regulated genes during zygotic embryo development in P. taeda and somatic embryo development/maturation in P. taeda and P. oocarpa. While somatic embryos were similar in relative gene expression patterns to their zygotic counterparts, some variation was encountered between zygotic and somatic embryos, and also between the two pine somatic embryo systems (Fig. 5).
Relative gene expression during P. taeda zygotic and somatic embryo developmental series.
We targeted genes for two of the storage proteins classes, and our results with P. taeda showed that during somatic and zygotic embryo development, both legumin-like and vicilin-like relative transcript levels followed the same pattern: the transcripts were minimal in the PE/ESM stages, increasing during subsequent development, with the maximum relative expression of both storage protein genes at the LC stage. These results mirrored storage protein accumulation in somatic embryos of P. taeda (Brownfield et al. 2007) and Pinus strobus (Klimaszewska et al. 2004), as well as transcript and storage protein accumulation in somatic and zygotic embryos of P. glauca (Flinn et al. 1993). We observed, major transcript accumulation during the cotyledonary phase in both somatic and zygotic embryos, and P. taeda somatic embryos showed a slightly more rapid increase in vicilin-like transcripts compared to zygotic embryos. Lippert et al. (2005) similarly noted that vicilin-like protein was detected early in somatic embryos after 7 d on maturation medium, and it accumulated gradually during embryo maturation. While storage protein transcript patterns were not assessed, Brownfield et al. (2007) did note that somatic embryos of P. taeda exhibited a shift in the ratio of their soluble to insoluble protein components, indicating some possible differences in somatic embryo metabolic activity. In some plants, the accumulation of storage proteins is not observed during somatic embryo development, and this feature has been used to distinguish “normal” somatic embryos (Stasolla and Yeung 2003). Thus, a high-quality somatic embryo includes the accumulation of storage proteins analogous to those of zygotic embryos (Merkle et al. 1995), and represents an excellent marker to check somatic embryo quality and fidelity.
In this study, P. taeda somatic embryos matured on a medium containing 40 μM ABA and 6 % (w/v) maltose appeared similar to zygotic embryos, with no precocious germination. As stated previously, the relative transcript levels of two storage proteins (legumin-like and vicilin-like) in these somatic embryo developmental samples followed a similar expression pattern than the counterpart zygotic samples. Thus, the actual maturation conditions used for P. taeda somatic embryo development were sufficient to stimulate similar expression patterns of the storage proteins. In P. glauca, a positive relationship was found between storage accumulation and ABA concentration, where low concentrations (10 μM) in the medium promoted precocious embryo germination, while higher concentration (40 μM) yielded normal embryos which contained significant amounts of storage protein (Roberts et al. 1990) and also promoted storage protein synthesis to similar levels found in their zygotic counterparts (Flinn et al. 1991). In the case of our group 4 LEA gene marker, the maximum transcript levels during both zygotic and somatic embryo development occurred at the LC stage, although expression was detected in pre-cotyledonary stages of both embryo systems. However, P. taeda somatic embryos exhibited a more prolonged and gradual accumulation of these transcripts than corresponding zygotic embryos. The continual exposure of our somatic embryos to ABA during maturation most likely accounted for the earlier and more gradual accumulation of LEA transcripts. In somatic embryos, a strong promotion of QrEM (LEA gene in Quercus robus) transcript accumulation was observed after induction of maturation on 6 % (w/v) sorbitol (no ABA) for 5 wk, then increased after partial desiccation (somatic embryos at 25 % loss of moisture) and declined when embryos lost more than 30 % of their moisture (Šunderlíková et al. 2009). In P. glauca, LEA gene transcripts accumulate after somatic embryo transfer to ABA (Dong and Dunstan 1996) and in the presence of polyethylene glycol (PEG) during the late stages of embryo maturation, which may enhance somatic embryo desiccation tolerance and thus improve postembryonic growth (Stasolla et al. 2003). These reports indicate the significance of ABA and/or osmotic agents in the stimulation of LEA gene expression. Hence, it is not surprising that modifications of the levels of these compounds in a tissue culture medium could influence in the LEA relative transcript levels. Our somatic embryo culture system maintained a prolonged culture on a constant level of 40 μM ABA, and we are unsure if this has any negative effects on the cultures. Thus, a modification that could be made during in vitro conditions would be to gradually increase the concentration of ABA or osmoticum during somatic embryo maturation and assess further impacts on embryo quality.
The LEA gene transcripts in zygotic P. taeda embryos accumulated later at the RG stage, showed little difference at the EC stage and then rapidly increased to maximum levels at the LC stage. Somewhat similar to our results, Leal and Misra (1993) noted LEA gene transcript accumulation in developing P. glauca zygotic embryos just prior to the LC phase, which reached maximal level in the fully mature embryo and was stable in dry seeds. Additionally, the isolation and characterization of QrEm (LEA gene in Q. robus) indicated that expression of this gene first occurred in the zygotic embryo EC stage, with gradual accumulation throughout mid-maturation and a subsequent decline toward the end of seed development (Šunderlíková et al. 2009).
The LEA proteins are more abundant during late embryogenesis than during mid-embryogenesis (Galau et al. 1986), their genes are expressed at the LC stage during zygotic embryogenesis, and they are considered a marker gene for the quality of the mature somatic embryo before undergoing germination (Zimmerman 1993). Although these genes were first identified from developing seeds, many LEA-like genes are induced by ABA or environmental stresses (Galau et al. 1986). In this study, we used a representative of the group 4 LEA genes for expression profiling during development. The group 4 LEA proteins are expressed during different developmental stages and plant organs in response to water deficit, and their deficiency also correlates with a reduced seed production, supporting a role in fruit and/or seed development (Olvera-Carrillo et al. 2010).
The genes described above are usually associated with the later stages of zygotic embryo maturation, so we also assessed the expression of a homeobox gene, HD-Zip I, a class of transcription factors in plants involved in the activation of other genes controlling tissue patterning (Chugh and Khurana 2002). We observed that HD-Zip I transcripts increased in somatic embryos following ESM transfer to maturation media containing ABA, but peaked at the RG stage and then declined. This was somewhat different from the zygotic embryos, which exhibited the highest relative expression of HD-Zip I in PEs and remained high until decreasing at the LC stage. Hence, there was a more prolonged expression pattern in the zygotic embryos than in our somatic embryos. HD-Zip I has been implicated in ABA responses in plants, and in water and light stress responses (Elhiti and Stasolla 2009). In our study, P. taeda HD-Zip I expression was somewhat similar to that previously reported for P. glauca (Tahir et al. 2008), who noted that PgHZ1 increased after 7 d on maintenance medium but remained high afterward during the maturation stage. However, no comparison of PgHZ1 expression during zygotic embryogenesis was addressed in that study. A phenotypic analysis of plants with higher levels of HD-Zip I gene expression showed that elevated levels could be related to cotyledon formation (Hanson et al. 2001) and that the expression was responsive to ABA and water deficit stress (Henriksson et al. 2005). While P. taeda zygotic and somatic embryos exhibited slightly different developmental transcript accumulation patterns, the key developmental stage difference between the somatic and zygotic embryos was the P. taeda PE/ESM sample; nevertheless, both zygotic and somatic embryos exhibited a progressive decrease from RG to LC during subsequent embryo maturation. While the mode of action of HD-Zips is poorly understood, these genes appear to be important regulators of plant development and differentiation in response to environmental factors, and in embryo-associated ABA responses (Deng et al. 2006). Here, we found that HD-Zip I was expressed during pine embryogenesis, perhaps increasing the sensitivity to ABA and promoting embryogenesis in vitro as previously suggested (Tahir et al. 2008).
In the case of the 26S Proteasome subunit S2 (RPN1), our results showed that expression of the P. taeda RPN1 gene was associated with the RG embryo stage transition of both somatic and zygotic embryos, with transcript levels increasing between PE/ESM and RG stage, and then declining with progression into later development. It is possible that the RG stage indicates a major morphological and physiological point, and thus the degradation of proteasome-targeted proteins is increased, with RPN1 participating in the target specificity and overall proteasome regulation at this crucial stage (Brukhin et al. 2005). Inactivation of subunit RPN1 arrests embryogenesis at the globular stage in Arabidopsis, suggesting that it is a key gene involved in controlling early embryo development and progress into maturation. Plant 26S subunit gene expression is regulated by growth regulator levels, which vary during plant development (Santner and Estelle 2010); thus, different embryo developmental stages (including somatic embryos) could require different proteasome activity levels controlled by endogenous growth regulators (Stasolla et al. 2003).
The final marker gene we considered, the clavata-like gene, was expressed during both somatic and zygotic embryo development. However, in P. taeda, differences were observed in the expression patterns between embryo samples, where more constant relative expression was observed during somatic embryogenesis. In contrast, zygotic embryos exhibited a peak level of expression during the RG and EC samples. The significance of these differences between zygotic and somatic embryos is unknown, as our somatic embryos exhibited SAM domes and cotyledonary development; however, we did not assess the SAM structure microscopically. The clavata-like gene regulates the activity of the shoot apical meristem, which is formed and develops during embryogenesis, and is required to produce new cells and tissues during post-germination growth (Tahir and Stasolla 2006). Previous studies have shown that SAM formation in somatic embryos of P. glauca (Tahir and Stasolla 2006) and P. pinaster (Tereso et al. 2007) followed the same pattern as their counterpart zygotic embryos. It is possible that the differences in relative expression levels observed in this study during somatic embryo development might be reflected in modified apical growth during subsequent germinative growth; this remains to be determined. If so, additional modifications of our current maturation medium, such as the addition of reduced glutathione (GSH; Belmonte et al. 2005) and PEG (Stasolla et al. 2003), may result in somatic embryo expression profiles more similar to zygotic embryo samples (Stasolla and Yeung 2003), and reflect on subsequent ex vitro performance.
Comparison between samples of somatic and zygotic embryos of P. taeda.
When the transcript levels of our marker genes were compared between similar zygotic and somatic embryo stages through normalization against the housekeeping control, we observed that in almost every case post-PE/ESM stage, transcript levels were higher in the somatic embryos than corresponding stage zygotic embryos, with the exception of the LEA transcripts, which were always higher in the zygotic embryos. Also, in the PE/ESM stage embryos, transcript levels were higher in the zygotic embryos, except for the legumin-like and RPN1 transcripts. In assessing these results, it is worth remembering that the total tissue of the P. taeda ESM was used for RNA extractions, and the ESM tissue is characterized by the continued multiplication of cell masses composed of aggregates of small, rapidly dividing cells and vacuolated non-dividing cells (Palovaara and Hakman 2008). These small, immature embryos highly resemble the early stages of zygotic embryos, designated in this study as PE, which were collected from seeds. Therefore, the transcript levels measured in the embryogenic cells of the ESM were diluted by the many surrounding accessory cells associated with the ESM mass, which could account for the lower relative transcripts observed for most of the genes in the ESM. Once the ESM tissues were transferred to maturation media, distinct individual somatic embryos were able to be identified and sampled for the later developmental stages; these later developmental sampled stages resembled their zygotic counterparts. During maturation from the RG stage and onward, differences in both somatic and zygotic embryos could be observed, but the basic developmental strategies are similar (Yeung 1995). It is possible that the direct exposure of the somatic embryos to 6 % (w/v) maltose and 40 μM ABA in vitro was sufficient to enhance gene transcript levels, as exposure to ABA and/or osmoticum can enhance embryo-associated gene expression in conifer somatic embryos (Flinn et al. 1993; Leal et al. 1995; Stasolla et al. 2003). It was interesting that the expression levels of the LEA gene were found to be higher in all of the zygotic embryo stages relative to the somatic embryo stages. The expression period exhibited less variation over the somatic embryo developmental series than the zygotic developmental series, which we attributed to the continual somatic embryo exposure to 40 μM ABA. This suggests that additional endogenous control mechanisms, apart from just applied ABA, are associated with overall LEA gene expression levels. Perhaps this may relate to differences in the embryogenic environments, such as other factors normally produced within the seed megagametophyte tissues, or to differences in osmotic conditions within the zygotic and somatic embryos.
Gene expression during somatic embryogenesis of P. oocarpa and comparison with P. taeda.
We previously described the significance of the marker genes used in this study and the overall gene expression similarities observed between the P. taeda zygotic and somatic embryo series, suggesting that the maturation protocol used was adequate to produce quality embryos. Our P. oocarpa somatic embryos, which were matured using the same media composition, exhibited a certain degree of similarity with the P. taeda somatic embryos in gene expression for some of the target genes. Overall, the legumin- and vicilin-like storage proteins exhibited low transcript levels at the ESM and RG stages, with major increases during the cotyledonary stage of development. However, the P. oocarpa somatic embryos, contrasting with the P. taeda somatic embryos, exhibited a significant decline in both storage protein transcripts at the later cotyledonary stage. Apart from storage protein transcripts, LEA gene transcripts exhibited a similar developmental expression profile between the somatic embryos of the two pine species, with the lowest levels at the ESM stage, increased levels at the RG and EC stages, and maximal levels at the LC stage. The HD-Zip I transcripts also displayed some similarity in developmental expression between P. oocarpa and P. taeda, with their lowest relative levels at the ESM stage followed by peak relative levels at the RG stage. However, while the P. oocarpa HD-Zip I transcript levels remained high, they subsequently declined during the P. taeda maturation series. Hence, it appeared that for these 4 marker genes, the variation in developmental expression patterns between the two pine somatic embryo series occurred during the later stages (cotyledonary) of maturation. In contrast to the above four genes, the RPN1 and clavata-like gene expression profiles were very dissimilar between the two pine somatic embryo systems. Both genes exhibited maximal or near maximal transcript abundance at the P. oocarpa ESM stage followed by a significant decline at the RG stage; but there was a minimal abundance in P. taeda at the ESM stage, followed by an increase at the RG stage. Taken together, all of these results suggest that under our current maturation protocol, the P. taeda somatic embryos may be of a higher quality than those of P. oocarpa, and that the P. oocarpa somatic embryo system requires further optimization.
Given that both pine species were matured using the same media, what might account for the observed differences? One initial difference between the two pine systems was that the ESM tissue was dispersed and plated out for P. taeda, while the P. oocarpa ESM was cultured as tissue clumps. Hence, the developing embryos for P. taeda would be closer to the nutritional medium, while the P. oocarpa embryos would most likely be exposed to nutritional/hormonal/oxygen gradients occurring within the clumps during development, which may impact on overall development and associated gene expression profiles. Others have reported that dispersion of suspended pine embryogenic tissue substantially enhances embryogenic tissue growth over the use of small tissue clumps (Aronen et al. 2009; Carneros et al. 2009). Additionally, the P. oocarpa embryos were matured for a longer period of time than the P. taeda embryos, which could impact on later stage developmental gene expression profiles. A similar observation relating to maturation time was made in which somatic embryos of P. strobus that matured for 9 wk accumulated storage proteins, but extension of the maturation period to 12 wk resulted in a significant decline in storage proteins, even on optimal maturation media (Klimaszewska et al. 2004). These observations may reflect depletions of tissue culture media components that have occurred during the culture period, or in the initiation of precocious germination. Storage protein transcript decreases have been reported in conifer somatic embryos during precocious germination in vitro (Flinn et al. 1993). Our P. oocarpa somatic embryos were not undergoing visual precocious germination, but it is possible that molecular processes associated with precocious germination had been initiated.
The maturation series reported here represented two different pine species. Therefore, the variations observed in overall gene expression profiles may reflect genotype effects. Genotypic variation has frequently been reported to help explain differences in somatic embryogenesis induction and development capacity within a species (Niskanen et al. 2004; Carneros et al. 2009). In this regard, a description of gene expression variation between normally maturing embryogenic cultures of P. abies was reported by Stasolla et al. (2004). In that report, similar transcription level patterns occurred for many genes, but some differential gene expression was observed, even though the embryogenic cultures developed normally through similar stages.
The present results describe the expression of 6 developmentally regulated genes during similar morphological stages of zygotic embryo development in P. taeda and somatic embryo development/maturation in P. taeda and P. oocarpa. This study also represents, to the best of our knowledge, the first analysis of somatic embryos of P. oocarpa and the molecular comparison of somatic embryo developmental series of 2 pine species matured under the same conditions. Our analyses indicated that all six marker genes were expressed during zygotic and somatic embryogenesis. Zygotic and somatic embryos of P. taeda exhibited similar developmental gene expression profiles, although some differences were observed. Gene expression levels tended to be higher in somatic embryos, in which transcripts were compared at similar zygotic embryo stages. In contrast, while somatic embryos of P. oocarpa exhibited some degree of similarity in developmental gene expression profiles with the P. taeda somatic embryos, there were also significant differences. In our study, the lower relative transcript levels observed with our P. oocarpa somatic embryos could explain their low germination success during somatic embryogenesis. Furthermore, they point to areas where modifications could be made to in vitro protocols using these and other markers.
Summary of relative expression of different target genes in four developmental stages of zygotic embryos of P. taeda and four developmental stages of somatic embryos of P. taeda and P. oocarpa. Heat map shading indicates relative values of gene expression (darker color represents higher expression), expressed as percentage of maximum gene expression.
The results from this study are in agreement with those obtained using other methods, such as microscopy of SAM development, biochemical quantification, and characterization of storage proteins and gene expression studies indicating general somatic embryo fidelity. Our study used a very small set of molecular markers for genes associated with key aspects of the embryo developmental process. The development of additional markers that could differentiate the quality of embryos from different cell lines and under different degrees of embryo maturation would facilitate the screening or identification of those somatic embryos cell lines that more closely resemble their zygotic counterparts. Theoretically, this could increase the potential for conversion of somatic embryos into plantlets by more closely following the program of zygotic embryo development. Metabolite analyses could be used to further characterize differences in the regenerative capacity of P. taeda embryogenic cultures (Robinson et al. 2009) and to differentiate between embryogenic and non-embryogenic cells in pine species (Park et al. 2011). From the latter study, approximately 50 compounds displayed significant differences between embryogenic and non-embryogenic cell lines (Park et al. 2011), while a model of 47 metabolites could predict the regenerative capacity of somatic embryogenic masses in P. taeda cell lines (Robinson et al. 2009). A metabolomics study at different embryo stages could provide new insights into the development and quality of somatic embryo during maturation. As suggested by Robinson et al. (2009), the identification of metabolite sets could aid in the monitoring of the physiological status of the embryo cultures from embryogenic suspensor masses through maturation/development stages. This may help to identify amendments needed in the culture conditions to improve the production of “normal” embryos.
References
Aronen T, Pehkonen T, Rynänen L (2009) Enhancement of somatic embryogenesis from immature zygotic embryos of Pinus sylvestris. Scand J For Res 24:372–383
Belmonte MF, Donald G, Reid DM, Yeung EC, Stasolla C (2005) Alterations of the glutathione redox state improve apical meristem structure and somatic embryo quality in white spruce (Picea glauca). J Exp Bot 56(419):2355–2364
Bonga J, Klimaszewska K, von Aderkas P (2010) Recalcitrance in clonal propagation, in particular of conifers. Plant Cell, Tissue Organ Cult 100(3):241–254
Brownfield D, Todd C, Stone S, Deyholos M, Gifford D (2007) Patterns of storage protein and triacylglycerol accumulation during loblolly pine somatic embryo maturation. Plant Cell, Tissue Organ Cult 88(2):217–223
Brukhin V, Gheyselinck J, Gagliardini V, Genschik P, Grossniklaus U (2005) The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17(10):2723–2737
Carneros E, Celestino C, Klimaszewska K, Park YS, Toribio M, Bonga JM (2009) Plant regeneration in Stone pine (Pinus pinea L.) by somatic embryogenesis. Plant Cell Tissue Organ Cult 98:165–178
Chugh A, Khurana P (2002) Gene expression during somatic embryogenesis-recent advances. Curr Sci 83(6):715–730
Deng X, Phillips J, Bräutigam A, Engström P, Johannesson H, Ouwerkerk P, Ruberti I, Salinas J, Vera P, Iannacone R (2006) A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Mol Biol 61(3):469–489
Dong JZ, Dunstan DI (1996) Expression of abundant mrnas during somatic embryogenesis of white spruce [Picea glauca (Moench) voss]. Planta 199(3):459–466
Dong JZ, Perras MR, Abrams SR, Dunstan DI (1997) Gene expression patterns, and uptake and fate of fed ABA in white spruce somatic embryo tissues. J Exp Bot 48(2):277–287
Dvorak W, Potter K, Hipkins V, Hodge G (2009) Genetic diversity and gene exchange in Pinus oocarpa, a mesoamerican pine with resistance to the pitch canker fungus. Int J Plant Sci 170(5):609–626
Elhiti M, Stasolla C (2009) Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav 4(2):86–89
Feher A, Pasternak T, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell, Tissue Organ Cult 74(3):201–228
Flinn BS, Roberts DR, Newton CH, Cyr DR, Webster FB, Taylor IEP (1993) Storage protein gene expression in zygotic and somatic embryos of interior spruce. Physiol Plant 89(4):719–730
Flinn BS, Roberts DR, Taylor I (1991) Evaluation of somatic embryos of interior spruce. Characterization and developmental regulation of storage proteins. Physiol Plant 82(4):624–632
Galau G, Hughes D, Dure L (1986) Abscisic acid induction of cloned cotton late embryogenesis-abundant (LEA) mRNAs. Plant Mol Biol 7(3):155–170
Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J, Morris P, Bouvier-Durand M, Vartanian N (1994) Current advances in abscisic acid action and signalling. Plant Mol Biol 26(5):1557–1577
Goldberg RB, De Paiva G, Yadegari R (1994) Plant embryogenesis: zygote to seed. Science 266(5185):605–614
Gupta PK, Durzan DJ (1987) Biotechnology of somatic polyembryogenesis and plantlet regeneration in loblolly pine. Nat Biotechnol 5(2):147–151
Hanson J, Johannesson H, Engström P (2001) Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol Biol 45(3):247–262
Henriksson E, Olsson A, Johannesson H, Johansson H, Hanson J, Engstrom P, Soderman E (2005) Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol 139:509–518
Ikeda M, Kamada H (2006) Comparison of molecular mechanisms of somatic and zygotic embryogenesis. In: Mujib A, Śamaj J (eds) Somatic embryogenesis. Springer, Berlin Heidelberg New York, pp 51–68
Klimaszewska K, Morency F, Jones-Overton C, Cooke J (2004) Accumulation pattern and identification of seed storage proteins in zygotic embryos of Pinus strobus and in somatic embryos from different maturation treatments. Physiol Planta 121(4):682–690
Lara-Chavez A, Flinn B, Egertsdotter U (2011) Initiation of somatic embryogenesis from immature zygotic embryos of Oocarpa pine (Pinus oocarpa Schiede ex Schlectendal). Tree Physiol 31(5):539–554
Leal I, Misra S (1993) Developmental gene expression in conifer embryogenesis and germination. III. Analysis of crystalloid protein mRNAs and desiccation protein mRNAs in the developing embryo and megagametophyte of white spruce (Picea glauca (Moench) Voss). Plant Sci 88(1):25–37
Leal I, Misra S, Attree SM, Fowke LC (1995) Effect of abscisic acid, osmoticum and desiccation on 11S storage protein gene expression in somatic embryos of white spruce. Plant Sci 106(2):121–128
Lelu-Walter MA, Bernier-Cardou M, Klimaszewska K (2006) Simplified and improved somatic embryogenesis for clonal propagation of Pinus pinaster (ait.). Plant Cell Rep 25(8):767–776
Lippert D, Zhuang J, Ralph S, Ellis DE, Gilbert M, Olafson R, Ritland K, Ellis B, Douglas CJ, Bohlmann J (2005) Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics 5(2):461–473
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta] Ct method. Methods 25(4):402–408
Merkle SA, Parrott WA, Flinn BS (1995) Morphogenic aspects of somatic embryogenesis. In: Thorpe TA (ed) In vitro embryogenesis in plants. Kluwer Academic Publishers, Dordrecht, pp 155–203
Newton R, Tang W, Jain S (2005) Slash Pine (Pinus elliottii Engelm.). In: Jain SM, Gupta PK (eds) Protocol for Somatic Embryogenesis in Woody Plants 77. Springer, The Netherlands, pp 1–10
Niskanen AM, Lu J, Seitz S, Keinonen K, Von Weissenberg K, Pappinen A (2004) Effect of parent genotype on somatic embryogenesis in Scots pine (Pinus sylvestris). Tree Physiol 24(11):1259–1265
Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias A (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 154(1):373–390
Palovaara J, Hakman I (2008) Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during Picea abies somatic embryogenesis. Plant Mol Biol 66(5):533–549
Park SY, Lee WY, Kim YW, Moon HK (2011) Characterization of metabolic differences between embryogenic and non-embryogenic cells in forest trees. BMC Proc 5(7):p14
Pullman GS, Chopra R, Chase KM (2006) Loblolly pine (Pinus taeda L.) somatic embryogenesis: Improvements in embryogenic tissue initiation by supplementation of medium with organic acids, vitamins B12 and E. Plant Sci 170(3):648–658
Pullman GS, Johnson S (2002) Somatic embryogenesis in loblolly pine (Pinus taeda L.): Improving culture initiation rates. Ann For Sci 59(5–6):663–668
Pullman GS, Johnson S, Peter G, Cairney J, Xu N (2003a) Improving loblolly pine somatic embryo maturation: Comparison of somatic and zygotic embryo morphology, germination, and gene expression. Plant Cell Rep 21(8):747–758
Pullman GS, Johnson S, Tassel SV, Zhang Y (2005) Somatic embryogenesis in loblolly pine (Pinus taeda) and Douglas fir (Pseudotsuga menziesii): Improving culture initiation and growth with MES pH buffer, biotin, and folic acid. Plant Cell, Tissue Organ Cult 80(1):91–103
Pullman GS, Montello P, Cairney J, Xu N, Feng X (2003b) Loblolly pine (Pinus taeda L.) somatic embryogenesis: Maturation improvements by metal analyses of zygotic and somatic embryos. Plant Sci 164(6):955–969
Quiroz-Figueroa FR, Rojas-Herrera R, Galaz-Avalos RM, Loyola-Vargas VM (2006) Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell, Tissue Organ Cult 86(3):285–301
Ratnaparkhe S, Egertsdotter E, Flinn B (2009) Identification and characterization of a matrix metalloproteinase (Pta1-MMP) expressed during loblolly pine (Pinus taeda) seed development, germination completion, and early seedling establishment. Planta 230(2):339–354
Roberts DR, Flinn BS, Webb DT, Webster FB, Sutton BCS (1990) Abscisic acid and indole-3-butyric acid regulation of maturation and accumulation of storage proteins in somatic embryos of interior spruce. Physiol Planta 78(3):355–360
Robinson AR, Dauwe R, Ukrainetz NK, Cullis IF, Mansfield SD, White R (2009) Predicting the regenerative capacity of conifer somatic embryogenic cultures by metabolomics. Plant Biotechnol J 7(9):952–963
Santner A, Estelle M (2010) The ubiquitin-proteasome system regulates plant hormone signaling. Plant J 61(6):1029–1040
Stasolla C, Bozhkov PV, Chu TM, Van Zyl L, Egertsdotter U, Suarez MF, Craig D, Wolfinger RD, von Arnold S, Sederoff RR (2004) Variation in transcript abundance during somatic embryogenesis in gymnosperms. Tree Physiol 24(10):1073–1085
Stasolla C, Kong L, Yeung EC, Thorpe TA (2002) Maturation of somatic embryos in conifers: Morphogenesis, physiology, biochemistry, and molecular biology. In Vitro Cell Dev Biol Plant 38(2):93–105
Stasolla C, Yeung EC (2003) Recent advances in conifer somatic embryogenesis: Improving somatic embryo quality. Plant Cell, Tissue Organ Cult 74(1):15–35
Stasolla C, Zyl L, Egertsdotter U, Craig D, Liu WB, Sederoff RR (2003) The effects of polyethylene glycol on gene expression of developing white spruce somatic embryos. Plant Physiol 131(1):49–60
Sterk P, de Vries S (1993) Molecular markers for plant embryos. In: Redenbaugh K (ed) Synseeds: Applications of synthetic seeds to crop improvement. CRC Press, Boca Raton, pp 115–132
Šunderlíková V, Salaj J, Matušíková I, Wilhelm E (2009) Isolation and characterization of an embryo-specific em-like gene of pedunculate oak (Quercus robur L.) and its temporal and spatial expression patterns during somatic and zygotic embryo development. Trees 23(1):135–144
Tahir M, Belmonte MF, Elhiti M, Flood H, Stasolla C (2008) Identification and characterization of PgHZ1, a novel homeodomain leucine-zipper gene isolated from white spruce (Picea glauca) tissue. Plant Physiol Biochem 46(12):1031–1039
Tahir M, Stasolla C (2006) Shoot apical development during in vitro embryogenesis. Botany 84(11):1650–1659
Tereso S, Zoglauer K, Milhinhos A, Miguel C, Oliveira M (2007) Zygotic and somatic embryo morphogenesis in Pinus pinaster: Comparative histological and histochemical study. Tree Physiol 27(5):661–669
van Gelder R, von Zastrow M, Yool A, Dement W, Barchas J, Eberwine J (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 87(5):1663–1667
von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L (2002) Developmental pathways of somatic embryogenesis. Plant Cell, Tissue Organ Cult 69(3):233–249
West MAL, Harada JJ (1993) Embryogenesis in higher plants: An overview. Plant Cell 5(10):1361–1369
Yang X, Zhang X (2010) Regulation of somatic embryogenesis in higher plants. Crit Rev Plant Sci 29(1):36–57
Yeung E (1995) Structural and developmental patterns in somatic embryogenesis. In: Thorpe TA (ed) In vitro embryogenesis in plants. Kluwer Academic Publishers, Dordrecht, pp 205–248
Yuan JS, Reed A, Chen F, Steward CN Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinforma 7:85
Zimmerman JL (1993) Somatic embryogenesis: A model for early development in higher plants. Plant Cell 5(10):1411–1423
Acknowledgments
This work represents a portion of A. Lara-Chavez’s doctoral dissertation. We would like to thank Dr. Scott Merkle for providing P. taeda cell cultures, Kyle Peer for collecting the immature P. taeda cones, Dr. Marty Fernandes for providing developing P. oocarpa cones, Anne Kathryn Dalton for help in embryo dissection and collection, Dr. Supriya Ratnaparkhe for providing the photo of the P. taeda pro-embryo stage in Fig. 1 and Guozhu Tang for support with RNA extraction and cDNA synthesis. Many thanks also to Drs. Scott Merkle, Thomas Fox and Jose Alexander Elvir for their comments during the course of this study. This work was funded through Special Grants (2003-38891-02112, 2008-38891-19353 and 2009-38891-20092) and HATCH funds (Project No. VA-135816) from the United States Department of Agriculture, and operating funds from the Commonwealth of Virginia to the Institute for Advanced Learning and Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editor: Thomas Clemente
Ulrika Egertsdotter and Barry S. Flinn contributed equally to project development, support, and supervision.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(JPEG 115 kb)
Rights and permissions
About this article
Cite this article
Lara-Chavez, A., Egertsdotter, U. & Flinn, B.S. Comparison of gene expression markers during zygotic and somatic embryogenesis in pine. In Vitro Cell.Dev.Biol.-Plant 48, 341–354 (2012). https://doi.org/10.1007/s11627-012-9440-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11627-012-9440-5