Abstract
Using specimens collected from subtropical pine-fagaceous mixed forests and phylogenetic analysis of DNA sequence data of ITS, 28S rDNA, rpb2 and tef1, we describe two new species, R. maguanensis and R. substriata, in R. subg. Heterophyllidia, subsect. Substriatinae subsect. nov. Russula maguanensis and R. substriata are similar to Indian R. shingbaensis in the tuberculate-striate pileus and spores with isolated warts but have more vividly coloured pileus and associate with pines and/or fagaceous trees rather than with Abies. In our multi-gene phylogeny, the new subsection and a representative of tropical African R. subsect. Aureotactinae compose one of the four major clades of R. subg. Heterophyllidia, the three remaining ones corresponding to R. sect. Heterophyllae, R. sect. Ingratae and R. subsect. Cyanoxanthinae. The overall characters of this new section combine those of some other sections in the same subgenus: mostly tuberculate-striate but more vivid pileus, spores with isolated warts, orthochromatic pileipellis with abundant erect aggregate mucronate pileocystidia in the suprapellis but absent in the subpellis and numerous cystidioid hyphae at the bottom of subpellis and trama beneath it. It differs from its sister clade R. subsect. Aureotactinae in lacking the intense yellowing of surface and context of their fruiting bodies and having pileal cystidioid elements clearly separated by the loose tissue of subpellis from the pileocystidia at the pileus surface. In order to compare our two new species with recently described Asian species and investigate their geographical distributions, we produced an ITS genealogy including also environmental sequences. This ITS genealogy suggests that R. subsect. Substriatinae includes at least seven potential species, shows an amphi-pacific distribution and its members associate with at least four families of host trees.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the fungal bar-code ITS region being used for identifying specimens and separating species of Russula Pers., one of the notoriously difficult groups in the taxonomy of agarics (Singer 1962), Asia is becoming an active region for Russula taxonomy. Since the first Chinese Russula species supported by ITS data (R. griseocarnosa X.H. Wang et al., Wang et al. 2009), 32 new taxa have been described from the country, all primarily based on ITS sequences for species recognition and systematic placement, except for R. zhejiangensis G.J. Li et al. with only morphology and R. vinosobrunneola G.J. Li & R.L. Zhao on multi-locus data (Li et al. 2011, 2012, 2013a, b, 2015a, b, 2018a, b; Ariyawansa et al. 2015; Zhao et al. 2015; Sang et al. 2016; Das et al. 2017; Jiang et al. 2017a; Tibpromma et al. 2017; Zhang et al. 2017; Li and Deng 2018; Song et al. 2018a, b). In contrast, hardly 13 Russula species had been described from China between 1935 and 2009 (Song et al. 2007). In addition, published ITS data confirmed the presence of one Indian (R. thindii K. Das & S.L. Miller), one Japanese (R. subnigricans Hongo) and 15 European species in China (Yin et al. 2008; Guo et al. 2014; Jiang et al. 2017b; Liu et al. 2017). These ITS-based Russula species are found from the northeastern, southern, eastern and southwestern parts of the country, in temperate, subtropical, tropical and subalpine coniferous and broad-leaved forests. Nevertheless, the large number of unnamed sequences from Chinese Russula samples in GenBank suggests that the diversity of the genus in China needs further exploring.
During recent forays in subtropical mixed forests dominated by fagaceous trees and Pinus kesiya var. langbianensis in Maguan County (Wenshan Prefecture, famous for its karst landscape) in Yunnan Province, China, we found several Russula specimens for which both morphology and ITS sequences suggested affinities to Indian R. shingbaensis K. Das & S.L. Miller. Although ITS sequence data suggested that R. shingbaensis belongs to R. subg. Heterophyllidia Romagn., an exact phylogenetic placement was pending as none of the known sections and subsections within the subgenus seemed appropriate to hold it. Our samples now re-address the problem regarding the phylogenetic placement of R. shingbaensis and its sibling R. verrucospora Y. Song & L.H. Qiu, recently published from southern China (Song et al. 2018b). Based on the most up to date Russula phylogeny using multi-gene data and worldwide sampling (Buyck et al. 2018), we used four loci, the nuc rDNA ITS1-5.8S-ITS2 (ITS), D1-D2 domains of nuc 28S rDNA (28S), part of the second largest subunit of the RNA polymerase II (rpb2) (6–7 region) and translation elongation factor 1-alpha (tef1) and morphological characters, to (1) delimit the potential new Chinese species from Indian R. shingbaensis and Chinese R. verrucospora, (2) find a satisfactory infrageneric placement for these Asian species using more conservative loci and (3) find out the distribution area of this species assemblage using all available ITS sequences retrieved from GenBank.
Material and methods
Morphological observation and taxonomic assignment
Descriptions of sporocarps were from fresh material, and micro-morphological study was performed on dried specimens. Basidiospores were observed and measured in Melzer’s reagent in side view, excluding ornamentation and apiculus. For each fruiting body, 20 spores were measured. All other micro-morphological structures were revived in 5% KOH then mounted with Congo red (aqueous solution). Pileipellis was examined for the presence of ortho- or metachromatic contents in Cresyl blue (aqueous solution) as explained in Buyck (1989). Sulphovanillin (SV) was used to test the reactions of cystidium contents. All drawings except those of the basidiospores were made with the aid of a drawing tube installed on a Nikon E400 microscope. Vertical sections of the pileipellis were cut approximately at the mid-radius of the pileus. Sections through the stipitipellis were taken from the middle part along the stipe length. Drawings of basidiospores were made by hand, using a 5000× amplification (1 cm on paper standing for 2 μm). The abbreviation for spore measurements (n/m/p) means n spores from m basidiocarps of p specimens. Measurements (and Q values) of spores are given as (a) b–m–c (d), in which “a” is the lowest value, “d” the biggest, “m” the mean and “b–c” covers a minimum of 90% of the values. “Q” stands for the ratio of length and width of a spore and “Q ± av” for the average Q of all spores ± sample standard deviation. Colour codes of fruiting bodies are from Kornerup and Wanscher (1961). Voucher specimens are deposited in Cryptogamic Herbarium, Kunming Institute of Botany, Chinese Academy of Sciences (HKAS section, KUN). Infrageneric classification follows Buyck et al. (2018).
DNA extraction, PCR amplifications and sequencing
DNA extraction and PCR protocols followed Wang et al. (2015). The following four regions were amplified: complete ITS (c. 700 bp), 28S (c. 900 bp), rpb2 (c. 800 bp) and tef1 (c. 920 bp). Primer pairs ITS1F + ITS4 or ITS5 + ITS4, LROR+LR5 and RPB2-6F + fRPB2-7cR were used to amplify the ITS, 28S and rpb2 regions/genes, respectively (Vilgalys and Hester 1990; White et al. 1990; Liu et al. 1999; Moncalvo et al. 2000). The tef1 gene was amplified in two pieces using the primer combinations 526F + EF-ir and 983F + 1567R (Rehner and Buckley 2005; Hansen et al. 2013). For problematic samples where the 526F–EF-ir region did not successfully amplify, a newly designed specific reverse primer EF-ir-Subs (5′-GAAATGCCTGCCTCGAATTCACC-3′) was used with the forward primer 526F. PCR products were visualised via UV light after electrophoresis on 1% agarose gels stained with ethidium bromide. Successful PCR products were sent to Sangon Biotech Limited Company (Shanghai, China) for Sanger sequencing using the same primers as PCR. When sequences have heterozygous INDELs or ambiguous sites, samples were sequenced bidirectionally to make contigs of the amplified regions or verify the ambiguous sites. Raw sequences were assembled with Sequencher v4.1.4 (Gene Codes Corporation, Ann Arbor, USA), and assembled sequences are deposited in GenBank (Table 1, Fig. 2).
Taxa sampling and phylogenetic analyses of multi-gene data
All our samples were sequenced to obtain 28S, rpb2 and tef1 sequences. Besides our two species with 18 sequences, 55 species (with at least two of the three loci available) were selected to cover all the seven subgenera of Russula recognised by Buyck et al. (2018) and major clades in Looney et al. (2016). Specifically, representative species of the various recognised subdivisions of R. subg. Heterophyllidia (in which the target group of this study is placed) used by Buyck et al. (2018) were included. These sequences formed a 28S-rpb2-tef1 combined dataset, which was used to determine the phylogenetic position of our two new species. Lactifluus aff. volemus and L. allardii were chosen as outgroup, following the phylogeny of Russulaceae of Buyck et al. (2008).
Alignments were made using the online version of the multiple sequence alignment program MAFFT v7 (Katoh and Toh 2008), applying the L-INS-I strategy, and were manually adjusted in BioEdit v.7.1.3.0 (Hall 1999). Introns of rpb2 and tef1 and an ambiguous section of rpb2 (ca 550 bp from the RPB2-6F primer) were removed from phylogenetic analyses. We followed the partitioning strategy of Buyck et al. (2018), i.e., seven partitions (28S, rpb2 1st, rpb2 2nd, rpb2 3rd codons, tef1 1st, tef1 2nd and tef1 3rd codons) as this partition gave higher bootstrap proportions (BP) in the maximum likelihood (ML) analysis of multi-gene phylogeny of the whole genus. ML phylogenetic analysis was conducted in RAxML v7.2x (Stamatakis 2006) and Bayesian inference (BI) in MrBayes v3.2.6 (Ronquist et al. 2012). ML analysis was executed applying the rapid bootstrap algorithm with 1000 replicates, followed by a ML tree search. For BI analysis, the best-fit model was selected by MrModeltest (Nylander 2004). The BI analysis was conducted using four runs with four chains each for one million generations sampling every 100th tree. Runs were inspected to make sure the average standard deviation of split frequencies went below 0.01 and effective sampling sizes were > 200 in Tracer v1.7.0 (Rambaut et al. 2018). A 50% majority rule consensus tree was built after discarding trees from a 25% burn-in. A ML-BP ≥ 70% and BI posterior probability (BI-PP) ≥ 95% were considered as significant support for a node to be monophyletic. The most likely tree generated in ML was viewed and exported in FigTree v1.3.1, with BI-PP added on the corresponding nodes. Matrix and the ML and BI trees are deposited in TreeBASE as study ID S23424.
Taxa sampling and phylogenetic analyses of ITS data
The six generated ITS sequences from our six samples (with the 18S and 28S ends trimmed) were submitted to BLASTn to find matches with high similarity (≥ 90%) and query cover (> 90%) in GenBank. The complete ITS, ITS1 and ITS2 were submitted, respectively, to retrieve sequences with different sizes. We retrieved sequences both from fruiting bodies and environmental samples to find out whether representatives of our new species and section have been recorded from other places. Following the phylogenetic implication of the 28S-rpb2-tef1 data, we included four ITS sequences of the R. brunneoannulata Buyck species complex. To show the genetic diversification among the two new species and some representative species or look-alikes in the same subgenus, especially those originally described from Asia and Africa, we included 61 ITS sequences of 38 species. Among these species, seven are from Europe. The purpose of including these European species is to indicate the traditional taxonomic classification, which is mostly based on European representatives. The ITS data were analysed to show how the two new species differ genetically from their relatives, the geographical distribution of the new section and the two new species and their putative hosts. We used four species of R. subg. Compactae (Fr.) Bon and R. subg. Archaea Buyck & V. Hofst. as outgroups, following the phylogeny of Buyck et al. (2018). The 18S and 28S ends of the ITS sequences were removed from phylogenetic analyses. Aligning and further analyses of the ITS data followed those of the 28S-rpb2-tef1 dataset, except that no partitioning was set and 200 million generations were ran for BI analysis. A ML-BP ≥ 70% and BI-PP ≥ 95% were considered as significant support for a node to be monophyletic. Matrix and the ML and BI trees are deposited in TreeBASE with study ID S23424, the same as 28S-rpb2-tef1 dataset.
Results
Multigene phylogeny
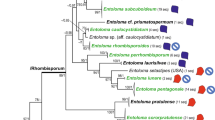
For BI analysis, GTR + I + G model was selected as the best-fit model by MrModeltest. ML and BI analyses of the 28S-rpb2-tef1 combined dataset produced nearly identical topologies with comparable support values (Fig. 1). Except that in the BI analysis the remaining of Russula excluding R. subg. Heterophyllidia was significantly supported to be a monophyly (BI-PP 0.97), the two analyses did not resolve the relationships among the major clades/subgenera of Russula. Nevertheless, except for R. subg. Crassotunicata Buyck & V. Hofst. (a singleton of R. farinipes Romell), all the subgenera were retrieved with high supports (ML-BP 89–100%, BI-PP 1.00). The six samples of the two new species formed a highly supported terminal clade with ML-BP 100% and BI-PP 1.00 with a long branch, sister to a singleton also with a long branch formed by African R. aff. brunneoannulata with ML-BP 71% and BI-PP 0.98. This Asian-African clade (clade B, Fig. 1) represented one of the four highly supported major clades of R. subg. Heterophyllidia (clades A–D). The phylogenetic relationships among these four clades did not receive supports either in the ML or BI analysis.
Most likely tree generated by maximum likelihood (ML) analysis of Russula species using 28S, rpb2 and tef1 sequence data (2299 bp), rooted with Lactifluus aff. volemus and L. allardii. Greyish-blue frame indicates the subgenus where the two new species R. maguanensis and R. substriata and the new subsection R. subsect. Substriatinae (in red) are placed. Infrageneric classification follows Buyck et al. (2018). Names of the infrageneric taxa under R. sect. Ingratae and R. sect. Heterophyllae are all of subsections except for Subvelatae of a section. We put the name Modestinae in quotation marks because the type specimen of R. modesta does not correspond to the current concept of the species. ML bootstrap values (ML-BP) and Bayesian posterior probabilities (BI-PP) are shown above and below the branches respectively. A ML-BP 100% or BI-PP 1.0 is replaced by an asterisk (*). Branches supported by both ML-BP ≥ 70% and BI-PP ≥ 95% are in black bold
ITS genealogy
The aligned ITS dataset includes 96 sequences and 811 characters (with the 18S and 28S ends trimmed), 311 bp of ITS-1, 156 bp of 5.8S and 343 bp of ITS-2. All sequences cover ITS-1, 5.8S and ITS-2 regions, except for AB509486 and AB509982 lacking ITS-1 region. ML analysis produced a genealogy without support for relationships among the six significantly supported major clades of the ingroup (clades A–F, Fig. 2). For BI analysis, HKY + I + G model was selected as the best-fit model by MrModeltest. BI analysis produced a highly similar typology with most supports comparable to the ML analysis. The only two differences between the two analyses are on the branch leading to the terminal clade formed by HKAS 102275, 102278 and 102279, which received significant support only in the ML analysis (ML-BP 85%) and two basal branches of clade F, which received significant supports only in the BI analysis (BI-PP 0.98 and 0.99). Our six samples from Maguan fell into one of the six major clades, clade A (ML-BP 100%, BI-PP 1.00) together with 21 mostly environmental samples from Australia, China, India, Japan, Malaysia, Mexico and USA which can be assigned into three additional highly supported subclades and two singletons. One of the singletons is the holotype of R. shingbaensis from India and the other a Malaysian sample (from Bornean island). In general, clade A shows an amphi-pacific distribution pattern and hosts of this clade involve Pinaceae (Abies and Pinus), Fagaceae (Castanopsis and Quercus), Jaglandaceae (Oreomunnea) and Myrtaceae (Eucalyptus). The assemblage of clade A is the new subsection we describe below. Within clade A, our five samples, together with five samples from eastern (Zhejiang), central-southern (Hunan) and southwestern (Sichuan) China, and two samples from central and southern Japan formed a fully supported terminal clade. Our last sample HKAS 102277 and a sample also from southern Japan formed another highly supported terminal clade with ML-BP 99% and BI-PP 1.00. These two terminal clades are the two new species that are described in this study.
Most likely tree generated by maximum likelihood (ML) analysis of Russula species using ITS sequence data (811 bp), rooted with four species of R. subg. Compactae (outgroup). Infrageneric classification follows Buyck et al. (2018). Sample names are provided in the order of species name (if available), GenBank accession number, origin of country and host (if available). For the sequences generated in this study, herbarium number is provided after the GenBank accession. For the two new species described, province name is provided after its country. ML bootstrap values (ML-BP) and Bayesian posterior probabilities (BI-PP) are shown above and below the branches, respectively. A ML-BP 100% or BI-PP 1.0 is replaced by an asterisk (*). Branches supported by both ML-BP ≥ 70% and BI-PP ≥ 95% are in black bold. Dashed part of the bar for R. subsect. Substriatinae indicates the uncertainty of the placement of R. verrucospora in the subsection
The successive clades of clade A are clade B, represented by a single species R. verrucospora and then a superclade formed by clade D (R. cf. brunneoannulata) and C (R. subsect. Cyanoxanthinae Singer). These sibling relationships, however, are without support in either analysis. Russula verrucospora was shown to be a sister of R. shingbaensis by Song et al. (2018b) with ML-BP 84% and R. cf. brunneoannulata was sibling to the clade formed by our two new species in the 28S-rpb2-tef1 phylogeny. Both species have spores with isolated warts and numerous pileocystidia and gloeoplerous elements in all tissues. The remaining two clades E and F are well in line with those in the 28S-rpb2-tef1 phylogeny, i.e. corresponding to R. sect. Heteropyllae Fr. and R. sect. Ingratae (Quél.) Maire in the infrageneric classification of Russula.
Taxonomy
Russula maguanensis J. Wang, X.H. Wang, Buyck & T. Bau, sp. nov., Figs. 3a and 4.
MycoBank: MB 827272.
GenBank: MH724918 (ITS), MH714537 (28S), MH939989 (rpb2), MH939983 (tef1), all from holotype.
Etymology: named after Maguan County, the type locality.
Holotype: China, Yunnan Prov., Wenshan Pref., Maguan Co., Dalishu Town, Xiangchang, 23°04′07.6″ N, 104°12′31.5″ E, 1633 m asl, in mixed forest of fagaceous trees and Pinus kesiya var. langbianensis, leg. X.H. Wang, 14 Oct. 2017, no. 4765 (HKAS 102277, KUN!).
Diagnosis: A species with medium-sized basidiocarps, sticky tuberculate-striate pileus with lilac red tinge, finely to coarsely cracked pileus cuticle, spores with isolated warts and suprapellis with tufts of pileocystidia.
Pileus 27–45 mm diam., concave with applanate margin, almost flat in the extreme centre, 60–70% of the radius strongly tuberculate-striate; surface finely cracked even when humid, reddish lilac to lilac (14B5-15B5), rapidly purplish white to purplish pink (close to 14A2 and 14A3) towards the margin and in the extreme centre, slimy-glutinous when wet, cuticle nearly completely separable. Stipe 30–45 × 7–7.5 mm, central, cylindrical, uniformly white, stuffed-spongy inside. Lamellae adnate, equal or with occasional lamellulae, 3–3.5 mm high, medium-crowded, strongly anastomosing in between and towards the stipe attachment, brittle, cream white. Context white, 1 mm thick in the pileus. Taste acrid. Odour none. Spore print not obtained.
Basidiospores (60/3/1/) (7.5) 8.0–8.5–9.5 × (6.5) 7.0–7.4–8.0 (8.5) μm [Q = (1.06) 1.08–1.23 (1.25), Q = 1.15 ± 0.04], subglobose to broadly ellipsoid; ornamentation amyloid, 0.5–1.5 μm high, composed of isolated warts never fused, conical or blunt, rarely truncate at the apex, 0.3–1.0 μm in diam.; suprahilar plage inamyloid, rarely slightly distally amyloid. Basidia 30–50 (60) × 9–15 μm, subclavate, 4-spored. Gloeocystidia common on sides and edges of the gills, 45–80 × 9–14 μm on sides, 32–52 × 7–12 μm on edges, mostly buried among the basidia, some projecting up to 20 μm beyond the basidia-layer on sides, fusiform, rarely lanceolate, often mucronate or even moniliform at the apex, with dense granular or crystalline contents turning to blackish grey in SV. Marginal cells not differentiated. Lamellar trama composed of rosettes and connective hyphae, sphaerocytes 15–30 μm in diam., connective hyphae 2–5 μm wide. Pileipellis orthochromatic in Cresyl blue, two-layered, covered with sparse slime, 500–800 μm thick, with a distinctly delimited suprapellis sitting on a well-delimited and strongly gelatinized subpellis of narrow hyphae that is abruptly separated from the voluminous sphaerocystes of trama; suprapellis a trichoderm, 50–70 μm thick, of abundant pileocystidia and inflated, septate, poorly branching, hyphal extremities composed of 1–4 cells 4–10 (12) μm wide, the terminal cells up to 37 μm long, usually tapering; pileocystidia (23) 30–60 × 5–10 μm, fusiform, sublanceolate, often mucronate or with a central knob at the apex, with strongly refractive granular or crystalline contents turning to greyish black or dark brown in SV; subpellis a loose tissue of very narrow hyphae lacking pileocystidia, 450–750 μm thick, hyphae 2–3 μm wide, hyaline, with clear outline in a gelatinized matrix, often branching, becoming repent and slightly wider (up to 4 μm) towards pileus trama, at the very bottom and adjacent trama with scattered to moderately numerous cylindrical cystidioid hyphae 80–260 × 7–13 μm, those with strongly refractive crystalline contents turning to greyish black in SV. Stipitipellis one-layered, 30–60 μm thick; caulocystidia common, 25–44 × 5–7 μm, fusiform, rarely sublanceolate or subcylindrical, with crystalline contents changing to greyish black in SV. Pileus and stipe trama with numerous rosettes and connective hyphae; sphaerocystes 13–70 μm in diam, connective hyphae 3–8 μm wide. Clamp connections absent.
Habit, habitat and distribution
Scattered, during fall, in mixed forest of fagaceous trees and Pinus kesiya var. langbianensis. Southwestern China and Japan (Mie, Yakushima Island).
Notes: Because of the pronounced, areolate-squamulose aspect of the pileus surface, our species is similar to those of R. subg. Heterophyllidia subsect. Virescentinae Singer, particularly those in the R. crustosa Peck complex and species in R. subsect. Aureotactinae R. Heim ex Buyck. When considering in addition the vividly coloured pileus with tuberculate-striate margin, our species could also easily be confused with some tropical African Heterophyllinae near R. roseoviolacea Buyck. Those African species, however, all have subreticulate to reticulate spores (Buyck 1994). For more notes, see under R. substriata and Discussion.
Russula substriata J. Wang, X.H. Wang, Buyck & T. Bau, sp. nov., Figs. 3b–d and 5.
MycoBank: MB 827273.
GenBank: MH724921 (ITS), MH714540 (28S), MH939992 (rpb2), MH939986 (tef1), all from holotype.
Etymology: named after the striate pileus.
Holotype: China, Yunnan Prov., Wenshan Pref., Maguan Co., Dalishu Town, Xiangchang, 23°03′59.28″ N, 104°12′41.31″ E, 1656 m asl, in mixed forest of fagaceous trees and Pinus kesiya var. langbianensis, leg. X.H. Wang, 14 Oct. 2017, no. 4766 (HKAS 102278, KUN!).
Diagnosis: A medium-sized species recognised by the sticky tuberculate-striate pileus with greyish rose tinge, finely to coarsely cracked pileus cuticle, spores with isolated warts and outmost pileipellis with aggregate pileocystidia.
Pileus 30–50 mm diam., hemispherical at first, later convex, finally concave with applanate margin, 50–70% of the radius strongly tuberculate-striate, finely to coarsely cracked even when humid, slimy-glutinous when wet, cuticle completely separable, greyish rose (11B3, 12B3), often much darker at the centre, margin cream-white or pinkish white (12A2) in age. Context of pileus 1–2 mm thick, white. Stipe 35–65 × 6–12 mm, central, equal or tapering upwards, with 6–10 cavities inside, neither extremely fragile nor very hard, cream white at the upper part, with purple-pinkish or pinkish tinge at the low part, finely scaly near base. Lamellae 2–6 mm high, much broader than flesh thickness, adnate, slightly attenuate towards pileus margin, medium-crowded (13–14 L/cm), shorter lamellulae not frequent to common, strongly anastomosing towards flesh, sometimes anastomosing and forming zone around stipe, elastic and greasy-buttery, cream white. Odour reminiscent of R. foetens. Taste mild. Spore print not obtained.
Basidiospores (200/10/5) 8.0–9.0–10.0 (11.0) × (6.5) 7.0–7.5–8.0 (9.0) μm [Q = (1.03) 1.11–1.30 (1.36), Q = 1.20 ± 0.06], broadly ellipsoid to ellipsoid, ornamentation amyloid, 0.5–1.8 μm high, composed of isolated warts with conical, round or truncate apex, suprahilar plage inamyloid. Basidia 28–63 × 11–16 μm, subclavate, 4-spored. Gloeocystidia common to numerous on sides and edges of the gills, 38–80 × 9–15 μm on sides, 23–45 × 6–11 μm on edges, mostly buried among the basidia, fusiform, mucronate or moniliform at the apex, with dense granular, crystalline or amorphous contents blackish grey in SV. Marginal cells not differentiated. Lamellar trama composed of rosettes and connective hyphae, sphaerocystes 8–50 μm in diam., connective hyphae 3–5 μm wide, oleiferous hyphae common, 5–9 μm wide, with granular contents. Pileipellis orthochromatic in Cresyl blue, mostly one-layered, locally two-layered, 220–400 μm thick, covered with sparse slime, where two-layered with a distinctly delimited suprapellis sitting on a well-delimited and strongly gelatinized subpellis of narrow hyphae that is abruptly separated from the voluminous sphaerocystes of trama; suprapellis (where two-layered) ± 50 μm thick, composed of pileocystidia and chains of 3–5 fusiform, clavate or cylindrical septate cells 4–10 (12) μm wide, the terminal cells up to 20 μm long, subclavate, cylindrical or slighly tapering; pileocystidia common to numerous, often aggregate into piles, 13–53 × 4–8 μm, fusiform, sublanceolate, often mucronate or with a central knob at the apex, with granular, crystalline or amorphous contents, nearly colourless, changing to greyish black or blackish brown in SV; subpellis (or whole pileipellis where one-layered) a loose tissue of very narrow hyphae lacking pileocystidia, 200–400 μm thick, hyphae 2–4 μm wide, often branching, strongly shrivelled, with clear outline in a gelatinized matrix, becoming repent and slightly wider (3–6 μm) towards pileus trama, at the very bottom and adjacent trama with scattered to moderately numerous cylindrical cystidioid hyphae 100–200 × 7–12 μm, those with strongly refractive crystalline contents turning to greyish black in SV. Stipitipellis one-layered, 10–55 μm thick, locally with repent or erect caulocystidia; caulocystidia common, 16–50 × 5–8 μm, subfusiform, subcylindrical, with crystalline contents turning to greyish black in SV. Pileus and stipe trama with numerous rosettes and connective hyphae; sphaerocystes 13–70 μm in diam, connective hyphae 3–8 μm wide, oleiferous hyphae common, with granular or amorphous contents. Clamp connections absent.
Additional specimens examined
China, Yunnan Prov., Wenshan Pref., Maguan Co., Dalishu Town, Xiangchang, 23°04′09″ N, 104°12′33″ E, 1650–1690 m asl, 14 Oct. 2017, leg. X.H. Wang, no. 4767 (HKAS 102279, KUN), no. 4749 (HKAS 102276, KUN); ibid., 14 Oct. 2017, leg. J. Wang, no. 292 (HKAS 102275); Maguan Co., Mabai Town, Yubo, 23°00′57.9″ N, 104°20′59.5″ E, 1364 m asl, 15 Oct. 2017, leg. X.H. Wang, no. 4785 (HKAS 102280, KUN).
Habit, habitat and distribution
Scattered, during fall, in mixed forest of fagaceous trees and Pinus kesiya var. langbianensis. Subtropical China and Japan (Mie, Yakushima Island).
Notes: The pileus of R. substriata is not as brightly coloured and lilac red as R. maguanensis, although in one specimen (HKAS 102276, Fig. 3d), the colour is close. The lamellae of R. substriata seem to be not as brittle as those of R. maguanensis, but this difference needs more testing with additional specimens of R. maguanensis. The spores of R. substriata are slightly longer than those of R. maguanensis. Absence of greenish tinge can easily distinguish these two new species from Indian R. shingbaensis and Chinese R. verrucospora (Das et al. 2014; Song et al. 2018b). The lamellulae of R. substriata are more frequent than those of R. maguanensis and R. shingbaensis (Das et al. 2014), but comparable with or fewer than in R. verrucospora. Russula verrucospora has the smallest spores (av. 5.7 × 5.0 μm) among the four Asian species.
Although R. substriata is here described as a new species, its mycorrhizae have been reported several times in Asia. In China, it was found in mixed forest of Pseudotsuga sinensis and Pinus spp. in Hunan, central China (as “Russula sp.12”, Wen et al. 2015), in mixed forest of Pinus massoniana and fagaceous trees in Zhejiang, eastern China (GenBank accession JQ991798) and in Castanopsis fargesii forest in Sichuan, southwestern China (Wang et al. 2011 and GenBank accessions JF273535 and JF273556). Matsuda et al. (2011) found that it was associated with Monotropastrum humile in oak forest in central Japan (Mie). It is one of the most common Russula species in Maguan County, Yunnan. We notice that R. substriata co-occurs with R. maguanensis both in Maguan, China and Yakushima island, Japan (Fig. 2). How two closely related species evolved sympatrically and dispersed would be an intriguing biogeographical topic.
Russula subsect. Substriatinae X.H. Wang & Buyck, subsect. nov.
MycoBank: MB 828233.
Type species: Russula substriata J. Wang, X.H. Wang, Buyck & T. Bau.
Diagnosis: Pileus sticky, tuberculate-striate or nearly so, greenish, olive, greyish rose or lilac red; lamellulae few to frequent, taste mostly mild, rarely acrid, brittle, rarely buttery; spores with isolated warts; hymenial gloeocystidia frequent, fusiform, contents SV+; pileipellis orthochromatic in Cresyl blue, two-layered, suprapellis composed of numerous to abundant pileocystidia and erect septate fusiform terminal cells, pileocystidia with contents SV+, subpellis a loose tissue of very narrow hyphae in a gelatinized matrix, lacking pileocystidia; cystidioid hyphae scattered to common at the bottom of subpellis and pileus trama, with contents SV+.
Species included: R. substriata, R. maguanensis, R. shingbaensis and four potential species from America, Australia and tropical Asia, possibly also R. verrucospora.
Discussion
The only described species that is closely related to our two new species for the moment is R. shingbaensis, a species recently discovered from Himalayan India, which is across the Chinese border with Tibet (Das et al. 2014). When considering also environmental ITS sequences, we find out that both our species and R. shingbaensis are part of a strongly supported larger clade (clade A, Fig. 2) with a distribution that encompasses not only China and India, but also Japan, the southern USA and Mexico and Australia. There are at least seven species in this clade, each occupying one particular continent and in general associated with such diverse hosts as Australian Eucalypts (Myrtacae), American and Asian Fagaceae and Pinaceae, as well as Juglandaceae in Mexico (Bastias et al. 2006; Wang et al. 2011; Das et al. 2014; Wen et al. 2015; data retrieved from Genbank). Moreover, our analysis of environmental sequences suggests that in Australia and Japan, species in this clade are subject to exploitation by heterotrophic plants to get access to the carbon produced by their host plants, and this both by orchids (Erythorchis cassythoides in Australian eucalypt stands) and ericaceous plants (Monotropastrum humile in Japanese oak stand) (Dearnaley 2006; Matsuda et al. 2011). This assemblage seems to be good representatives in ectomycorrhizal communities.
The widely distributed assemblage above, however, was never recognised as a distinct infrageneric entity in R. subg. Heterophyllidia. Using ITS data, Das et al. (2014) placed R. shingbaensis in R. subg. Heterophyllidia but could not give further assignment. Also using ITS data but a wider taxa sampling, Song et al. (2018b) found that R. shingbaensis was sister to their new species R. verrucospora and then a successive sister to R. subsect. Cyanoxanthinae, still in R. subg. Heterophyllidia. Song et al. (2018b) suggested that R. verrucospora might have affinity to tropical African R. subsect. Aureotactinae, but did not assign it to either Cyanoxanthinae or Aureotactinae due to clear morphological differences. To test the relationship with R. subsect. Aureotactinae, we included four sequences of R. cf. brunneoannulata. Our ITS phylogeny (Fig. 2) suggests that the big assemblage above (including R. shingbaensis), R. verrucospora, R. cf. brunneoannulata and R. subsect. Cyanoxanthinae fell into the same clade, although without significant support. Our 28S-rpb2-tef1 phylogeny confirmed the sibling relationship between our two species (implicitly also R. shingbaensis) and a Malagasy species close to African mainland R. brunneoannulata [R. aff. brunneoannulata in Buyck et al. 2018] and clearly suggested they represented one of the four major clades of R. subg. Heterophyllidia (Fig. 2).
Following our 28S-rpb2-tef1 phylogeny, the multi-gene phylogeny of Buyck et al. (2018) and Looney et al. (2016) and hierarchical classification, it is reasonable to split R. subg. Heterophyllidia into four sections, corresponding to the four major clades A–D (Fig. 1). In such a classification, R. subsect. Substriatinae and R. subsect. Aureotactinae compose one of the four sections (clade B), whereas the remaining three correspond to R. sect. Heterophyllae (clade A), R. sect. Ingratae (clade C) and R. subsect. Cyanoxanthinae (clade D). Morphologically, R. subsect. Substriatinae does share spores with isolated warts and pileipellis with numerous pileocystidia with its sister group R. subsect. Aureotactinae. However, it clearly differs from all African/Malagasy species of R. subsect. Aureotactinae in lacking the intense yellowing of surface and context of their fruiting bodies (see Buyck 1994). In addition, the distribution patterns of pileal cystidioid elements are different between the two groups: in R. subsect. Substriatinae, typical fusiform pileocystidia are only at the outermost surface, i.e. suprapellis, clearly separated from the long cylindrical cystidioid hyhpae at the bottom of subpellis and trama by the thick loose cystidia-free subpellis, whereas in R. subsect. Aureotatinae, as well as in R. sect. Ingratae, R. subsect. Cyanoxanthinae and most species of R. sect. Heterophyllae, there is a gradual transition from short sometimes mucronate at the surface to longer cylindrical with a blunt apex when going down to the trama (Buyck 1990; Wang et al. 2018). Such clear separation of pileal cystidioid elements is reminiscent of R. crustosa complex, which has subreticulate spores and is distantly placed in R. subsect. Virescentinae in clade A. In fact, the overall characters of R. subsect. Substriatinae look very much like a mixture up of all other sections of R. subg. Heterophyllidia, e.g. the tuberculate-striate pileus margin and the multi-chambered stipe cortex of some specimens are strongly reminiscent of R. sect. Ingratae, the short-celled branching hyphal endings in the pileipellis of R. subsection Griseinae, the finely cracked pileus cuticle (at least in R. maguanensis and R. substriata) of R. crustosa group (R. subsect. Virescentinae) and spores with isolated warts of some species of R. sect. Heterophyllae. Recognising this assemblage as an infrageneric subdivision will be of importance to highlight the diverse combinations of morphological characters within Russula.
After R. subsect. Substriatinae is described in R. subg. Heterophyllidia, two questions will be left: (1) what is the section for subsections Substriatinae and Aureotactinae? Unfortunately, there is no available valid section name for them. There are two candidate names, i.e. R. sect. Aureotactae and R. sect. Radicantes (Fig. 5). However, neither is valid (Buyck 1990). Nevertheless, we feel it is premature to validate the name in this study because the type species from Madagascar, R. aureotacta R. Heim and R. radicans R. Heim have never been found again since their description and no sequences are available to verify their true affinities to our representative of R. subsect. Aureotactinae. Moreover, the possibility that subsections Substriatinae and Aureotactinae may merit two independent sections cannot be easily excluded. Genetically, even in the phylogeny constructed with three conservative loci (with introns and ambiguous sites removed), the branches leading to subsections Substriatinae and Aureotactinae both are very long, in sharp contrast to the short branch grouping them together and the support values are relatively low in the ML analysis (ML-BP 71%) in comparison with other three equivalent lineages. In the ITS genealogy, the monophyletic relationship of the two subsections could even not be re-trieved (Fig. 2). (2) Does R. verrucospora belong to R. subsect. Substriatinae? Although Song et al. (2018b) grouped R. verrucospora with R. shingbaensis with high support, the sibling relationship of R. verrucospora and our new subsection in the ITS genealogy did not receive significant support. On top of this, the extremely small spores with typical round rather than conical or truncate warts, the presence of pileocystidia in the subpellis and the obscurely striate pileus of R. verrucospora make this species not fully fulfil the morphological criterion of R. subsect. Substriatinae. We leave these two questions as open until more representatives of “R. sect. Aureotactae” and our new subsection are sequenced and multi-gene data are available for R. verrucospora and its potential allies, to avoid any possible artfact in phylogenetic analyses (e.g. long branch attraction). This study as well addresses nomenclatural issue regarding R. subsect. Cyanoxanthinae as it merits a section in this updated classification. We leave it for a thorough update of infrageneric classification of Russula with a broader sampling.
References
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT et al (2015) Fungal diversity notes 111–252 — taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75:27–274
Bastias B, Xu ZH, Cairney JWG (2006) Influence of long-term repeated prescribed burning on mycelial communities of ectomycorrhizal fungi. New Phytol 172:149–158
Buyck B (1989) Utilité taxonomique du bleu de crésyldans le genre Russula Persoon. Bull Trimest Soc Mycol Fr 95:1–6
Buyck B (1990) Nouveaux taxons infragénériques dans le genre Russula Persoon en Afrique centrale. Bull Jard Bot Nat Belg 60:191–211
Buyck B (1994) Russula II (Russulaceae). In: Rammeloo J, Heinemann P (eds) Flore illustreé des Champignons d’ Afrique Centrale, vol 16, pp 411–542, pl 69–87
Buyck B, Hofstetter V, Eberhardt U, Verbeken A, Kauff F (2008) Walking the thin line between Russula and Lactarius: the dilemma of Russula subsect. Ochricompactae. Fungal Divers 28:15–40
Buyck B, Zoller S, Hofstetter V (2018) Walking the thin line…ten years later: the dilemma of above versus below-ground features to support phylogenies in the Russulaceae (Basidiomycota). Fungal Divers 89:267–292
Das K, Dowie NJ, Li GJ, Miller SL (2014) Two new species of Russula (Russulales) from India. Mycosphere 5:612–622
Das K, Ghosh A, Chakraborty D, Li JW, Qiu LH et al (2017) Fungal biodiversity profiles 31-40. Cryptogam Mycol 38:353–406
Dearnaley J (2006) Molecular identification of fungal endophytes in Australian myco-heterotrophic orchids. In: 8th International Mycological Congress (IMC8), 20–25 Aug 2006, Cairns, Australia
Guo J, Karunarathna SC, Mortimer PE, Xu J, Hyde KD (2014) Phylogenetic diversity of Russula from Xiaozhongdian, Yunnan, China, inferred from internal transcribed spacer sequence data. Chiang Mai J Sci 41:811–821
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hansen K, Perry BA, Dranginis AW, Pfister DH (2013) A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol Phylogenet Evol 67:311–335
Jiang XM, Li YK, Liang JF, Wu JR (2017a) Russula brunneovinacea sp. nov. from northeastern China. Mycotaxon 132:789–797
Jiang XM, Li YK, Liang JF, Wu JR (2017b) Two new Russula species in China. Journal of Fujian Agriculture and Forestry University (Natural Science Edition) 46:103–108
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Kornerup A, Wanscher JH (1961) Farver i Farver. Politikens Forlag, København
Li F, Deng QL (2018) Three new species of Russula from South China. Mycol Prog 17:1305–1321
Li GJ, Li SF, Wen HA (2011) Russula zhejiangensis sp. nov. from East China. Cryptogam Mycol 32:127–123
Li GJ, Zhao D, Li SF, Yang HJ, Wen HA, Liu XZ (2012) Russula jilinensis sp. nov. (Russulaceae) from Northeast China. Mycotaxon 120:49–58
Li GJ, Zhao Q, Zhao D, Yue SF, Li SF, Wen HA, Liu XZ (2013a) Russula atroaeruginea and R. sichuanensis spp. nov. from Southwest China. Mycotaxon 124:173–188
Li GJ, Zhao D, Li SF, Yang HJ, Wen HA, Liu XZ (2013b) Russula changbaiensis sp. nov from Northeast China. Mycotaxon 124:269–278
Li YK, Zhang X, Yuan Y, Cao Z, Liang JF (2015a) Morphological and molecular evidence for a new species of Russula (Russulaceae) from southern China. Phytotaxa 202:94–102
Li GJ, Zhao D, Li SF, Wen HA (2015b) Russula chiui and R. pseudopectinatoides, two new species from southwestern China supported by morphological and molecular evidence. Mycol Prog 14(6):33
Li GJ, Zhang CL, Lin FC, Zhao RL (2018a) Hypogeous gasteroid Lactarius sulphosmus sp. nov. and agaricoid Russula vinosobrunneola sp. nov. (Russulaceae) from China. Mycosphere 9:838–858
Li GJ, Zhang CL, Zhao RL, Lin FC (2018b) Two new species of Russula from Northeast China. Mycosphere 9:431–443
Liu YJ, Whelen S, Benjamin DH (1999) Phylogenetic among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808
Liu XL, Bau T, Wang XH (2017) Species diversity of Russula from the greater and lesser Hinggan Mountains in Northeast China. Mycosystema 36:1355–1368
Looney BP, Ryberg M, Hampe F, Sánchez-García M, Matheny PB (2016) Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol Ecol 25:630–647
Matsuda Y, Okochi S, Katayama T, Yamada A, Ito S-I (2011) Mycorrhizal fungi associated with Monotropastrum humile (Ericaceae) in central Japan. Mycorrhiza 21:569–576
Moncalvo JM, Lutzoni F, Rehner SA, Jhonson J, Vilgalys R (2000) Phylogenetic relationships of agaric fungi based on nuclear large subunit ribosomal DNA sequences. Syst Biol 49:278–305
Nylander JAA (2004) MrModeltest 2.3. Program distributed by the author. Evolutionary Biology Center, Uppsala University
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using tracer 1.7. Syst Biol 67:901–904
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A et al (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Sang XY, Li XD, Wang YW, Fan L (2016) Four new sequestrate species of Russulaceae found in China. Phytotaxa 289:101–117
Singer R (1962) The Agaricales in modern taxonomy. 2nd Edition. J. Cramer, Weinheim
Song B, Li TH, Wu XL, Li JJ, Shen YH, Lin QY (2007) Known species of Russula from China and their distribution. Journal of Fungal Research 5:20–42
Song Y, Buyck B, Li JW, Yuan F, Zhang ZW, Qiu LH (2018a) Two novel and a forgotten Russula species in sect. Ingratae (Russulales) from Dinghushan Biosphere Reserve in southern China. Cryptogam Mycol 39:341–357
Song Y, Li J, Buyck B, Zheng J, Qiu LH (2018b) Russula verrucospora sp. nov. and R. xanthovirens sp. nov. two novel species of Russula (Russulaceae) from southern China. Cryptogam Mycol 39:129–142
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SSN, Liu JK et al (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 83:1–261
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Wang XH, Yang ZL, Li YC, Knudsen H, Liu PG (2009) Russula griseocarnosa sp. nov. (Russulaceae, Russulales), a commercially important edible mushroom in tropical China: mycorrhiza, phylogenetic position, and taxonomy. Nova Hedwigia 88:269–282
Wang Q, Gao C, Guo LD (2011) Ectomycorrhizae associated with Castanopsis fargesii (Fagaceae) in a subtropical forest, China. Mycol Prog 10:323–332
Wang XH, Buyck B, Verbeken A, Hansen K (2015) Revisiting the morphology and phylogeny of Lactifluus with three new lineages from southern China. Mycologia 107:941–958
Wang XH, Das K, Horman J, Antonin V, Baghela A et al (2018) Fungal biodiversity profiles 51-60. Cryptogam Mycol 39(2):1–47
Wen ZG, Murata M, Xu ZY, Chen YH, Nara K (2015) Ectomycorrhizal fungal communities on the endangered Chinese Douglas-fir (Pseudotsuga sinensis) indicating regional fungal sharing overrides host conservatism across geographical regions. Plant Soil 387:189–199
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. A guide to methods and applications. San Diego, Academic, pp. 315–322
Yin JH, Zhang P, Gong QF, Chen ZH (2008) Sequence analysis of the internal transcribed spacer of gene coding for rDNA in Russula subnigricans and R. nigricans. Mycosystema 27:237–242
Zhang JB, Li JW, Li F, Qiu LH (2017) Russula dinghuensis sp. nov. and R. subpallidirosea sp. nov., two new species from southern China supported by morphological and molecular evidence. Cryptogam Mycol 38:191–203
Zhao Q, Li YK, Zhu XT, Zhao YC, Liang JF (2015) Russula nigrovirens sp. nov. (Russulaceae) from southwestern China. Phytotaxa 236:249–256
Funding
This study is funded by the CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences (project no. LPB201501) and the project “Investigation of Macrofungi of Maguan County” issued by Ministry of Ecology and Environment of the People’s Republic of China to XHW.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Zhu-Liang Yang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, J., Buyck, B., Wang, XH. et al. Visiting Russula (Russulaceae, Russulales) with samples from southwestern China finds one new subsection of R. subg. Heterophyllidia with two new species. Mycol Progress 18, 771–784 (2019). https://doi.org/10.1007/s11557-019-01487-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-019-01487-1