Abstract
During studies on Mucorales in semiarid and littoral dune areas in the northeast of Brazil, two cultures of an Absidia-like species were isolated from soil. They were characterized based on morphological, physiological and molecular data (5.8S and LSU rDNA sequences). The phylogenetic analyses of the isolates revealed that they belong to the Lichtheimiaceae and are closely related to species of Lichtheimia. The two isolates produced simple or branched, erect and circinate sporophores, occasionally with a septum under the sporangia, characteristics also common in Lichtheimia species. However, different from the described Lichtheimia species, the columellae of our isolates were mainly short hemispherical, never spatulate or elliptical and without projections. Sometimes, a long conical or bell shaped apophysis was found. Both isolates grew better at 30–35 °C, with no development at 42 °C, and giant cells were not observed. Based on the evidence of the analyzed datasets a new species of Lichtheimia is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Absidia Tiegh. (Mucoromycotina, Mucorales) includes species commonly isolated from soil, characterized by the production of stolons, rhizoids and apophysate sporangia with deliquescent walls. One single septum is formed in the sporangiophore and the zygospores are formed from suspensor cells that contain several appendices (Benny 2010). Sporophores of Absidia are never produced opposite to the rhizoids like those of Apophysomyces P.C. Misra and Rhizopus Ehrenb. (Misra et al. 1979; Zheng et al. 2007). In general, taxa of Absidia present fast growth between 25 and 34 °C, although some representatives are capable of growing between 12 and 37 °C (Hoffmann et al. 2007).
Since Absidia was described (van Tieghem 1876), some species of this genus were transferred to other genera proposed later, such as Tieghemella Berl. & De Toni, Mycocladus Beauverie, Proabsidia Vuill., Pseudo-Absidia Bainier, Lichtheimia Vuill. and Protoabsidia Naumov. However, in time, with the exception of Lichtheimia, the other five genera were synonymyzed with Absidia (Hesseltine and Ellis 1964; Schipper 1990; Kirk et al. 2008).
Until 2007, the delimitation of Absidia species was mainly based on morphology criteria, such as size and form of vegetative structures and those of sexual and asexual reproduction. Beauverie (1900) proposed the genus Mycocladus Beauverie, containing only M. verticillata, which is morphologically identical to some species of Absidia. Mirza et al. (1979) also recognized the genus Mycocladus to group a few species of Absidia that grew well at 37 °C, never produced sporophores in whorls and had optimum temperature to form zygospores at 31 °C. Thus, A. blakesleeana Lendn., A. corymbifera (Cohn) Sacc. & Trotter, A. hyalospora (Saito) Lendn., A. parricida Muskat and A. ramosa (Zopf) Lendn. were transferred to Mycocladus. However, these species (already within Mycocladus) were considered invalid, according to Art. 36.1 of the International Code of Botany (ICBN) (Benny 2010). Therefore, in 2007, 27 species and nine varieties of Absidia were accepted (Benny 2010). Hoffmann et al. (2007), in a study that combined molecular techniques (with a phylogenetic approach) with physiology and micromorphology studies of Absidia, showed that known species presented differentiated growth under different temperatures, and divided them into three groups: thermotolerant—optimum growth temperatures above 37 °C (between 37 and 45 °C); mesophilic—optimum growth temperatures between 25 and 34 °C; mycoparasite—species potentially mycoparasitic on other Mucorales with optimum growth temperatures below 30 °C. Thus, Hoffmann et al. (2007) reintroduced the genus Mycocladus and proposed that those thermotolerant species of Absidia presenting zygospores with suspensor cells without appendices, and forming equator rings on the zygospores (Absidia-like species), were transferred again to this genus, as follows: M. blakesleeanus (Lendn.) J.H. Mirza, M. corymbifer (Cohn) J.H. Mirza, M. ramosus (Zopf) J.H. Mirza and M. hyalospora (Saito) J.H. Mirza. The two mycoparasite species (A. parricida Renner & Muskat ex Hesseltine & J.J. Ellis and A. zychae Hesseltine & J.J. Ellis) were transferred to the genus Lentamyces Kerst. Hoffm. & K. Voigt by Hoffmann and Voigt (2009), including L. parricida (Renner & Muskat ex Hesselt. & J.J. Ellis) Kerst. Hoffm. & K. Voigt and L. zychae (Hesselt. & J.J. Ellis) Kerst. Hoffm. & K. Voigt.
After a meticulous review of the original description of M. verticillata, Hoffmann et al. (2009) verified that this species is not thermotolerant, as it does not grow above 40 °C, having its optimum growth at 30 °C. Besides, the zygosporangia’s suspensor cells of M. verticillata present appendices and the zygosporangial wall is ornamented with scales (or tubercles), sexual characteristic corresponding to Lentamyces parricida (Hoffmann and Voigt 2009). Therefore, Hoffmann et al. (2009), considering the elevated probability of the M. verticillata culture observed by Beauvérie to be a mesophilic co-culture of Absidia sp. with Lentamyces sp., transferred the thermotolerant species to the genus Lichtheimia Vuill. (Vuillemin 1903), of which the type species is L. corymbifera (Cohn) Vuill. (Vuillemin 1903). Lichtheimia blakesleeana (Lendn.) Kerst. Hoffm., Walther & K. Voigt, L. corymbifera, L. hyalospora (Saito) Kerst. Hoffm., Walther & K. Voigt and L. ramosa (Zopf) Vuill. have been validated and included in the new Lichteimiaceae K. Hoffm., G. Walther & K. Voigt family. Next, Alastruey-Izquierdo et al. (2010) transferred A. ornata A.K. Sarbhoy to Lichtheimia (L. ornata [A.K. Sarbhoy] A. Alastruey-Izquierdo & G. Walther), described L. sphaerocystis A. Alastruey-Izquierdo & G. Walther and placed L. blakesleeana in synonymy with L. hyalospora.
Although morphology and mating behaviour are important characteristics in traditional concepts of species recognition, delimitation of species within mucoralean fungi needs to be combined with molecular data to ensure a reliable delimitation. Despite several proposed concepts for species recognition (Mayden 1997), the genealogical concordance phylogenetic species recognition (Taylor et al. 2000) seems to be the one to most likely recognize natural species (e.g., Alastruey-Izquierdo et al. 2010). One additional interesting finding is the correlation between sexual incompatibility and the occurrence of compensatory base changes (CBC) in the secondary structure of ITS2 sequences (Müller et al. 2007). CBCs are changes of paired nucleotides in the double-stranded regions of the secondary structure. With a confidence of 93 %, a single observed CBC seems to be meaningful in the detection of species boundaries (Coleman and Vacquier 2002; Müller et al. 2007).
During diversity studies of Mucoromycotina in soils from the semiarid region (Caatinga) and the Atlantic Forest in Brazil, we isolated two specimens of Lichtheimia presenting optimum growth temperatures below 37 °C. The isolates varied morphologically in relation to the other species within this genus. Based on the morphophysiology and molecular (LSU and 5.8S rDNA) analyses carried out, a new species of Lichtheimia is being proposed.
Materials and methods
Sampling sites
Soil samples were collected in the municipalities of Araripina (7º27′58″S, 40º24′53″W), Pernambuco State, and Mataraca (6°28′20″S, 34°57′10″W), Paraíba State, both located in the Northeast of Brazil. In Araripina, soils were collected in a preservation area of Caatinga belonging to the Instituto Agronômico de Pernambuco, in September 2008. The area is covered by hyperxerophilic Caatinga vegetation with stretches of deciduous forest and the climate is tropical semiarid. The rainy season begins in November and ends in April. The average annual rainfall is 431.8 mm (MME 2005a). In Matacara, soil sampling was carried out in July 2010, in sand dune areas of the Millennium Inorganic Chemicals Mining, a Cristal Company. The climate in this area is tropical rainy with dry summer. The rainy season extends from February to October. The average annual rainfall is 1634.2 mm. The vegetation is predominantly sub-evergreen forest, with sub-deciduous forest and Cerrado inclusions (MME 2005b).
Isolation of Lichtheimia
A volume of 5 mg of soil were added to Petri dishes containing culture medium wheat germ agar (Benny 2008) amended with chloramphenicol (NeoFenicol—Neo Química) (100 mg.L−1). The dishes were left over the laboratory bench at room temperature (26 ± 2 °C) for 7 days, in alternate periods of light and dark. Sporangiospores were transferred directly from the colonies by touching a single sporangium using a sterile needle on a stereomicroscope (Leika EZ4), and placing the spores onto a fresh malt extract agar (MEA) plate (Benny 2008).
Experiments
Pure/axenic cultures from specimens were cultured in triplicate, in MEA and PDA, and incubated at 15, 20, 25, 30, 35, 37, 40 and 42 °C, during 15 days. Fragments of mycelia were removed from cultures, placed on microscope slides with KOH (3 %) and observed under a light microscope (Carl Zeiss Axioscope 40). The color designation of colonies was established according to Maerz and Paul (1950). Mating experiments were carried out on three SMA and PDA plates at 25, 33 and 35 °C, on which one 5 mm colony disk of each isolate was placed in opposite sides of the plates.
Molecular analyses
Fungal biomass was obtained from MEA cultures in test tubes kept at 28 °C for up to 6 days. All mycelia was removed from the test tube with a platinum loop and transferred to 2 mL screw caped microtubes containing 0.5 g of glass beads (1:1 [w:w], acid-washed, 150–212 μm and 425–600 μm; Sigma, U.S. sieve). The fungal material was macerated by vigorous agitation using a FastPrep homogenizer. Genomic DNA extraction was carried out with previously macerated material according to Góes-Neto et al. (2005), that includes a washing in chloroform/isoamyl alcohol (24:1) followed by homogenization of the material in 2 % CTAB buffer, isopropanol precipitation, 70 % ethanol washing and re-suspension in 50 μL of ultrapure water.
To amplify the ITS region, the primers ITS1 and ITS4 (White et al. 1990) were used and the LR1 (van Tuinen et al. 1998) and LSU2 (5′- GGTCCGTGTTTCAAGACGGGTCG- 3′) were employed to amplify the LSU rDNA. PCR reactions were carried out in a 50 μL volume containing 75 mM Tris–HCl pH 8.8, 200 mM (NH4)2SO4, 0.01 % Tween 20, 2 mM MgCl2, 200 μM each dNTP, 1 μM of each primer and 2 units of TaqTM DNA polymerase (Fermentas, Maryland, USA); cycling parameters were 5 min at 95 °C (1 cycle), 45 s at 94 °C, 1 min at 60 C, 1 min at 72 °C (39 cycles), and a final elongation of 7 min at 72 °C followed the last cycle.
The final amplicons were purified with the PureLink PCR Purification Kit (Invitrogen), sequenced directly or cloned with a CloneJETTM PCR Cloning Kit (Fermentas; Carlsbad, USA), following the manufacturer’s instructions, and sequenced. Sequencing was provided by the Human Genome Research Center (São Paulo, Brazil). Sequence data were compared to gene libraries (EMBL and GenBank) using BLASTn. The sequences derived from the new species were deposited in the NCBI database under the accession numbers KC740484–KC740489.
Phylogenetic analyses
The phylogeny was reconstructed using sequences of the 5.8S and partial sequences of the LSU rDNA gene. Fungal sequences were aligned in CLUSTALX (Larkin et al. 2007) and edited using BioEdit (Hall 1999). Prior to phylogenetic analysis, the model of nucleotide substitution was estimated using Topali 2.5 (Milne et al. 2004). Bayesian (two runs over 1 × 106 generations with a burning value of 2500) and maximum likelihood (1000 bootstrap) analyses were performed, respectively, in MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003) and PhyML (Guindon and Gascuel 2003), launched from Topali 2.5, using the K2P + G model for the matrix generated for the 5.8S rDNA and GTR + G model for the matrix generated for LSU rDNA. Neighbor-joining (established with the models cited above) and maximum parsimony analyses were performed using PAUP*4b10 (Swofford 2003) with 1000 bootstrap replications. The sequence alignment files were deposited in TreeBASE and are available at http://purl.org/phylo/treebase/phylows/study/TB2:S14094 and http://purl.org/phylo/treebase/phylows/study/TB2:S14099.
Sequence similarities based on alignments of the LSU, 5.8S rDNA or ITS1-5.8S rDNA-ITS2 sequences were calculated using BioEdit (Hall 1999) after shortening them to same length and exclusion of sequences containing indefinite bases.
Prediction of secondary structures and CBC estimations
Analyses for compensatory base changes (CBCs), as indicators for species boundaries, were performed after prediction of the putative secondary structures of the ITS2 sequences. Secondary structures for L. brasiliensis and L. ramosa were predicted using RNAfold (Vienna RNA website) (Gruber et al. 2008). Structures were used as templates for the other sequences of Lichtheimia using the ‘model’ option in the ITS2 Database (Schultz et al. 2006; Koetschan et al. 2012). CBCs were estimated using 4SALE (Seibel et al. 2006, 2008). Since no experimentally proven secondary structure for Lichtheimiaceae is known, the predicted structures used for CBC estimation remain highly putative. Applying the same parameters to different predictions may result in comparable structures and CBC values.
Results
Phylogenetic analyses
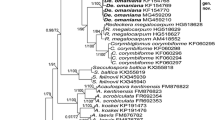
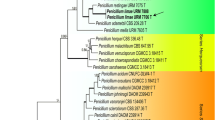
Phylogenetic analyses of the 5.8S and LSU rDNA sequences placed the two isolates in the family Lichtheimiaceae (bootstrap values > 80 %). The specimens of Lichtheimia form a separate clade close to Dichotomocladium Benny & R.K. Benj. Lichtheimia and Dichotomocladium were supported with bootstrap values above 90 % in all analyses (Figs. 1 and 2).
Phylogenetic tree of the Lichtheimiaceae constructed using 5.8S rDNA sequences. Fennellomyces linderi was used as the outgroup. Sequences are followed by their respective access number. Bootstrap values are for neighbor-joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian analysis, respectively. Sequences from this study are in bold letters. Consistency index = 0.86; Retention index = 0.96. Right site shows sequence similarity matrices for ITS1-5.8S rDNA-ITS2 (left matrix) or 5.8S rDNA (right matrix) alone. Similarities are summarized in Table 2
Phylogenetic tree of the Lichtheimiaceae and related taxa constructed using LSU rDNA sequences. Representatives of the genus Absidia were used as the outgroup. Sequences are followed by their respective access number. Bootstrap values are for neighbor-joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian analysis, respectively. Sequences from this study are in bold letters. Consistency index = 0.5r; Retention index = 0.91. Right: Sequence similarity matrix for LSU sequences. Similarities are summarized in Table 1
In the BLASTn analysis of the LSU rDNA sequences, the closest related species to the two isolates were L. hyalospora (CBS 102.36 and CBS 173.67), Lichtheimia sp. AA-2009 (CBS 958.68) and Lichtheimia sp. AA-2009a (CBS 291.66) with 90 % identity. In relation to the 5.8S rDNA sequences, several isolates of L. corymbifera and L. hyalospora showed 96 % identity with the new species. The 5.8S rDNA is a highly conserved region and can be used to separate genus and above, but does not give good resolution at the species level. The ITS 1 and ITS 2 regions were not used in the analyses due to the high level of variation between species and genera, resulting in difficulties to properly align the sequences. Based on LSU sequences, similarities between the different species of Lichtheimia and other genera include ranges overall from 60 % to 77 % and 81–100 % within Lichtheimia itself (Table 1). Sequence similarities of full-length ITS sequences (including 5.8S rDNA) range from 38 % to 45 % identity between the investigated genera Fennellomyces, Dichotomocladium and Lichtheimia and 44–65 % between species within the genus Lichtheimia itself (Table 2). Lichtheimia brasiliensis shows similar, but slightly lower interspecies similarities (44–52 %) when compared to the other Lichtheimia species (50–65 %). Yet, the highly conserved 5.8S rDNA shows 87–90 % intergeneric and 93–100 % interspecies similarities (Table 2 ). Comparing the secondary structures of ITS2 and analyzing them for CBCs, the species appears to be well delimited (Table 3). The number of observed CBCs for L. brasiliensis is comparable to other CBCs observed in this genus, supporting its proposed unique status as new species. The observed CBCs should be considered carefully, since different parameters used for the prediction of the secondary structures will lead to different structures and therefore to different values of CBCs, although in similar comparable relations (data not shown). Nevertheless, estimation of CBCs could be considered as supportive information in full accordance with all observed morphological features, including the phylogenetic analyses and the so far unsuccessful crossings.
Taxonomy
Lichtheimia brasiliensis A.L. Santiago, Lima & Oliveira, sp. nov. Fig. 3
Lichtheimia brasiliensis. a Colony surface; b Sporophores and sporangia; c Simple sporangiophore with a bell-shaped apophysis (arrow) under the sporangium; d, e Simple sporangiophore with a sporangium; f Branched sporangiophore with sporangium and columella; g, h Circinate sporangiophore; i Two sporangiophores with short hemispherical columellae and collars (arrow); j Rhizoids; k Sporangiospores. Bars: B, C, D, E, K = 50 μm; F, G, H, I = 25 μm; J =100 μm
MycoBank803795
White colonies at first then turning light grey (MP 11A1), higher in the central portion, covering the entire Petri plate and touching the plate lid in the central region and in some points in the periphery of the colony after 15 days in MEA at 25 °C. Reverse yellow (MP 11I2). Coenocytic or irregularly septate stolons, with few septa, mainly in proximity to the place of sporophore origin. Rhizoids branched and with bulbous dilatation in some points. Sporophores simple, few branched, erect, bent or circinate, raising from aerial hyphae or from stolons (the latter, in several points and terminally), solitary or in pairs, never in whorls, occasionally with one septum below the sporangium, colorless and becoming grey near the columella (2.6)3.6–7.2(12) μm diameter (diam.). Yellowish sporangia, turning brown, multispored, sphaerical or sub-pyriform, apophysate, sometimes with long conical or bell shaped apophysis, 19–55 μm diam. with a smooth, transparent wall. Columellae short hemisphaerical, sometimes almost non-existing, subglobose, never spatulate nor conical, smooth and without projections, 7.2–26.5 × 2.5–26.5 μm. Collar present or absent. Sporangiospores hyaline with smooth wall, ellipsoid, subglobose, 2.8–5.0 × 2.4–3.4. Giant cells absent. Zygospores not observed.
Holotype URM 6910.
Media, temperature tests and mating experiments: On MEA. At 15 °C—slow growth (4.2 cm in 120 h). Few sporophores formed; poor sporulation. At 20 °C—slow growth (5.3 cm in 96 h); medium sporulation. At 25 °C—faster growth than at 20 °C (9 cm in 96 h); good sporulation. At 30 °C—better growth than at 25 °C (9 cm in 72 h); excellent sporulation. At 35 °C—similar growth to 30 °C (9 cm in 72 h); excellent sporulation. At 37 °C—slower growth than at 35 °C (9 cm in 96 h); good sporulation. At 40 °C—limited growth (6.5 cm in 168 h); rare sporophores produced and sporulation poor; colonies with irregular borders and many peripheral colonies. At 42 °C—lack of growth and sporulation. The growth of L. brasiliensis on PDA was similar to that on MEA for the temperatures of 15, 20, 25, 30, 37 and 40 °C. At 35 °C the growth was faster in PDA than in MEA (9 cm in 48 h). The sporangiospores of URM 6911 are mostly subglobose or subglobose to ellipsoid, differing from URM 6910 that are mostly ellipsoid. Colonies in the former are gray to deep grey, differing from the latter that is light grey. Plates from mating experiments were analyzed at 5, 7, 14, 21, 28 and 35 d and zygosporangia were not observed.
Influence of light: not detected.
Specimen examined: Brazil, Pernambuco, Araripina, Instituto Agronômico de Pernambuco (IPA), 7º27′58″S and 40º24′53″W. Soil, 09. 2009, R.J. Oliveira. Holotype (URM 6910).
Other examined specimen. Brazil, João Pessoa, Mataraca, dune areas of the Millennium Inorganic Chemicals Mining, a Cristal Company, 6°30′00″S and 34°57′10″W, Soil, 03. 2010, D.X. Lima, (URM 6911).
Etymology: Brasiliensis. Referring to country where the species was first isolated.
Habitat: Soil
Distribution: Araripina (Pernambuco, Brazil) and Mataraca (Paraíba, Brazil).
Discussion
Several studies have reported the variability of nucleotide sequences of the ITS region (Iwen et al. 2002; Schwarz et al. 2006; Hoffmann et al. 2009) and LSU rDNA (Álvarez et al. 2011; de Souza et al. 2012) as reliable indication of taxon differentiation in Mucorales. According to our phylogenetic and morphophysiological analysis, L. brasiliensis is genetically distinct from other described species within the genus. Evidences from the analyzed datasets support the delimitation of a new species. The analysis of either the 5.8S region or the LSU rDNA showed the two sequenced specimens grouping strongly in the family Lichtheimiaceae, in the genus Lichtheimia and close to Dichotomocladium. Topology and statistical support of the phylogenetic trees, as well as genetic differences (maximum identity) between the studied specimens and representatives in Lichtheimiaceae confirm the proposal of a new species for this family.
Morphologically, L. brasiliensis resembles the described species of Lichtheimia as it presents erect or circinate sporophores emerging from aerial hyphae or from stolons, occasionally with a septum below the sporangium, besides producing apophysed sporangia that are common characteristics of the genus Lichtheimia (Hoffmann et al. 2007, 2009). However, the presence of short and very short hemispheric columellae that are never spatulate and lack projections, besides the absence of giant cells in the mycelium also differentiate L. brasiliensis from the described Lichtheimia species. Some bell shaped apophyses, sometimes very dilatated can eventually be observed in L. brasiliensis.
According to Santos et al. (2003) and Hoffmann et al. (2007, 2009), production of giant cells is considered an important morphological characteristic in Lichtheimia. However, giant cells are not produced by all isolates of this genus. Alastruey-Izquierdo et al. (2010) cited that on MEA, at 24 °C, many samples (not all) of all known species of Lichtheimia developed giant cells. These structures were also common on PDA, with higher degree of differentiation at 24 °C and lower degree of differentiation at 33 and at 37 °C. Hoffmann et al. (2007) reported the presence of giant cells as an important characteristic together with thermotolerance to include L. hyalospora in the family Mycocladiaceae K. Hoffmann, S. Discher & K. Voigt (= Lichtheimiaceae), as the zygospores in this species were never observed. These structures were cited by Ellis and Hesseltine (1966) to L. ramosa (cited as A. ramosa [Lindt] Lendn) and to L. corymbifera (cited as A. corymbifera [Cohn] Sacc. & Trotter). In the former species, the giant cells were common on PDA and absent on Synthetic Mucor Agar (SMA) and, to the latter, giant cells were common on SMA and PDA. Hesseltine and Ellis (1966) cited the presence of giant cells in L. hyalospora (cited as A. hyalospora [Saito] Lendn. and as A. blakesleeana Lendn.). In this latter species, giant cells were observed in cultures grown on SMA and MEA. Alastruey-Izquierdo et al. (2010) observed the presence of giant cells in L. sphaerocystis and in L. ornata on MEA and yeast extract agar (YEA), respectively, and used the globose form of these structures, associated with molecular information, to separate L. sphaerocystis from the other species within the genus. Thus, as some isolates of Lichtheimia do not produce giant cells, the total absence of these structures in L. brasiliensis, either on MEA or PDA, at the tested temperatures, cannot be seen as an indication to exclude it from Lichtheimia.
The presence of projections in the columellae has been cited to all known species of Lichtheimia. Hesseltine and Ellis (1966) reported projection up to 3.5 μm diam. in columellae of L. hyalospora, while Ellis and Hesseltine (1966) observed that columellae of L. ramosa and L. corymbifera presented, frequently, one to several projections of 2.5 and 4.5 μm length, respectively. Alastruey-Izquierdo et al. (2010) cited occasional production of projections of 8.5–29 μm length in L. sphaerocystis. As these structures were not observed in L. brasiliensis, the absence can be considered as distinctive characteristic between our isolate and the other Lichtheimia species. Likewise, globose, elliptical, elongated and spatulate columellae, besides the hemispheric, were described for Lichtheimia species. In L. brasiliensis only hemispheric (short and very short) columellae and few subglobose (never spatulate) were observed. Moreover, some columellae are so small that they seem inconspicuous, differently from the observed in Lichtheimia species so far.
Besides the presence of giant cells, growth temperature has been an important physiological characteristic to separate Lichtheimia species from other “Absidia-like” species. According to Hoffmann et al. (2007), growth capacity at 37 °C, or at temperatures above this, distinguish species included in the group of thermolerant (Lichtheimia) from the mesophilic (Absidia) group, being permissive to the former and suppressive to the latter group. These authors showed that L. hyalospora (as A. hyalospora and A. blakesleeana), and L. corymbifera (as A. corymbifera) presented optimum growth temperatures between 37 and 45 °C, and were capable of growing at 42, 50 and 53 °C, respectively. However, Alastruey-Izquierdo et al. (2010) observed that specimens of L. sphaerocystis presented optimum growth temperatures on MEA at 33 °C and, among the three studied specimens, two presented maximum growth temperatures at 37 °C and only one was capable of growing at 40 °C. Regarding L. brasiliensis, the two specimens studied in this work were able to grow at 37 °C, with optimum growth temperature at 30–35 °C, and very limited growth at 40 °C. The growth temperatures observed to L. sphaerocystis and L. brasiliensis indicate that these species of Lichtheimia do not exhibit strict thermotolerance.
In conclusion, none of the described species of Lichtheimia have all the characteristics as found in L. brasiliensis, therefore it is described as new. Although both specimens of L. brasiliensis have been grouped in a subclade slightly distant from the other species of the genus, when considering the two phylogenetic trees shown (5.8S region and the LSU rDNA), we did not find morphological characteristics sufficiently strong to propose a new genus.
References
[MME] Ministério das Minas e energia (2005a) Projeto cadastro de fontes de abastecimento por água subterrânea. Pernambuco. Diagnóstico do Município de Araripina. Secretaria de Geologia, Mineração e Transformação Mineral, Brasil
[MME] Ministério das Minas e energia (2005b) Projeto cadastro de fontes de abastecimento por água subterrânea. Paraíba. Diagnóstico do Município de Mataraca. Secretaria de Geologia, Mineração e Transformação Mineral, Brasil
Alastruey-Izquierdo A, Hoffmann K, de Hoog GS, Rodriguez-Tudela JL, Voigt K, Bibashi E, Walther G (2010) Species recognition and clinical relevance of the Zygomycetous Genus Lichtheimia (syn. Absidia Pro Parte, Mycocladus). J Clin Microbiol 48:2154–2170
Álvarez E, Cano J, Stchigel AM, Sutton DA, Fothergill AW, Salas V, Rinaldi MG, Guarro J (2011) Two new species of Mucor from clinical samples. Med Mycol 49:62–72
Beauverie J (1900) Mycocladus verticillatus (gen. nov. sp. nov.). Ann Univ Lyon Sér 2 Sci Méd 3:162–180
Benny GL (2008) The methods used by Dr. R. K. Benjamin, and other mycologists, to isolate Zygomycetes. Aliso 26:37–61
Benny GL (2010) Zygomycetes. Published on the internet at www.zygomycetes.org
Coleman AW, Vacquier VD (2002) Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis). J Mol Evol 54:246–257. doi:10.1007/s00239-001-0006-0
de Souza JI, Pires-Zottarelli CLA, dos Santos JF, Costa JP, Harakava R (2012) Isomucor (Mucoromycotina): a new genus from a Cerrado reserve in state of São Paulo, Brazil. Mycologia 104:232–241. doi:10.3852/11-133
Ellis JJ, Hesseltine CW (1966) Species of Absidia with ovoid sporangiospores. II. Sabouraudia 5:59–77
Góes-Neto A, Loguercio-Leite C, Guerrero RT (2005) DNA extraction from frozen field-collected and dehydrated herbarium fungal basidiomata: performance of SDS and CTAB-based methods. Biotemas 18:19–32
Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL (2008) The Vienna RNA websuite. Nucl Acids Res 36:70–74. doi:10.1093/nar/gkn188
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi:10.1080/10635150390235520
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hesseltine CW, Ellis JJ (1964) The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 56:568–601. doi:10.2307/3756362
Hesseltine CW, Ellis JJ (1966) Species of Absidia with ovoid sporangiospores. I. Mycologia 58:761–785. doi:10.2307/3756851
Hoffmann K, Voigt K (2009) Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol 10:537–554
Hoffmann K, Discher S, Voigt K (2007) Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, the Mycocladiaceae fam. nov. Mycol Res 111:1169–1183. doi:10.1016/j.mycres.2007.07.002
Hoffmann K, Walther G, Voigt K (2009) Mycocladus vs. Lichtheimia: a correction (Lichtheimiaceae fam. nov., Mucorales, Mucoromycotina). Mycol Res 113:277–278
Iwen PC, Hinrichs SH, Rupp ME (2002) Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol 40:87–109. doi:10.1080/mmy.40.1.87.109
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth & Bisby’s dictionary of the fungi, 10th edn. CAB International, Wallingford
Koetschan C, Hackl T, Müller T, Wolf M, Förster F, Schultz J (2012) ITS2 database IV: Interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Mol Phylogenet Evol 63:585–588. doi:10.1016/j.ympev.2012.01.026
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Maerz A, Paul MR (1950) A dictionary of color. McGraw-Hill book Company, USA
Mayden RL (1997) A hierarchy of species concepts: the denouement in the saga of the species problem. In: Claridge MF, Dawah HA, Wilson MR (eds) Species: the units of biodiversity. Chapman & Hall, London, pp 381–424
Milne I, Wright F, Rowe G, Marshal DF, Husmeier D, McGuire G (2004) TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20:1806–1807. doi:10.1093/bioinformatics/bth155
Mirza JH, Khan SM, Begum S, Shagufta S (1979) Mucorales of Pakistan. University of Agriculture, Faisalabad
Misra PC, Srivastava KJ, Lata K (1979) Apophysomyces, a new genus of the Mucorales. Mycotaxon 8:377–382
Müller T, Philippi N, Dandekar T, Schultz J, Wolf M (2007) Distinguishing species. RNA 13:1469–1472. doi:10.1261/rna.617107
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi:10.1093/bioinformatics/btg180
Santos MJS, de Oliveira PC, Trufem SFB (2003) Morphological observations on Absidia corymbifera and Absidia blakesleeana strains preserved under mineral oil. Mycoses 46:402–406. doi:10.1046/j.0933-7407.2003.00923.x
Schipper MAA (1990) Notes on Mucorales—I. Observations on Absidia. Persoonia 14:133–149
Schultz J, Müller T, Achtziger M, Seibel PN, Dandekar T, Wolf M (2006) The internal transcribed spacer 2 database–a web server for (not only) low level phylogenetic analyses. Nucleic Acids Res 34:704–707. doi:10.1093/nar/gkl129
Schwarz P, Bretagne S, Gantier JC, Garcia-Hermoso D, Lortholary O, Dromer F, Dannaoui E (2006) Molecular identification of Zygomycetes from culture and experimentally infected tissues. J Clin Microbiol 44:340–349. doi:10.1128/JCM.44.2.340-349.2006
Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M (2006) 4SALE—a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics 7:498
Seibel PN, Müller T, Dandekar T, Wolf M (2008) Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res Notes 1:91. doi:10.1186/1756-0500-1-91
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and Other Methods). Sinauer Associates, Sunderland
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32. doi:10.1006/fgbi.2000.1228
van Tieghem P (1876) Troisième mémoire sur les Mucorinées. Ann Sci Nat Bot sér VI 4:312–399
van Tuinen D, Zhao B, Gianinazzi-Pearson V (1998) PCR in studies of AM fungi: from primers to application. In: Varma AK (ed) Mycorrhizal manual. Springer, Berlin, pp 387–399. doi:10.1007/978-3-642-60268-9_24
Vuillemin P (1903) Le genre Tieghemella et la série de Absidées. Bull Soc Mycol France 19:119–127
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Zheng R-Y, Chen G-Q, Huang H, Liu X-Y (2007) A monograph of Rhizopus. Sydowia 59:273–372
Acknowledgments
The authors express their gratitude to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for Master scholarships provided to Rafael José Vilela de Oliveira and Diogo Xavier de Lima, respectively. We are also thankful to Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE) and to CNPq for a Post Doctoral fellowship and research grants (Protax Proc. 562330/2010-0, INCT-Herbario Virtual Proc. 573883/2008-4, Sisbiota Proc. 563342/2010-2) to the first author and to Leonor C. Maia, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de A. Santiago, A.L.C.M., Hoffmann, K., Lima, D.X. et al. A new species of Lichtheimia (Mucoromycotina, Mucorales) isolated from Brazilian soil. Mycol Progress 13, 343–352 (2014). https://doi.org/10.1007/s11557-013-0920-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-013-0920-8