Abstract
Two new species were isolated during a survey on the mycobiota of soil of sugarcane fields in a deforested area of Atlantic Forest in Pernambuco, northeastern Brazil. Using a polyphasic approach, combining ITS, partial β-tubulin, calmodulin, and RPB2 gene sequences and morphological features, we described the new species Penicillium barbosae sp. nov. (URM 7705T) and Penicillium limae sp. nov. (URM 7706T), both belonging to section Sclerotiorum, series Adametziorum. Descriptions based on morphological features are provided and these data show that the species differ from their phylogenetically closely related relatives. Both new species produce monoverticillate conidiophores and globose to subglobose shaped conidia. Penicillium barbosae is phylogenetically related to P. bilaiae; however, P. barbosae attains a colony diameter of 37–38 mm at 25 °C on Czapek yeast extract agar (CYA), is unable to grow at 30 and 37 °C, and produces sclerotia on oatmeal agar. In contrast, the colony diameter of P. bilaiae on CYA at 25 °C is 25–33 mm and is able to grow at 30 °C and 37 °C, and sclerotia production is not reported. Penicillium limae is related to P. restingae. The former species does not grow at 37 °C, in contrast to the latter. Furthermore, P. limae grows faster on CYA (22–32 mm vs 18–27 mm), malt extract agar (23–32 vs 16–23 mm), and dichloran 18% glycerol agar (35–40 vs 9–16 mm). The description of these new species increases our knowledge on Penicillium biodiversity in tropical agricultural soils.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil is one of nature’s most complex ecosystems and one of the most diverse substrates on earth, resulting from the interactions of climate, relief, organisms, and organic material, being an excellent microbial habitat, housing several microorganisms, including fungi (Prade et al. 2007; Abreu and Pfenning 2008; Ponge 2015). Fungi are found in communities usually ranging from 104 to 106 colony forming units per gram of soil (Blackwell 2011) and experience less competition in acidic environments (Brandão 1992).
Brazil is the largest producer of sugarcane (Saccharum officinarum L.) in the world and this crop is mainly used for sugar and ethanol production (Cheavegatti-Gianotto et al. 2011; Bordonal et al. 2018). In the northeastern region of Brazil, areas of the Atlantic Forest biome were deforested to give rise to the cultivation of sugarcane (Moraes et al. 2016) and the coastal zone of the Pernambuco state is an ideal region for this monoculture due to its favorable climatic and soil condition (Pereira and Alves 2007).
A limited number of studies have analyzed the fungi present in soil of sugarcane plantations in Brazil. Barros (2012) evaluated the fungal community present in soils of sugarcane plantations in the Northeast region and reported the presence of the genera Aspergillus, Fusarium, Penicillium, Phytophthora, and Trichoderma. Romão-Dumaresq et al. (2016) studied fungi in roots and the rhizosphere of two varieties of sugarcane in the state of São Paulo, and observed that the genus Penicillium was one of the most prevalent genera in the soil analyzed. Ramos et al. (2018) evaluated the species diversity of Penicillium and Talaromyces in sugarcane soils in Pernambuco state, in the Northeast region of Brazil. On a total of 1344 isolates, 1108 belonged to Penicillium and 236 to Talaromyces.
Penicillium has an important role in various natural processes and its ubiquitous presence has been reported in several studies. Penicillium species occupy a wide range of habitats, including soil environments (such as forest soil, beach soil, cultivated soil, desert soil), and species are also associated with plants and food. Several new species were discovered during ecological and biodiversity studies of specific substrates around the world (e.g., Houbraken et al. 2016; Barbosa et al. 2018, 2020). Fungal spores, like those of Penicillium, are air dispersed and can be deposited in the soil. However, due to their nutritional requirements and large enzymatic apparatus, Penicillium species have been frequently isolated from various soils, including many diverse tropical ecosystems (e.g., Cruz et al. 2013; Barbosa et al. 2016), where they actively participate in biogeochemical cycles (Visagie et al. 2014).
The genus Penicillium has been extensively revised in the last decade, due to new taxonomic insights and the introduction of a single name nomenclature system in fungi (Houbraken and Samson 2011; Visagie et al. 2014; Houbraken et al. 2020). In the most recent overview of the order Eurotiales, the genus Penicillium was subdivided into two subgenera, 32 sections and 89 series, among them section Sclerotiorum (Houbraken et al. 2020). This section was introduced by Houbraken and Samson as section Sclerotiora (2011) and typified with P. sclerotiorum. Most species in this section share the production of yellow and/or orange mycelium and have an orange or reddish colony reverse and bright-colored sclerotia (Visagie et al. 2013). The number of species described in this section has increased in the last decade and currently 36 species are accepted (Houbraken et al. 2020; Choi et al. 2021). Accurate identification of Penicillium species nowadays relies on partial β-tubulin (BenA) sequencing and these data are ideally supplemented with phenotypic and extrolite data (Visagie et al. 2014).
The aim of the present study was to describe two new Penicillium species belonging to section Sclerotiorum. The strains were isolated during a study of the mycobiota from sugarcane soil in the Northeast region of Brazil (Ramos et al. 2018). The phylogenetic position of the species in section Sclerotiorum is determined using a multigene phylogeny and species descriptions are provided.
Materials and methods
Strains
Soil samples were collected in 2014 and 2015 in an agricultural area of the Engenho Trapiche (8° 35′ 21″ S, 35° 6′ 55″ W), located in the municipality of Sirinhaém, along the southern coast of Pernambuco (Fig. 1). The soil was classified as dystrophic yellow latosol/very clayey (Saldanha et al. 2007) and analysis of the samples was performed as described in Ramos et al. (2018). The strains (Table 1) that represent the new species described in the present study were reported previously by Ramos et al. (2018). Following the recommendation of Barbosa et al. (2020), the strains were deposited in the Micoteca URM culture collection and the holotypes (slide preparation) in the URM Mycology Herbarium, both housed at the Federal University of Pernambuco, Recife, Brazil.
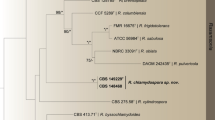
Phylogenetic position of new the Penicillium species in the section Sclerotiorum based on a combined dataset containing ITS, and partial BenA and CaM sequences. The new species P. barbosae sp. nov. (URM 7705T) and P. limae sp. nov. (URM 7706T) are shown in bold and indicated with arrow in the green clade of series Adametziorum
Morphological analysis
For morphological analysis, the strains were inoculated in three equidistant points on creatine sucrose agar (CREA) medium, Czapek yeast extract agar (CYA), CYA supplemented with 5% NaCl (CYAS), dichloran 18% glycerol agar (DG18), malt extract agar (MEA), oatmeal agar (OA), and yeast extract sucrose agar (YES). All agar media were incubated at 25 °C for 7 days. The composition of media followed Samson et al. (2010). Additional CYA and MEA plates were incubated at 5, 30, and 37 °C. Colony diameters were measured after 7 days of incubation and colony characteristics were recorded. Microscopic observations were made from colonies grown on MEA. The presence of a sexual stage was investigated in cultures incubated on CYA, MEA, and OA and incubated for at least 40 days at 25 °C. Lactic acid (60%) was used as a mounting fluid and 96% ethanol was used to remove excess conidia. A Nikon SMZ25 dissecting microscope and a Zeiss AxioImager.A2 with differential interference contrast (DIC) microscope, both equipped with Nikon DS-Ri2 cameras, were used to capture digital images using the software NIS-Elements D v4.50. The size, shape, and pigmentation of microscopic features were recorded. The mycological color chart by Rayner (1970) was used.
DNA isolation, PCR, and sequencing
Genomic DNA extractions were made from 7-day-old colonies grown on MEA using the Promega DNA isolation kit (Wizard Genomic DNA Purification Kit). Polymerase chain reaction (PCR) amplification of the ITS barcode (ITS1, 5.8S rDNA, and ITS2), BenA, calmodulin (CaM), and RPB2 gene regions was performed using the methods described by Visagie et al. (2014). PCR products were purified using the Exosap illustrative enzyme ExoProStar™ 1-Step (GE Healthcare Life Sciences) and sequenced on a LABCEN/CB sequencing platform at UFPE (Recife, Brazil) using the same primers. The electropherograms were analyzed using the BioEdit software version 7.2.5 (https://bioedit.software.informer.com/7.2/) from which the consensus nucleotide sequences were obtained.
Phylogenetic analysis
Sequence datasets were generated by combining the newly generated sequences with reference (preferably ex-type) sequences from previous studies deposited and stored the nucleotide database at NCBI (GenBank) (Table 1). The sequences were aligned using MAFFT v.7 (Katoh and Standley 2013) and manually optimized using MEGA v. 6.06 (Tamura et al. 2013). Initially, the positioning of the new species in section Sclerotiorum was analyzed with a concatenate dataset with ITS, BenA, and CaM sequences (TreeBase 28142). After this initial analysis, more comprehensive ITS, BenA, CaM, and RPB2 sequence datasets for series Adametziorum were generated and analyzed (TreeBase 28143). The combined datasets for section Sclerotiorum and series Adametziorum were made by concatenating the individual alignments using Mesquite v. 3.04 (Maddison and Maddison 2016). Phylogenetic trees were constructed using maximum likelihood analyses (ML) using RAxML-HPC v. 8.2.8 (Stamatakis 2014) BlackBox with 1000 rapid bootstrap inferences via the CIPRES science gateway (http://www.phylo.org/) (Miller et al. 2010) adopting default parameters. Bayesian inference (BI) analysis was performed in MrBayes 3.2.2 (Ronquist et al. 2012). In the Bayesian analyses, every 1000 generations were sampled and the first 25% of the samples were discarded. The most suitable substitution model was determined separately for each gene region using jModelTest v. 2.1.7 (Posada 2008). Trees were visualized in FigTree v. 1.1.2 (Rambaut 2009) and edited in Adobe Illustrator v. 5.1. Bayesian inference (BI) posterior probabilities (pp) and bootstrap (bs) values are labelled at the nodes. Branches with full support in Bayesian and ML analyses are thickened. Values below 0.95 pp and 70% bootstrap support are not shown or indicated with a hyphen.
Results
The phylogenetic relationship between the strains and the accepted species of section Sclerotiorum was determined by analysis of a concatenated sequence dataset of three loci (ITS, BenA, and CaM). According to this analysis, both new species belong to series Adametziorum (Fig. 1). The phylogenetic relationship within series Adametziorum and the concordance between the generated single-gene phylograms was studied using a more comprehensive set of strains (Fig. 2). An overview of each dataset and the most optimal substitution model is given in Table 2.
Strains URM 7705 and URM 7824, both belonging to the new species described here as P. barbosae, form a sister clade related to P. bilaiae with high statistical support in the single-gene phylogenies (ITS 1.00 pp, 99% bs; BenA 1.00 pp, 100% bs; CaM 1.00 pp, 99% bs; RPB2 1.00 pp, 100% bs) (Fig. 2) and the combined phylogram (1.00 pp, 99% bs) (Fig. 3). URM 7706 and URM 7808 (Penicillium limae sp. nov.) are in all phylogenies positioned as a sister to P. restingae with high statistical support (ITS 0.99 pp, 75% bs; BenA 1.00 pp, 98% bs; CaM 1.00 pp, 97% bs; RPB2 1.00 pp, 100% bs) (Figs. 2 and 3).
After the introduction of the two new species in this study, the number of accepted species in the section rises to 38. The morphology of the new species was compared with the phylogenetically closely related species, and details are given in Table 3 and in the “Discussion” section. Descriptions containing details of the distinguishing characteristics are provided in the “Taxonomy” section below.
Taxonomy
Penicillium barbosae S. Ramos, R. Cruz, R.N. Barbosa, Houbraken, sp. nov. Fig. 4.
MycoBank MB 837908
Etymology: In honor of Eliane Barbosa, a mycologist from the Micoteca URM, for her contributions to fungal identification.
Type: Brazil: Pernambuco: Sirinhaém, ex sugarcane soil at Trapiche - sugarcane mill factory, December 2014, collected and isolated by S. Ramos. Holotype URM 94474 (slide preparation), deposited in the URM fungarium (Recife, Brazil); ex-type strain URM 7705.
ITS barcode: MW191494. Alternative markers: BenA = MG452818; CaM = MW183245; RPB2 = LR898886.
Colony diam, 7 days (mm): CYA 37–38; MEA 39–40; YES 36–37; DG18 35–37; OA 28–30; CYAS 34–35; CREA 17–19; CYA 5 °C no growth; CYA 30 °C no growth; 37 °C no growth.
Colony characters: CYA, 25 °C, 7 days: colonies radially sulcate; margins entire, low, narrow; mycelium white; colony texture slightly floccose; sporulation weak to moderate; conidial color en masse pale greenish gray (110); exudate and soluble pigment absent; reverse buff (45) to umber (9). MEA, 25 °C, 7 days: colonies radially sulcate; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation weak to moderate; conidial color en masse pale grayish green (110); exudate absent or present as clear droplets; soluble pigment absent, reverse pale luteous (11) to orange (7). YES, 25 °C, 7 days: colonies radially sulcate slightly, raised at center; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation moderate; conidial color en masse grayish green (110); exudate absent; soluble pigment absent, reverse pale luteous (11) to orange (7). DG18, 25 °C, 7 days: colonies plane, slightly raised at center; margins low, entire; mycelium white; colony texture slightly floccose; sporulation weak; conidial color en masse grayish green (110); exudate absent; soluble pigment absent; reverse straw (46). OA, 25 °C, 7 days: Colonies radially sulcate, entire; margins regular; mycelium smoke gray (105); colony texture velvety; sporulation sparse; conidial color en masse mouse gray (118); exudate absent; soluble pigment absent; reverse straw (46) to umber (9). CYAS, 25 °C, 7 days: colonies radially sulcate slightly raised at center; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation moderate; conidial color en masse grayish green (110); exudate absent; soluble pigment absent, reverse straw (46) to umber (9). CREA, 25 °C, 7 days: good growth, acid production absent.
Micromorphology: Conidiophores monoverticillate. Stipes smooth-walled, (20–)80–130(–170) × 2–3 μm, vesiculate, up to 7 μm, sometimes non-vesiculate. Phialides ampulliform, (8–10–11) × 2–3 μm. Conidia globose to subglobose, echinulate, 2.5–3 μm. Sclerotia abundantly produced on OA, near substrate and covered by conidiophores, hard, pale brown, globose to subglobose, (100–)200–350 μm.
Additional material examined: URM 7824 (ITS: MW191495, BenA: MG452819, CaM: MW183246, RPB2: LR898887), from sugarcane soil at Trapiche - sugar cane mill factory, December 2014, S. Ramos.
Notes: Penicillium barbosae is phylogenetically most closely related to P. bilaiae. Both species predominantly produce monoverticillate conidiophores and roughened, globose to subglobose conidia. However, these species can be distinguished by their growth rates on CYA incubated at 30 and 37 °C. The former species is unable to grow at these temperatures, while the latter can (Visagie et al. 2013). Furthermore, sclerotia are produced by P. barbosae on OA, and these structures are not reported in P. bilaiae.
Penicillium limae S. Ramos, R. Cruz, C. Souza-Motta, N. Tinti, sp. nov. Fig. 5.
MycoBank MB 837909
Etymology: In honor of Prof. Dr. Nelson Lima, head of the fungal culture collection, Micoteca da Universidade do Minho (MUM), Braga — Portugal, for his contribution to mycology in Brazil, and especially to the Mycology Department of UFPE.
Type: Brazil: Pernambuco: Sirinhaém, ex sugarcane soil at Trapiche - sugarcane mill factory, November 2014, collected and isolated by S. Ramos. Holotype 94475 (slide preparation), deposited in the URM fungarium (Recife, Brazil); ex-type strain URM 7706.
ITS barcode: MW191493. Alternative markers: BenA = MG452820; CaM = MW183244; RPB2 = LR898888.
Colony diam, 7 days (mm): CYA 22–32; MEA 23–32; YES 36–37; DG18 35–40; OA 30–48; CYAS 29–30; CREA 12–20; CYA 5 °C no growth; CYA 30 °C 31–33; CYA 37 °C no growth.
Colony characters: CYA, 25 °C, 7 days: colonies radially sulcate; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation sparse to moderate; conidial color en masse pale greenish gray (110); exudate and soluble pigment absent; reverse straw (46). MEA, 25 °C, 7 days: colonies plane, radially sulcate; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation sparse to moderate; conidial color en masse greenish gray (110); exudate present as clear droplets; soluble pigment absent, reverse straw (46) to umber (9). YES, 25 °C, 7 days: colonies radially sulcate; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation sparse; conidial color en masse indeterminate; exudate absent; soluble pigment absent, reverse straw (46). DG18, 25 °C, 7 days: colonies plane, radially sulcate raised at center; margins low, entire; mycelium white; colony texture floccose; sporulation sparse to moderate; conidial color en masse pale greenish gray (110); exudate absent; soluble pigment absent; reverse buff (45). OA, 25 °C, 7 days: colonies radially sulcate, entire; margins regular; mycelium white; colony texture floccose; sporulation sparse; conidial color en masse pale greenish gray (110); exudate absent; soluble pigment absent; reverse buff (45). CYAS, 25 °C, 7 days: colonies radially sulcate slightly raised at center; margins entire, low, narrow; mycelium white; colony texture floccose; sporulation sparse; conidial color en masse indeterminate; exudate absent; soluble pigment absent, reverse buff (45). CREA, 25 °C, 7 days: good growth, acid production absent. No growth on CYA at 5 °C and 37 °C.
Micromorphology: Conidiophores monoverticillate. Stipes smooth walled, 10–30(–70) × 1.5–2.5 μm, mostly non-vesiculate, sometimes present, up to 6 μm. Phialides ampulliform, 6.5–7.5 × 1.7–2.5 μm. Conidia globose to subglobose, finely roughened, 2–2.5(–3) μm. Sclerotia or ascomata not observed.
Additional material examined: URM 7808 (ITS: FR997883, BenA: FR997876, CaM: FR997877, RPB2: FR997878), from sugarcane soil at Trapiche - sugar cane mill factory, December 2014, S. Ramos.
Notes: Penicillium limae is phylogenetically most closely related to P. restingae. These species (P. limae vs P. restingae) differ by the fast growth on CYA (22–32 vs 18–27 mm), MEA (23–32 vs 16–23 mm) (Crous et al. 2014), DG18 (35–40 vs 30–35) and CYAS (29–30 vs 33–35). Penicillium limae does not grow on CYA at 37 °C, while P. restingae does (23–33 mm).
Discussion
The genus Penicillium is of great importance to several scientific fields, is one of the most used fungi in biotechnology, and has great economic impact on human life, e.g., by the production of bioactive compounds such as antibiotics (penicillin, Houbraken et al. 2011) and mycotoxins (Perrone and Susca 2017). Species of this genus occur in a wide variety of habitats including soil, air, indoor environments, and food (Visagie et al. 2014). An infrageneric classification system is traditionally used in Penicillium, and currently 32 sections and 89 series are accepted (Houbraken et al. 2020). Section Sclerotiorum belongs to the subgenus Aspergilloides and includes three series (Adametziorum, Herqueorum, and Sclerotiorum). This section is phylogenetically well-defined, with several recent studies introducing new species (e.g., Visagie et al. 2013; Wang et al. 2017; Barbosa et al. 2018; Wanasinghe et al. 2018; Crous et al. 2019; Hyde et al. 2019; Choi et al. 2021). With the exception of the species of series Herqueorum (P. choerospondiatis, P. herquei, P. malachiteum, P. sanshaense, and P. verrucisporum), all other species belonging to this section produce predominantly monoverticillate conidiophores. Specific phenotypic characters that define the three series of the section could not be identified; however, species classified in series Sclerotiorum generally produce orange colonies and lack strongly colored, soluble pigments such as those generally observed in the species of series Adametziorum. The new species P. barbosae and P. limae belong to series Adametziorum and produce monoverticillate conidiophores, colored colonies, and sclerotia, indicating a relationship with other taxa of section Sclerotiorum. No sexual state was observed in the new species described in our study.
Series Adametziorum species are commonly isolated from soil around the world. In Brazil, three taxa of this series have been recorded, P. mellis from honey samples collected in Pernambuco state (Barbosa et al. 2018), P. restingae from soil (Crous et al. 2014), and P. reconvexovelosoi from leaf litter (Crous et al. 2019), the last two were collected from sand dunes in Guaibim, Bahia state. It is interesting to mention that these three reports, and also the two new species described here, were isolated in states located in the Northeast region of Brazil. Cruz et al. (2013) and Barbosa et al. (2016, 2020) reported several species of Pencillium section Sclerotiorum species in soils in Brazil; thus, our record reinforces the idea that soil is a good substrate for the isolation of Penicillium species. Some species can actively participate in biogeochemical cycles (Visagie et al. 2014). On the other hand, others might just be dispersed and latently present in the substrate. It is important to highlight that studies on the diversity and ecology of Penicillium and other genera in Brazilian tropical soils are still scarce, despite the country harboring an important component of global biodiversity (Cruz et al. 2013).
The diversity and interactions between fungi and sugarcane, one of the most important crops in Brazil, have rarely been studied. Romão-Dumaresq et al. (2016), based on a culture dependent strategy, determined the structure and diversity of the fungal community (root endophytes and rhizosphere) associated with two varieties of sugarcane in Brazil. They isolated 2236 fungal colonies, which were subsequently identified using ITS barcoding, and these data showed that the phylum Ascomycota was predominated present, and the most frequent genera were Penicillium (33.3%), Fusarium (16.9%), Aspergillus (7.2%), and Trichoderma (4.4%). These strains were unfortunately not confidentially identified at the species (or series) level. The interactions among microorganisms and plant roots are essential for the nutrition, growth, and productivity of the plant (Ortíz-Castro et al. 2009). The description of these new species adds to our knowledge of biodiversity and distribution of these organisms in soils of tropical ecosystems.
Data Availability
Strains deposited in URM Collection, sequences in GenBank/ENA/NCBI, alignments of phylogenies in TreeBASE, and names in MycoBank.
Code availability
Not applicable.
References
Abreu LM, Pfenning LH (2008) Diversidade de microfungos em solos tropicais. In: Moreira FMS, Siqueira JO, Brussaard L (eds) Biodiversidade do solo em ecossistemas brasileiros. Editora UFLA, Lavras, pp 445–481
Barbosa RN, Bezerra JDP, Costa PMO et al (2016) Aspergillus and Penicillium (Eurotiales: Trichocomaceae) in soils of the Brazilian tropical dry forest: diversity in an area of environmental preservation. Rev Biol Trop 64:45–53
Barbosa RN, Bezerra JDP, Souza-Motta CM et al (2018) New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie Van Leeuwenhoek 111:1883–1912
Barbosa RN, Bezerra JDP, Santos ACS et al (2020) Brazilian tropical dry forest (Caatinga) in the spotlight: an overview of species of Aspergillus, Penicillium and Talaromyces (Eurotiales) and the description of P. vascosobrinhous sp. nov. Acta Bot Bras 34:409–429
Barros RP (2012) Diversidade de fungos em um vertissolo com adição de vinhaça na cultura de cana-de-açúcar (Saccharum officinarum L.). Revista Uniabeu 5:181–196
Blackwell M (2011) The Fungi: 1, 2, 3…5, 1 million species? Am J Bot 98:426–238
Bordonal RO, Carvalho JLN, Lal R et al (2018) Sustainability of sugarcane production in Brazil. A review. Agron Sustain Dev 38:13. https://doi.org/10.1007/s13593-018-0490-x
Brandão EM (1992) Os componentes da comunidade microbiana do solo. In: Cardoso EJBN, Tsai SM, Neves MCP (eds). Microbiologia do solo. Campinas, Sociedade Brasileira de Ciência do Solo, pp 1–15
Cheavegatti-Gianotto A, de Abreu HMC, Arruda P et al (2011) Sugarcane (Saccharum X officinarum): a reference study for the regulation of genetically modified cultivars in Brazil. Trop Plant Biol 4:62–89
Choi DH, You YH, Lee IS, Hong SB, Jung TY, Kim JG (2021) Penicillium ulleungdoense sp. nov. from Ulleung Island in Korea. Mycobiology 49(1):46–53
Crous PW, Shivas RG, Quaedvlieg W et al (2014) Fungal Planet description sheets: 214-280. Persoonia 32:184–306
Crous PW, Wingfield MJ, Lombard L et al (2019) Fungal Planet description sheets: 951-1041. Persoonia 43:223–425
Cruz R, Santos C, Lima JS et al (2013) Diversity of Penicillium in soil of Caatinga and Atlantic Forest areas of Pernambuco, Brazil: an ecological approach. Nova Hedwigia 97:543–556
Houbraken J, Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70:1–51
Houbraken J, Frisvad JC, Samson RA (2011) Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2:87–95
Houbraken J, Samson RA, Yilmaz N (2016) Taxonomy of Aspergillus, Penicillium and Talaromyces and its significance for biotechnology. In: de Vries RP, Gelber IB, Andersen MR (eds) Aspergillus and Penicillium in the post-genomic era. Caister Academic Press, Norfolk, pp 1–15
Houbraken J, Kocsubé S, Visagie CM et al (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Stud Mycol. https://doi.org/10.1016/j.simyco.2020.05.002
Hyde HD, Tennakoon DS, Jeewon R et al (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers 96:1–242
Katoh KL, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Maddison WP, Maddison DR (2016) Mesquite: a modular system for evolutionary analysis, version 3.11. http://mesquiteproject.org
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: IEEE Gateway Computing Environments workshop (GCE), pp 1–8
Moraes ER, Domingues LAS, Medeiros MH et al (2016) Produtividade e características agronômicas da cana-de-açúcar em diferentes sistemas de preparo do solo. Revista de Agricultura Neotropical 3:27–32
Ortíz-Castro R, Contreras-Cornejo HA, Macías-Rodríguez L, López-Bucio J (2009) The role of microbial signals in plant growth and development. Plant Signal Behav 4:701–712
Pereira MS, Alves RRN (2007) Composição Florística de um remanescente de Mata Atlântica na Área de Proteção Ambiental Barra do Rio Mamanguape, Paraíba, Brasil. Revista de Biologia e Ciências da Terra 7:1–10
Perrone G, Susca A (2017) Penicillium species and their associated mycotoxins. In: Moretti A, Susca A (eds) Mycotoxigenic Fungi. Methods in molecular biology, vol 1542. Humana Press, New York
Ponge JF (2015) The soil as an ecosystem. Biology and fertility of soils, vol 51. Springer Verlag, pp 645–648
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Prade CA, Matsumura AT, Ott AP, Porto ML (2007) Diversidade de fungos do solo em sistemas agroflorestais de citrus com diferentes tipos de manejo no município de Roca Sales, Rio Grande do Sul, Brasil. Biociências 15:73–81
Rambaut A (2009) FigTree version 1.3.1 [computer program] Available from: http://tree.bio.ed.ac.uk
Ramos SMS, Silva LR, Barbosa RN, Machado AR, Costa AF, Souza-Motta CM, Oliveira NT (2018) Penicillium and Talaromyces communities of sugarcane soils (Saccharum officinarum L.): ecological and phylogenetic aspects. J Agric Sci 10:335–350
Rayner RW (1970) A mycological colour chart. Commonwealth Mycological Institute and British Mycological Society. Kew, Surrey, UK
Romão-Dumaresq AS, Dourado MN, Fávaro LCDL et al (2016) Diversity of cultivated fungi associated with conventional and transgenic sugarcane and the interaction between endophytic Trichoderma virens and the host plant. PLoS ONE 11:e0158974. https://doi.org/10.1371/journal.pone.0158974
Ronquist F, Teslenko M, Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Saldanha ECM, Rocha AT, Oliveira ECA, Nascimento CWA, Freire FJ (2007) Uso do gesso mineral em latosssolo cultivado com cana de açúcar. Caatinga 20:36–42
Samson RA, Houbraken J, Thrane U et al (2010) Food and indoor fungi. CBS Laboratory Manual Series 2. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands
Stamatakis A (2014) RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Visagie CM, Houbraken J, Rodriques C et al (2013) Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31:42–62. https://doi.org/10.3767/003158513X667410
Visagie CM, Houbraken J, Frisvad JC et al (2014) Identification and nomenclature of the genus Penicillium. Stud Mycol 78:343–372
Wanasinghe DN, Phukhamsakda C, Hyde KD et al (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers 89:1–236
Wang XC, Chen K, Zeng ZQ, Zhuang WY (2017) Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci Rep 7:8233. https://doi.org/10.1038/s41598-017-08697-1
Acknowledgements
The authors are grateful for the technical support offered by the Post-graduate Program in Fungal Biology of UFPE. We also thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for scholarships and Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE).
Funding
FACEPE (APQ-0350-2.12/19).
Author information
Authors and Affiliations
Contributions
SMSR, RC, and NTO prepared the project and guided the experiment; SMSR and RC collected the material and executed the specified methodology; SMSR, RC, and RNB wrote the text; SMSR, RC, RNB, and JH identified the species; ARM and RNB made the phylogenetic trees; all authors revised the text.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Roland Kirschner
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ramos, S.M.S., Cruz, R., Barbosa, R.d.N. et al. Two new Penicillium section Sclerotiorum species from sugarcane soil in Brazil. Mycol Progress 20, 823–835 (2021). https://doi.org/10.1007/s11557-021-01705-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11557-021-01705-9