Abstract
Purpose
Precise localization of cystic bone lesions is crucial for osteolytic bone tumor surgery. Recently, there is a move toward ultrasound imaging over plain radiographs (X-rays) for intra-operative navigation due to the radiation-free and cost-effectiveness of the modality. In this process, the intra-operative bone model reconstructed from the segmented ultrasound image is registered with the pre-operative bone model. Deep learning approaches have recently shown remarkable success in bone surface segmentation from ultrasound images. However, to train deep learning models effectively with limited dataset size, data augmentation is essential. This study investigates the applicability of the generative approach for data augmentation as well as identifies standard data augmentation approaches for bone surface segmentation from ultrasound images.
Methods
The generative approach we used in our work is based on Pix2Pix image-to-image translation network. We have proposed a multiple-snapshot approach, which mitigates the uni-modal deterministic output issue in the Pix2Pix network without using any complex architecture and training process. We also identified standard data augmentation approaches necessary for ultrasound bone surface segmentation through experiments.
Results
We have evaluated our networks using 800 ultrasound images from trained regions (humerus bone) and 1200 ultrasound images from untrained regions (tibia and femur bones) using four different augmentation approaches. The results show that the generative augmentation approach has a positive impact on accuracy in both trained (+ 4.88%) and untrained regions (+ 25.84%) compared to using only standard augmentations. Moreover, compared to standard augmentation approaches, the addition of the generative augmentation approach also showed a similar trend in both trained (+ 8.74%) and untrained (+ 11.55%) regions.
Conclusion
Generative approaches are very beneficial for data augmentation, where limited dataset size is prevalent, such as ultrasound bone segmentation. The proposed multiple-snapshot Pix2Pix approach has the potential to generate multimodal images, which enlarges the dataset considerably.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Surgery for osteolytic bone tumors requires precise localization of cystic bone lesions. The localization process is performed by registering the intra-operative bone surface to a pre-operative 3D bone model using magnetic resonance imaging (MRI) images. X-ray imaging has been commonly used to extract intraoperative bone surfaces [1]. However, recent studies have proposed the use of ultrasound images to eliminate the risk of radiation overdosing, especially where real-time navigation is required [2]. Ultrasound imaging has gained considerable interest as an intra-operative modality due to its cost-effective, non-invasive, and radiation-free nature. The bone surfaces segmented from ultrasound images can be used to reconstruct the intra-operative bone model in real-time for registration with pre-operative MRI 3D bone models. Because the MRI 3D bone model includes cystic bone lesions, this approach can localize the lesion in intra-operative settings without the use of X-ray imaging.
However, bone surface segmentation from ultrasound images is challenging due to speckle noise, shadow noise, low contrast, and the presence of imaging artifacts. Numerous approaches have been reported to address these challenges, including dynamic programming [3], random forest [4], local phase feature [5], and others [6]. Recently, deep learning-based methods have shown remarkable success [7]. The success of any deep learning approach greatly depends on the availability of a quality training dataset with a large number of samples. Nevertheless, it is difficult to obtain a large, publicly available dataset for ultrasound images for bone segmentation due to patient information security, data privacy, and lack of data sharing practices among institutes, etc. Moreover, most medical imaging datasets require domain experts for data labeling. Therefore, appropriate data augmentation is essential in the training of a discriminative deep learning model for a small training dataset.
Data augmentation has long been used for training deep learning algorithms [8], mostly to encourage generalization of the trained network to unseen data and to improve the results obtained from a smaller training dataset. Since then, rotation, reflection, translation, scaling, and cropping are some of the most prominent data augmentation techniques that have been used to train deep learning algorithms. However, the choice of data augmentation operation is very context-specific, and it is crucial to determine which data augmentation operations are required for the given problem at hand. In addition to standard augmentation practices, generative adversarial networks (GAN) [9] have recently joined data augmentation [10, 11] approaches because of their ability to find inherent data distributions to produce realistic synthetic samples. Recently, Shin et al. [12] reported the use of GAN to produce synthetic brain MRI slices, which gave improved segmentation results. Moreover, Frid-Adar et al. [13] used GAN-based augmentation for classifying liver lesions, which showed improved performance when used together with standard augmentation techniques.
In ultrasound imaging for bone surface segmentation, and for medical imaging in general, patient-specific non-dominant anatomical features can cause the trained model to produce inaccurate results, as standard data augmentation approaches very rarely account for these differences. However, generative approaches can come off improved performance in this regard through their ability to tweak these types of non-dominant features to produce realistic samples. These approaches are often described as image-to-image translation networks [14] and mostly use conditional GANs [15].
However, in image-to-image translation networks, the generated output is very deterministic, producing only one variation of a synthetic sample from one input image, which restricts the diversity of the augmented data. In contrast, traditional augmentation operations can produce vast numbers of augmented samples. Although some very successful architectures, such as BicycleGAN [16], have been proposed that can offer partial solutions to the issue, the complexity of their architecture and substantial expensive training process represent barriers for their usefulness in our case.
The primary objective of this study is to investigate the applicability of the generative approach for training-data augmentation in deep learning-based ultrasound bone surface segmentation. Moreover, we sought to identify the standard augmentation techniques required for ultrasound imaging of bone surfaces.
Our main contribution is that we adopted a generative approach for data augmentation for ultrasound bone surface segmentation and evaluated the performance gain in the context of various augmentation approaches. Besides, we propose a straightforward approach to solve uni-modal deterministic output issues for image-to-image translation architecture used in our case. We also empirically determine the standard data augmentation approaches necessary for ultrasound imaging for bone surface segmentation.
Methods
Deep learning architecture for ultrasound bone segmentation
Data
We acquired 10,165 ultrasound images from 25 healthy volunteers (2 females and 23 males) from humerus, femur, and tibia regions with approval from the institutional review board (IRB) [KNUH 2018-08-015]. In-house domain experts performed the labeling for all acquired ultrasound images. For this work, we trained our networks with 3104 slices of humerus ultrasound images from 17 volunteers. Architecture validation was then performed on 560 slices of humerus ultrasound images from three volunteers, and 870 slices of humerus ultrasound images from the remaining five volunteers were used to evaluate the network performance. We also used 1200 slices of tibia and femur ultrasound images from the same five volunteers to evaluate the performance in untrained ultrasound imaging regions.
CNN architecture for segmentation
Image segmentation is a process where each pixel of an image is provided with a class label, generating the same-sized output image. In this work, we used a standard U-Net architecture [17], where the input image is passed through several contracting layers followed by several expanding layers to provide an output image of the same size, as shown in Fig. 1. In addition to supplying information to the next layer, the contracting layer-group also provides information to subsequent layers in the expanding layer-group through long skip-connections.
Each frame of the ultrasound sequence was cropped and/or padded to a size of \( 512 \times 512 \) for training. Each pixel of the ultrasound images was labeled as a bone/non-bone pixel. We used binary cross-entropy as the loss function for optimization with the Adam optimizer [18] with a batch size of 8 and an initial learning rate of \( 1{\text{e}}^{ - 4} \) for 100 epochs.
Standard approaches for data augmentation
In the case of the ultrasound bone segmentation problem, we carefully experimented to determine the types of augmentation required, which we have observed in real-world contexts. Although most of the observations were made using the Philips HD11 XE Ultrasound system, they are valid for almost all off-the-shelf ultrasound systems used for bone imaging.
Time gain compensation
Time gain compensation (TGC) is the most-used feature for ultrasound bone imaging and is used to overcome the ultrasound attenuation caused by the echo traveling back from the deeper layers. Using the TGC, weaker signals from deeper layers are compensated by boosting their gain. In our experiment, the TGC control demonstrated changes in brightness and/or contrast in the ultrasound images, as shown in Fig. 2, which could be augmented in the deep learning training process by random brightness and/or contrast enhancement of the ultrasound images without affecting the related ground truth labels.
Effect of TGC control on ultrasound images, which shows a change in brightness and contrast. TGC line represents the amount of gain compensation used in different depth levels. a The line is set to low for the whole image, b the line is set to high on the upper half of the image, and c the line is set to high for almost the whole image
Depth control
Bones in ultrasound images show small changes in shape and depth from the top due to the depth control parameter in the ultrasound system. The phenomenon is shown in Fig. 3, where the white dotted window represents the final ultrasound image after cropping and/or zero-padding the image to a pixel size of \( 512 \times 512 \). Moreover, a small change in brightness and/or contrast can also be observed, where the shallower images tend to be brighter than the deeper images. A small degree of shearing can be randomly introduced during the training to augment the shape change behavior. The change in depth of the bone from the top can be augmented either by random cropping or random translation of the image. The shearing, random cropping, and random translation should also be applied to the related ground truth labels.
Roll and pitch motion of the ultrasound probe w.r.t. bone surface
Roll motion of the ultrasound probe w.r.t. longitudinal bone surface direction produces a rotated bone surface response in the output, as shown in Fig. 4. Random rotation of the image to a certain degree can represent the roll motion adequately for our purpose.
The pitch motion of the ultrasound probe w.r.t. bone surface longitudinal direction produces a thicker and softer bone response in the output, as shown in Fig. 5. In the inclined position, the bone surface from adjacent regions w.r.t. the ultrasound probe center plane is also registered in the probe while showing different levels of signal attenuation due to varying depth of signals traveling back to the probe. This effect produces a softer and thicker bone response, whereas in the probes’ perpendicular position registers a thinner and sharper bone response. As discussed in [6], ultrasound scanning in the elevational direction also produces a similar effect on the bone response. The effect of the pitch motion and elevational directional scanning can be augmented by adding a Gaussian blurring effect on the ultrasound images without affecting the related ground truth labels.
Generative approach for data augmentation
Generative adversarial network [9] (GAN) is a deep learning approach to identify the inherent distribution of a dataset and leveraging that to generate realistic synthetic samples. GANs have long been used for data augmentation [10, 11]. Recently, GANs have also been used for data augmentation for deep learning algorithms in the medical imaging domain [12, 13] to solve image-to-image translation problems. Although many approaches have been reported for image-to-image translation, the Pix2Pix architecture [14] is one of the most popular choices due to its simplicity. We used this architecture for the generative data augmentation approach.
Pix2Pix architecture
While GANs generate an output image \( y \) from a random noise vector \( z, G: z \to y \), in Pix2Pix architecture, the output image \( y \) is generated from input image \( x \) in addition to the noise vector \( z, G:\left\{ {x,z} \right\} \to y \). However, the authors of Pix2Pix architecture argued that the noise vector \( z \) does not contribute to the diversity of the output image and is usually ignored by the network [14]. Therefore, the noise vector \( z \) should be discarded from the equation, leading to \( G:x \to y \). The loss function of the network becomes-
where the discriminator \( D \) tries to maximize the log-likelihood between real image-label pair and minimize the log-likelihood between fake image-label pair. The authors of Pix2Pix architecture also added \( L1 \) distance between the generated fake image and the real image—
The final objective of the whole network becomes—
where \( \lambda \) is the weighting factor between adversarial loss \( {\mathcal{L}}_{cGAN} \left( {G,D} \right) \) and pixel loss \( {\mathcal{L}}_{L1} \left( G \right) \). In our implementation, we set it to a value of 1, so that both losses can influence the network on their own.
The generator \( G \) of the network uses a standard U-Net architecture [17], which proved very effective for medical image segmentation with its long skip connections between contracting and expanding layers. The discriminator \( D \) is a PatchGAN discriminator architecture introduced by Pix2Pix architecture [14], where the discriminator \( D \) tries to classify whether each of the \( \left( {N \times N} \right) \) patches of the image are real or fake and then averages the outcome to produce the final \( D \) score.
Multiple snapshot Pix2Pix
Pix2Pix networks are known to produce deterministic output images from the input images, thus providing only one output ultrasound image from one segmented label image [14]. However, the primary reason for using data augmentation is to augment small datasets. While the Pix2Pix network can augment an original dataset by only two-fold, classic data augmentation approaches can provide virtually unlimited augmentation when done on the fly by using different combinations of random flip, brightness and contrast enhancement, rotation, and blurring. Although BicycleGAN proposed by Zhu et al. [16] can be used to generate more than one output image from the same input image, its architecture complexity and expensive training process render it a very unattractive option.
In this work, we proposed to use a multiple snapshot Pix2Pix approach based on the standard Pix2Pix network. In this approach, after training the network for one epoch, we visually inspected the quality of the rendered ultrasound images from the input labels. If the rendered ultrasound images are visually realistic, we stored the network weights as snapshots. The process is then repeated for a predetermined number of training epochs, and this process is illustrated in Fig. 6. Because each of the handpicked Pix2Pix snapshots have different network weights and parameters, they produce different ultrasound images from a single labeled image, thus providing an effective multimodal image-to-image translation capability without the use of complex GAN architecture or an expensive training routine.
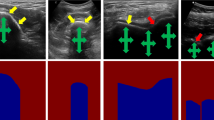
The result of the multiple snapshot Pix2Pix method can be visualized in Fig. 7, where eight handpicked snapshots from the training process were used to generate eight different ultrasound images from the same label. It is noteworthy that, even though the overall structure is very similar, there are very minute changes among the generated images, which demonstrates that generative approaches can model patient-specific non-dominant anatomical features, a challenging task using standard data augmentation approaches.
Semi-automatic snapshot selection
Visual inspection of each and every snapshot performance can be a daunting task, especially when the number of training epochs to the number of manually selected snapshots ratio is very low. Mode collapse in some training epochs renders the problem worse. Therefore, we proposed a semi-automatic approach where each snapshot is assigned a snapshot-selectability-score (SSS), which is the average distance score between periodically generated images and their ground-truth counterpart.
where \( D \) is the selected distance metric, \( R_{i} \) is the GAN-rendered image and \( G_{i} \) is the associated ground truth image. After the score assignment, the potential snapshots are selected based on the SSS score and then go through visual inspection for final selection. Any image similarity algorithm could be used as the distance metric; however, the mutual information between the two images used best for our case. The results are shown in Fig. 8.
It is notable that, using the semi-automatic approach, we were able to minimize the number of visual inspection of snapshots from 80 to 20. Although the approach still requires human intervention, it has the potential to greatly reduce the amount of said intervention.
Experimental results
To train the GAN Pix2Pix network, the 25 available volunteers were divided into training (n = 17), validation (n = 3), and testing (n = 5) subsections. For the training, we used only humerus ultrasound images. For testing, we used ultrasound images from both the trained region (i.e., humerus bone region) and untrained regions (i.e., tibia and femur regions). According to our multiple-snapshot Pix2Pix approach, as described in ``Multiple snapshot Pix2Pix” section, we handpicked 15 snapshots based on their realistic ultrasound image generation capability after training the network for 100 epochs. During the U-Net training process, we used four different data-augmentation approaches, as described in Table 1.
After training the U-Net network on the humerus ultrasound images with different augmentation approaches, we tested the network performance on the humerus ultrasound test set with 870 slices. We also tested the network with 1200 tibia and femur ultrasound image slices from the same volunteers from the test set, which was used to demonstrate how the trained networks perform on ultrasound images from unknown regions. We have used the dice similarity score to evaluate the performance of the networks. Figure 9 shows the performance analysis of the data augmentation approaches applied to our test set data. We performed our statistical analysis using SPSS (version 25.0, IBM, Chicago, IL, USA). ANOVA was used to analyze the data difference between the four groups. Afterward, we used the post hoc test (Tukey) to find out which group showed a significant difference \( \left( {p \le 0.05} \right) \).
For evaluating the usability of the multiple-snapshot Pix2Pix approach, we trained one network for each of the handpicked snapshots using the GAN + CLASSIC augmentation approach. Each of the snapshots was handpicked for their ability to render visually realistic ultrasound images from labels. Each of the trained networks was then tested on both trained and untrained regions. Figure 10 illustrates the performance of different snapshots on the test set both from the trained region and untrained regions.
Discussion
From our experiments, it is evident that the accuracy of the trained model increases when GAN-generated images are used. We observed a performance gain of \( + 8.7\% \) over the trained region and \( + 11.55\% \) over the untrained region when the GAN augmented images were used in addition to the standard augmentation approaches. Moreover, when only the GAN augmented images were used without applying any other augmentation, the performance was very similar to using conventional augmentation approaches in both trained and untrained regions. Compared to NO AUG approach where only the networks were trained without using any data augmentation approach, the GAN + NO AUG approach is \( 4.88\% \) and \( 25.83\% \) more effective in trained and untrained regions respectively. Statistical analysis confirms that there is a significant difference between GAN + CLASSIC and any of the three other approaches, which supports our hypotheses. Moreover, CLASSIC and NO AUG + GAN showed no significant difference in our analysis, which suggests that using only GAN as the augmentation approach has similar effect of using standard augmentation approaches. The analysis showed similar trend in both trained and untrained regions.
Besides, the effect of GAN augmentation is more prominent in untrained regions. The anatomical features of the humerus region, on which the network was trained, are quite different from the tibia and femur regions, which can make non-dominant anatomical features in the ultrasound image very distinct. We believe that the non-dominant anatomical features played a major role in reduced accuracy for all of the networks. However, GAN-augmented networks have a slight advantage in this regard because the GAN architectures encourage minute changes in non-dominant image features.
The multiple-snapshot Pix2Pix approach showed that the performance is very stable over different snapshots, which demonstrates that each of the snapshots can produce realistic synthetic ultrasound samples for use as augmented training samples. Therefore, the multiple snapshot approach can be used as multi-modal image-to-image translation architecture. The simplicity of the architecture and training process makes it a desirable option compared to other state-of-the-art multimodal image-to-image translation approaches, primarily where the image-to-image translation approach is used as the data augmentation scheme. Moreover, the proposed semi-automatic snapshot selection approach greatly reduce the amount of human intervention in the whole process.
In our experiments to determine the standard augmentations necessary for ultrasound bone imaging, we encountered some common issues that the segmentation algorithm could face in the real world. We have found that most of the variations can be augmented using simple operations such as contrast and brightness enhancement, rotation, translation, and Gaussian blur, etc. Although we did not suggest horizontal flip operation, since the bone ultrasound images are not symmetric, we have added horizontal flip operation in our standard augmentation approaches.
Conclusion
In this work, we have shown that images generated using GAN-based image-to-image translation architecture can be used as an additional source of augmented data for the training of deep learning algorithms, especially for ultrasound image segmentation purposes, where a limited dataset size is typical. Moreover, GAN-based architectures can generate ultrasound images with very minute changes in patient-specific non-dominant anatomical features, which otherwise can be very difficult to achieve using standard data augmentation approaches. The experimental results show that the GAN-augmented training dataset has an overall positive effect on the performance of the network. It has been previously proven that a larger training dataset always has a positive impact on the generalizability of deep learning algorithms [19]. Because the addition of GAN-augmented images offers the same advantage as the addition of more training images, it is expected to have a positive effect on network performance.
In addition, we have proposed a multiple snapshot approach for the Pix2Pix architecture used for image-to-image translation. This approach is useful for generating realistic multi-modal ultrasound images from a single input image without using an overly complicated architecture or expensive training procedure. This approach has the potential to minimize the deterministic output issue for image-to-image translation architectures. This approach can also be applied to many other similar situations. Our evaluation of the multiple snapshot approach demonstrates that the performance is very stable for different snapshots. It is possible to use the augmented datasets from each of the single snapshots differently, or multiple snapshots can be stacked together in different ratios to build a larger augmented dataset. The whole process is entirely subject-specific and should be determined by the user. The augmentation approaches we suggested are based on our observation of the ultrasound bone imaging process. Therefore, the suggested augmentation approaches could be regarded as the baseline standard augmentations for medical imaging applications where ultrasound imaging is used.
However, although our system is able to generalize over untrained bone region, since our dataset only contained ultrasound images from one specific ultrasound imaging system (Philips HD11 XE Ultrasound system), we could not show its generalization capability over different ultrasound imaging system. Our future goal would be to acquire more data using a range of imaging systems and evaluate our approach. Moreover, we would like to compare our multiple-snapshot approach to more sophisticated BicycleGAN approach which can produce similar effect as ours.
References
Soeharno H, Povegliano L, Choong PF (2018) Multimodal treatment of bone metastasis–A surgical perspective. Front Endocrinol 9:518. https://doi.org/10.3389/fendo.2018.00518
Jung EM, Friedrich C, Hoffstetter P, Dendl LM, Klebl F, Agha A, Wiggermann P, Stroszcynski C, Schreyer AG (2012) Volume navigation with contrast enhanced ultrasound and image fusion for percutaneous interventions: first results. PLoS ONE 7(3):e33956. https://doi.org/10.1371/journal.pone.0033956
Jia R, Mellon SJ, Hansjee S, Monk AP, Murray DW, Noble JA(2016) Automatic bone segmentation in ultrasound images using local phase features and dynamic programming. In: 2016 IEEE 13th international symposium on biomedical imaging (ISBI), pp 1005–1008. IEEE. https://doi.org/10.1109/ISBI.2016.7493435
Baka N, Leenstra S, van Walsum T (2017) Random forest-based bone segmentation in ultrasound. Ultrasound Med Biol 43(10):2426–2437. https://doi.org/10.1016/j.ultrasmedbio.2017.04.022
Quader N, Hodgson A, Abugharbieh R (2014) Confidence weighted local phase features for robust bone surface segmentation in ultrasound. In: Workshop on clinical image-based procedures. Springer, Cham, pp 76–83 https://doi.org/10.1007/978-3-319-13909-8_10
Hacihaliloglu I (2017) Ultrasound imaging and segmentation of bone surfaces: a review. Technology 5(02):74–80. https://doi.org/10.1142/S2339547817300049
Salehi M, Prevost R, Moctezuma JL, Navab N, Wein W (2017). Precise ultrasound bone registration with learning-based segmentation and speed of sound calibration. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 682–690 https://doi.org/10.1007/978-3-319-66185-8_77
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems. pp. 1097–1105. https://doi.org/10.1145/3065386
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Bengio Y (2014) Generative adversarial nets. In: Advances in neural information processing systems. pp 2672–2680
Antoniou A, Storkey A, Edwards H (2017) Data augmentation generative adversarial networks. arXiv preprint arXiv:1711.04340
Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, Rueckert D (2018) GAN augmentation: augmenting training data using generative adversarial networks. arXiv preprint arXiv:1810.10863
Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, Michalski M (2018) Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In: International workshop on simulation and synthesis in medical imaging. Springer, Cham, pp 1–11 https://doi.org/10.1007/978-3-030-00536-8_1
Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H (2018) GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 321:321–331. https://doi.org/10.1016/j.neucom.2018.09.013
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 1125–1134 https://doi.org/10.1109/CVPR.2017.632
Mirza M, Osindero S (2014) Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784
Zhu JY, Zhang R, Pathak D, Darrell T, Efros AA, Wang O, Shechtman E (2017) Toward multimodal image-to-image translation. In: Advances in neural information processing systems (pp 465–476)
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241 https://doi.org/10.1007/978-3-319-24574-4_28
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Sun C, Shrivastava A, Singh S, Gupta A (2017) Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE international conference on computer vision. pp 843–852 https://doi.org/10.1109/ICCV.2017.97
Acknowledgements
This material is based upon work supported by the Ministry of Trade Industry & Energy (MOTIE, Korea), Ministry of Science & ICT (MSIT, Korea), Ministry of Health & Welfare (MOHW, Korea) under Technology Development Program for AI-Bio-Robot-Medicine Convergence (20001234), and Kyungpook National University, Daegu, South Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zaman, A., Park, S.H., Bang, H. et al. Generative approach for data augmentation for deep learning-based bone surface segmentation from ultrasound images. Int J CARS 15, 931–941 (2020). https://doi.org/10.1007/s11548-020-02192-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-020-02192-1