Abstract
Purpose
Segmentation of organs from chest X-ray images is an essential task for an accurate and reliable diagnosis of lung diseases and chest organ morphometry. In this study, we investigated the benefits of augmenting state-of-the-art deep convolutional neural networks (CNNs) for image segmentation with organ contour information and evaluated the performance of such augmentation on segmentation of lung fields, heart, and clavicles from chest X-ray images.
Methods
Three state-of-the-art CNNs were augmented, namely the UNet and LinkNet architecture with the ResNeXt feature extraction backbone, and the Tiramisu architecture with the DenseNet. All CNN architectures were trained on ground-truth segmentation masks and additionally on the corresponding contours. The contribution of such contour-based augmentation was evaluated against the contour-free architectures, and 20 existing algorithms for lung field segmentation.
Results
The proposed contour-aware segmentation improved the segmentation performance, and when compared against existing algorithms on the same publicly available database of 247 chest X-ray images, the UNet architecture with the ResNeXt50 encoder combined with the contour-aware approach resulted in the best overall segmentation performance, achieving a Jaccard overlap coefficient of 0.971, 0.933, and 0.903 for the lung fields, heart, and clavicles, respectively.
Conclusion
In this study, we proposed to augment CNN architectures for CXR segmentation with organ contour information and were able to significantly improve segmentation accuracy and outperform all existing solution using a public chest X-ray database.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Chest radiography is a diagnostic method for detecting pathological changes in the chest, organs of the thoracic cavity and nearby anatomical structures. Two-dimensional chest X-rays (CXRs) remain the most commonly acquired diagnostic images, and its computerization can significantly reduce diagnostic cost and potentially improve diagnostic accuracy [1]. An important stage of computer-assisted CXR image analysis is the automated segmentation of the chest organs. Recently, Candemir and Antani [2] conducted a comprehensive review on the topic and demonstrated that CXR segmentation is an active research topic and such segmentation can significantly facilitate accurate diagnosis and quantification of chest pathologies. For example, pleural effusion and emphysema distort the healthy lung appearance and can be diagnosed from lung field segmentation [3]. The combined segmentation of lung fields and heart from CXR opens a pathway for early diagnosis of hypertension, systemic atherosclerosis, automated estimation of cardiothoracic ratio for cardiomegaly quantification, and morphometry of aortic valve boundary for diagnosis of other heart pathologies [4, 5]. Measuring the shape and size of the lung fields is a step toward the localization of pulmonary nodules and other abnormalities [6]. Segmentation of clavicles can improve the differentiation of normal and pathological structures that visually collide in the apical lung region.
The field of computerized segmentation of CXRs has been greatly facilitated by the availability of a public JSTR database [7] with manual segmentations released by van Ginneken et al. [8], who compared the performance of existing shape-based and intensity-based segmentation methods. Machine learning approaches with predefined appearance features also demonstrated potential on segmentation of CXR images [9, 10]. Recently, image segmentation based on machine learning shifted from predefining appearance features to automated feature learning through deep neural networks. In-depth training approaches achieved expert-level performance in interpreting natural and medical images [11]. Different deep learning approaches for the CXR segmentation were proposed and evaluated on the JSTR database, with the reported Jaccard coefficient reaching 0.963 for the lung fields segmentation [12,13,14,15,16,17,18,19]. The maximum-to-date accuracy was achieved by Ngo and Carneiro [20] on the JSTR database, but the authors unfortunately did not use the common evaluation protocol [8].
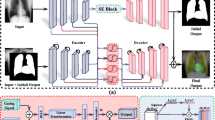
In this study, we propose a framework for contour-aware multi-label CXR organ segmentation (Fig. 1). There are the following contributions of our study. First, we analyze the benefits of augmenting deep CNNs with object contours with the aim to improve the segmentation of chest organs. We leverage the recent work on contour-aware cell segmentation [21, 22] to investigate possibilities of moving from single-object-type to multi-object-type segmentation, and check whether the improvements observed for cell segmentation continue to be present for respiratory organs with low image intensity, e.g., lungs, soft tissues with poorly visible boundaries, e.g., heart, and bones, e.g., clavicles. Second, we augmented three state-of-the-art segmentation CNNs to comprehensively evaluate contour-aware multi-label segmentation methodology. Finally, we validated the obtained results against the public JSTR database [8] and compared segmentation accuracy to 20 algorithms presented in the literature.
Methodology
After deep CNNs proved successful in solving image classification problems, they have been also adopted for image segmentation problems [23]. Two main challenges were addressed during this transition. First, the CNN pooling layer that adds local translation invariance to its input and reduces the computational complexity also progressively reduces the size of the input. While this size reduction is beneficial for classification, where a high-resolution input image is down-sampled to form an output prediction vector, it is not required for segmentation, where the output image resolution is expected to be the same as for the input image. Second, the preservation of image resolution results in a potentially rapid growth of network parameters, which may reduce the CNN generalization abilities, slow down the training phase, and affect the segmentation performance. Modern CNN architectures for image segmentation are based on mathematical concepts that are able to address both challenges, and typically consist of an encoder model followed by a decoder model (“Proposed augmented networks” section). To improve the segmentation performance, we propose to augment such architectures with organ contours (“Contour-aware multi-label segmentation” section), and consequently request the last CNN layer to return both segmentation masks and the corresponding contours (Fig. 1).
An aggregation block of a ResNet (an earlier version of ResNeXt) and b ResNeXt, both with approximately the same computational complexity in terms of floating-point operations and a similar number of parameters [24]. Each network layer is described by the number of input channels m, filter size of \(n\,{\times }\,n\), and number of output channels of p
Proposed augmented networks
In this paper, we investigate the following three state-of-the-art architectures within the proposed contour-aware approach to determine the best performing model for organ segmentation from CXR images:
The UNet architecture [23] augmented with the ResNeXt encoder [24] pre-trained on the ImageNet database [11] (See “UNet” section).
The LinkNet architecture [25] augmented with the ResNeXt encoder pre-trained on the ImageNet database (See “LinkNet” section).
The Tiramisu architecture [26] augmented with the fully convolutional DenseNet [27] (See “Tiramisu” section).
The LinkNet architecture [25]. The residual skip connections that go from early encoder to late decoder blocks are based on the layer summation operation, which, in contrast to the layer concatenation operation, does not increase network parameters in subsequent layers
UNet
The UNet architecture introduced skip connections between the down-sampling encoder and up-sampling decoder paths [23], which help to propagate features from early layers that preserve fine input details to deeper layers that aggregate high-level information but lose small image details due to a long sequence of intermediate pooling layers [28]. The UNet-based approach was shown to be efficient even when trained on relatively small databases [29], and won several public computational challenges [30]. In our experiments, we augmented the UNet architecture by replacing the original encoder with a 50-layer ResNeXt encoder [24] pre-trained on the ImageNet database [11] and adapting the corresponding decoder to the new encoder. The ResNeXt50 encoder introduces a building block that aggregates a set of transformations with the same topology, uses residual connections that augment blocks of multiple convolution layers, and generates gradient shortcuts that reduce the risk of gradient vanishing or explosion, therefore allowing to train deeper network architectures (Fig. 2) [31].
a A diagram of the Tiramisu architecture [26]. The “transition-down” operation consists of a \(1\,{\times }\,1\) convolution operation and \(2\,{\times }\,2\) pooling operation that reduce the size of the operation input. The “transition-up” operation is the transposed convolution layer that upscales its input to eventually restore the original size of the input image. b A diagram of a dense block of four layers as part of the Tiramisu architecture
LinkNet
Similarly to UNet, the LinkNet architecture [25] focuses on utilizing the parameters of network efficiency by introducing residual skip connections that bypass the features from the encoder to the decoder and applying the summation of corresponding down-sampling and up-sampling features (Fig. 3). In contrast to layer concatenation in UNet skip connections, summation does not increase the number of input channels for the subsequent layer and therefore does not result in the same growth of the number of network parameters as concatenation. Similarly to UNet, we augmented the LinkNet architecture with a 50-layer ResNeXt encoder pre-trained on the ImageNet database and adapted the corresponding decoder to the new encoder.
Tiramisu
The Tiramisu architecture [26] combines the encoder–decoder concept and the idea of densely connected CNNs–DenseNets [27]. It utilizes UNet skip connections by feature concatenation with additional feature extraction in dense blocks of the up-sampling path. The DenseNet component consists of dense blocks and pooling layers, and of a relatively small number of parameters in comparison with a regular stacked CNN that result from using direct connections from any layer to all subsequent layers (Fig. 4). By reusing the features, such an architecture becomes very efficient in terms of parameters and convergence. A Tiramisu version with 103 layers was applied to accurately segment brain tumors [32]. In our experiments, we combined the Tiramisu with a 56-layer DenseNet.
Contour-aware multi-label segmentation
The recent work on histopathological image segmentation with deep CNN architectures [21, 22] has shown certain benefits of analyzing the contours of cell nuclei jointly with their corresponding masks. In our work, we extend the idea of contour-aware segmentation into the segmentation of multiple object types. For each training CXR image, we have three binary masks representing lung fields, heart, and clavicles and we compute three contour masks by applying morphological operations to the corresponding binary masks. The segmentation CNN is trained to map an input CXR to \(N\,{=}\,6\) output channels, i.e., three channels representing segmentation masks and three channels representing contours of the lung fields, heart, and clavicles. All of the output channels are of the same size as the input CXR. The presence of the contour masks in the output will impose additional costs to the errors made at organ boundaries as not only mask channel but also the corresponding contour channel will be negatively affected by such errors. By arming a CNN with contour information, we can explicitly indicate that contour pixels preserve more valuable information than internal mask pixels, instead of assuming that the CNN will automatically recognize the information richness of contour pixels. Although contours are not used to evaluate the segmentation performance, requesting them at the CNN output is required to accommodate the corresponding CNN architecture. The idea of targeting CNNs to specific image regions has shown potential in other applications of computer-aided diagnosis, e.g., targeting CNNs on ventricular walls helps to quantify myocardial infarction [33] while targeting CNNs on anatomical landmarks helps to diagnose orthodontics abnormalities [34].
Loss function
The loss function for our networks is based on a combination of two functions, namely the Dice coefficient loss D(x, y) [35], which copes well with the cases when the foreground area is relatively small in comparison with the background area, and the binary cross-entropy B(x, y), which is preferred for classification tasks:
where x is the mask predicted by a network, y is the corresponding ground-truth mask, and P is the set of pixel indexes in the mask x (and y). A combination of binary cross-entropy and Dice coefficient losses is shown to be efficient for the segmentation of medical structures. It was utilized by the top-scoring and winning teams at the 2018 Data Science Bowl and 2019 Kidney and Kidney Tumor Segmentation Challenge [36,37,38]. The final loss function L(X, Y) is defined as:
where X denotes the output of a network that consists of N channels and Y is the corresponding ground truth that also consists of N channels. It is important to note that the CNN output has individual channels for each organ segmentation instead of uniting all organ segmentations into one multi-label channel. The reason for such an algorithm design is the projective nature of CXRs. In contrast to natural view images, where each pixel belongs to one segmentation class, organs in CXRs intersect and pixels may belong to multiple classes simultaneously. From Fig. 5, it can be seen that most of the pixels defining clavicles also belong to lung fields, etc. We, therefore, use the binary cross-entropy loss with multiple output channels instead of the categorical cross-entropy loss.
Experiments and results
Experiments
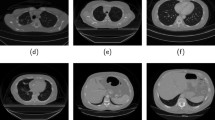
The proposed contour-aware multi-label segmentation framework was evaluated on the segmentation of lung fields, heart, and clavicles from CXR images from JSRT database [7, 8]. The JSRT database consists of 247 posteroanterior CXR images with and without lung nodules, with a resolution of \(2048\,{\times }\,2048\) pixels and pixel size of 0.175 mm (Fig. 5). To obtain the organ contours required by the proposed segmentation framework, we applied morphological edge detection by first eroding the original masks using an all-ones \(3\,{\times }\,3\) matrix and then subtracting the eroded mask from the original mask (Fig. 5c).
The images, segmentations masks and contours were subsampled to a resolution of \(512\,{\times }\,512\) pixels and partitioned into two folds as proposed by van Ginneken et al. [8]. In the twofold cross-validation scheme, we first trained the networks on the first fold and performed evaluation on the second fold and then repeated the procedure by inverting the folds. The networks used the Adam optimization algorithm [39], with the initial learning rate set to 0.001 that was reduced each time when the training processes reached a plateau, and the batch size set to 16. We also used an early stopping technique and a set of image augmentation approaches (Table 1) to reduce the risk of potential overfitting and enrich the network training phase. For the output network layer, the sigmoid function \(\sigma (x)\,{=}\,1/(1+\mathrm{{e}}^{-x})\) was used as the activation function.
Example of segmentation results (light-colored regions) in comparison with the ground truth (dark-colored contours) superimposed on the chest X-ray image. a Mask-only segmentation (UNet_ResNeXt50_Masks). b Contour-aware segmentation (UNet_ResNeXt50_Masks+Contours). The arrows indicate the regions where segmentation improvement is observed
The final segmentation masks were obtained by thresholding the probabilistic output of the networks at a 0.5 level, and the quality of the segmentation was evaluated by computing the Jaccard coefficient against the corresponding ground-truth masks.
Activation maps, obtained from the third to last layer of the UNet architecture with the ResNeXt50 decoder. a Training for the mask-only segmentation (UNet_ResNeXt50_Masks). b Training for the contour-aware segmentation (UNet_ResNeXt50_Masks+Contours), which results in sharper organ contours, e.g., lung fields, heart, and clavicles. (Note: Although activation maps in a and b correspond to the image of the same subject, there is no pairwise correspondence between them because their order is unpredictable during network training)
Results
Table 2 shows the segmentation results, achieved by the proposed augmented networks on the JSRT database, where we compared architectures with and without taking into account the contours. The best performing architecture was the UNet architecture augmented with the ResNeXt50 encoder that incorporated organ contours (i.e., UNet_ResNeXt_Masks+Contours), which reached the highest mean Jaccard coefficient for each observed organ, i.e., \(0.971 \pm 0.007\) for the lung fields, \(0.933 \pm 0.024\) for the heart and \(0.903 \pm 0.022\) for the clavicles. The incorporation of contours improved the performance of every tested network architecture. In Table 3, the results obtained by incorporating contours are compared to existing approaches evaluated on the JSRT database according to a common evaluation protocol [8]. An example of typical segmentation results is shown in Fig. 6.
Discussion
The analysis of CXR is one of the important topics in computer-aided diagnosis, which has been receiving more attention with the rapid expansion of deep learning [2]. The deep learning architectures may diagnose chest pathologies in the end-to-end fashion, i.e., directly from CXRs without a need for intermediate image processing steps [50]. It is, however, a premature conclusion to suggest that end-to-end solutions eliminated the need for organ segmentation. Lung field segmentation improves pathology localization as it was shown by some methods on the recent RSNA Kaggle Pneumonia Detection Challenge [51]. The shape features of segmented lungs can improve the accuracy of tuberculosis diagnosis [52] and can augment end-to-end solutions. Moreover, computer-aided chest pathology diagnosis is not the only problem of interest; segmentation is needed for longitudinal chest disease monitoring and standardized radiological reporting. In general, segmentation and landmark detection have shown exceptional applicability on various diagnostic challenges, including cephalometry [30], spinal structure analysis [53], and heart morphometry [54].
Log-scaled histograms of inner-boundary pixel values after the sigmoid activation function (inner-boundary pixels are all pixels within a distance of 5 pixels inward from the corresponding ground-truth mask boundary). a Mask-only segmentation (UNet_ResNeXt50_Masks). b Contour-aware segmentation (UNet_ResNeXt50_Masks+Contours)
In this study, we investigated the benefits of augmenting deep CNN segmentation architectures by including advanced feature extraction and taking into account, besides segmentation masks, also the corresponding contours [55]. We selected three state-of-the-art CNNs (i.e., UNet, LinkNet and Tiramisu) and modified them to include advanced feature extraction backbones (i.e., ResNeXt and DenseNet). The augmentation by contours architectures was evaluated on segmentation of the lung fields, heart, and clavicles from a public database of CXR images. The idea behind the proposed contour-aware segmentation is to explicitly force the CNNs to focus on organ boundaries so that during the training phase the boundary appearance features are always learned. Contour-aware segmentation performance was evaluated against existing segmentation solutions (Tables 2 and 3).
In this section, we analyzed the results of contour-aware segmentation in terms of segmentation accuracy and CNN properties. From the observed segmentation results, we can see that augmenting CNNs with contours resulted in improved accuracy for all structures, namely lung fields, heart, and clavicles, and all tested networks (Table 2, Fig. 6). It is important to note that our raw UNet_ResNeXt50_Masks resulted in a very similar performance to the UNet implementation of [16]. This observation supports the conclusion that there is minimal platform dependency in our findings. Requesting the organ contours as the network output requires it to learn the appearance of organ borders, which is expected to be manifested in the activation maps of the network. To visually confirm this theoretical expectation, we generated and compared the activation maps for the UNet_ResNeXt50_Masks and UNet_ResNeXt50_Masks+Contours networks (Fig. 7). The UNet_ResNeXt50_Masks+Contours activation maps are sharper at borders for lung fields (6th and 14th maps of Fig. 7b), heart (9th and 13th maps of Fig. 7b), and clavicles (4th and 10th maps of Fig. 7b) in comparison with activation maps for UNet_ResNeXt50_Masks returning fuzzier borders for lung fields (12th and 14th maps of Fig. 7a), heart (2nd and 10th maps of Fig. 7a), and clavicles (4th map of Fig. 7a). It is important to indicate that the activation maps for UNet_ResNeXt50_Masks+Contours highlighted the upper and lower borders of the heart. Such heart border decomposition is of high practical value and needed to compute the 1D cardiothoracic ratio, defined as the ratio between the maximum transverse cardiac diameter and the maximum thoracic diameter, and 2D cardiothoracic ratio, defined as the ratio between heart and lung perimeters [4].
In addition to visual comparison of activation maps of the mask-only and contour-aware network versions (Fig. 7), we also performed a numerical analysis of activations at organ boundaries. We computed the log-scaled histograms (Fig. 8) to estimate the proportion of organ boundary pixels correctly assigned to the corresponding organ for contour-aware and mask-only segmentations. From the histograms, we can, for example, observe that around 13% of lung boundary pixels are classified as background for UNet_ResNeXt50_Masks, whereas this number drops to around 3% for UNet_ResNeXt50_Masks+Contours. The mean pixel activation values were also statistically compared using the one-sided nonparametric Mann–Whitney test, showing that they were statistically higher for the contour-aware than for the mask-only segmentation (Table 4). These experiments statistically confirm more accurate segmentation at organ boundaries for UNet_ResNeXt50_Masks+Contours.
Example of heart segmentation results (light-colored regions) for the contour-aware segmentation (UNet_ResNeXt50_Masks+Contours) in comparison with the ground truth (dark-colored contours) superimposed on the chest X-ray image. a Contour prediction. b Mask prediction. The arrows indicate the regions with poorly recognized heart contours and poor segmentation results
To further validate the proposed concepts of contour-aware CNNs for CXR segmentation, we evaluated the best performing architecture UNet_ResNeXt50_Masks+Contours on a Montgomery public database with lung field segmentations [56]. The database consists of 138 CXR (80 are normal, 58 are abnormal with tuberculosis) with the pixels size of 0.0875 mm. We performed fivefold cross-validation on 138 CXRs and obtained segmentation results of 0.966 and 0.967 in terms of the Jaccard coefficient for UNet_ResNeXt50_Masks and UNet_ResNeXt50_Masks+Contours, respectively. Augmentation with contours resulted in improved CXR segmentations for the Montgomery database; however, the improvements are less pronounced than for the JSRT database. One of the potential explanations for slightly lower segmentation accuracy is the fact that the Montgomery database has more cases with pathologies resulting in poorly visible boundaries. Candemir et al. [4] also observed small deterioration of lung segmentation accuracy for the Montgomery database in comparison with the JSRT database. We finally evaluated how potential patient mispositioning may affect segmentation accuracy. To emulate the situation when the patient is not perfectly upright, we introduced artificial rotations to the testing CXRs with the \(10^{\circ }\), \(20 ^{\circ }\), and \(30 ^{\circ }\) magnitude. The segmentation results for rotated CXRs are summarized in Table 5. We can observe that rotations of \(10 ^{\circ }\) do not result in performance deterioration due to the fact that \([-15^{\circ }, +15^{\circ }]\) rotations we added to the input CXRs as the training data augmentation for CNNs.
Further improvements in segmentation performance can be achieved by imposing additional anatomical constraints on contour definitions, as the current contour detection still faces challenges in the case of poorly visible boundaries (Fig. 9). One of the strategies is to additionally integrate fuzzy contour information into the loss function, as it was performed in the original UNet paper [23]. The authors computed the distances between image pixels and the target object borders and added a distance-based loss component in order to penalize errors near the object borders. Such a loss function may strengthen the segmentation algorithm and improve the robustness of the resulting masks and contours. At the same time, that loss function requires the introduction and tuning of two additional algorithm parameters per object type.
Conclusion
In this study, we evaluated an end-to-end contour-aware CNN framework for the segmentation of the lung fields, heart and clavicles from a public database of CXR images. The contour information improved the performance of three state-of-the-art CNN architectures. Moreover, we numerically demonstrated that contour information helps CNNs to learn useful features about both the segmentation mask and contour of chest organs, therefore improving the quality of the predicted segmentation mask along the corresponding contour.
References
Chen S, Zhong S, Yao L, Shang Y, Suzuki K (2016) Enhancement of chest radiographs obtained in the intensive care unit through bone suppression and consistent processing. Phys Med Biol 61:2283
Candemir S, Antani S (2019) A review on lung boundary detection in chest X-rays. Int J Comput Assist Radiol Surg 14:563–576
Miniati M, Coppini G, Monti S, Bottai M, Paterni M, Ferdeghini E (2011) Computer-aided recognition of emphysema on digital chest radiography. Eur J Radiol 80:169–175
Candemir S, Jaeger S, Lin W, Xue Z, Antani S, Thoma G (2016) Automatic heart localization and radiographic index computation in chest X-rays. In: SPIE medical imaging, 9785
Finnegan R, Dowling J, Koh E-S, Tang S, Otton J, Delaney G, Batumalai V, Luo C, Atluri P, Satchithanandha A (2019) Feasibility of multi-atlas cardiac segmentation from thoracic planning CT in a probabilistic framework. Phys Med Biol 64:085006
Gordienko Y, Gang P, Hui J, Zeng W, Kochura Y, Alienin O, Rokovyi O, Stirenko S (2018) Deep learning with lung segmentation and bone shadow exclusion techniques for chest x-ray analysis of lung cancer. In: International conference on ICAFS, pp 638–647
Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K, Matsui M, Fujita H, Kodera Y, Doi K (2000) Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. AJR Am J Roentgenol 174:71–4
van Ginneken B, Stegmann MB, Loog M (2006) Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Med Image Anal 10:19–40
Vittitoe NF, Vargas-Voracek R, Floyd CF Jr (1998) Identification of lung regions in chest radiographs using Markov random field modeling. Med Phys 25:976–85
Shi Z, Zhou P, He L, Nakamura T, Yao Q, Itoh H (2009) Lung segmentation in chest radiographs by means of Gaussian kernel-based FCM with spatial constraints. In: 6th international conference on fuzzy systems and knowledge discovery (FSKD). IEEE, pp 428–432
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) ImageNet: a large-scale hierarchical image database. In: Computer vision and pattern recognition (CVPR). IEEE, pp 248–55
Wang C (2017) Segmentation of multiple structures in chest radiographs using multi-task fully convolutional networks. In: Scandinavian conference on image analysis (SCIA). Volume 10270 of Lecture notes in computer science. Springer, Cham, pp 282–289
Hwang S, Park S (2017) Accurate lung segmentation via network-wise training of convolutional networks. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, vol 10553 lecture notes in computer science. Springer, Cham, pp 92–99
Dai W, Dong N, Wang Z, Liang X, Zhang H, Xing EP (2018) SCAN: structure correcting adversarial network for organ segmentation in chest X-rays. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, volume 11045 of lecture notes in computer science. Springer, Berlin, pp 263–73
Bi L, Feng D, Kim J (2018) Dual-path adversarial learning for fully convolutional network (FCN)-based medical image segmentation. Vis Comput 34:1043–52
Novikov AA, Lenis D, Major D, Hladůvka J, Wimmer M, Bühler K (2018) Fully convolutional architectures for multi-class segmentation in chest radiographs. IEEE Trans Med Imaging 37:1865–76
Mittal A, Hooda R, Sofat S (2018) LF-SegNet: a fully convolutional encoder-decoder network for segmenting lung fields from chest radiographs. Wirel Pers Commun 101:511–29
Frid-Adar M, Ben-Cohen A, Amer R, Greenspan H (2018) Improving the segmentation of anatomical structures in chest radiographs using U-Net with an ImageNet pre-trained encoder. In: Image analysis for moving organ. breast, and thoracic images, volume 11040 of lecture notes in computer science. Springer, Cham, pp 159–68
Bonheur S, Stern D, Payer C, Pienn M, Olschewski H, Urschler M (2019) Matwo-capsnet: a multi-label semantic segmentation capsules network. MICCAI 2019:664–672
Ngo T A, Carneiro G (205) Lung segmentation in chest radiographs using distance regularized level set and deep-structured learning and inference. In: International conference on image processing (ICIP). IEEE, pp 2140–2143
Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A (2017) A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans Med Imaging 36:1550–60
Cui Y, Zhang G, Liu Z, Xiong Z, Hu J (2018) A deep learning algorithm for one-step contour aware nuclei segmentation of histopathological images. arXiv, p 1803.02786
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Medical image computing and computer-assisted intervention (MICCAI), volume 9351 of lecture notes in computer science. Springer, Cham, pp 234–241
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Computer vision and pattern recognition (CVPR). IEEE, pp 5987–5995
Chaurasia A, Culurciello E (2017) LinkNet: exploiting encoder representations for efficient semantic segmentation. In: Visual communications and image processing (VCIP). IEEE
Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y (2017) The one hundred layers tiramisu: fully convolutional DenseNets for semantic segmentation. In: Computer vision and pattern recognition workshops (CVPRW). IEEE, pp 1175–1783
Huang G, Liu Z, van der Maaten L, Weinberger K Q (2017) Densely connected convolutional networks. In: Computer vision and pattern recognition (CVPR). IEEE, pp 2261–2269
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Computer vision and pattern recognition (CVPR). IEEE, pp 3431–3440
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: a nested U-Net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support (DLMIA / ML-CDS), volume 11045 of lecture notes in computer science. Springer, Cham, pp 3–11
Wang CW, Huang CT, Lee JH, Li CH, Chang SW, Siao MJ, Lai TM, Ibragimov B, Vrtovec T, Ronneberger O, Fischer P, Cootes TF, Lindner C (2016) A benchmark for comparison of dental radiography analysis algorithms. Med Image Anal 31:63–76
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Computer vision and pattern recognition (CVPR). IEEE, pp 770–788
Shaikh M, Anand G, Acharya G, Amrutkar A, Varghese A, Krishnamurthi G (2018) Brain tumor segmentation using dense fully convolutional neural network. Brainlesion: Glioma. In: Multiple sclerosis, stroke and traumatic brain injuries (BrainLes), volume 10670 of lecture notes in computer Science. Springer, Cham, pp 309–319
Xu C, Xu L, Brahm G, Zhang, Li S (2018) Mutgan: simultaneous segmentation and quantification of myocardial infarction without contrast agents via joint adversarial learning. In: Conference on Medical Image Computing and Computer-Assisted Intervention, pp 525–534, 2018
Arik SO, Ibragimov B, Xing L (2017) Fully automated quantitative cephalometry using convolutional neural networks. J Med Imaging 4:014501
Milletari F, Navab N, Ahmadi S-A (2016) V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 3D vision (3DV). IEEE, pp 565–571
DSBowl2018 (2018) 2018 data science bowl. https://www.kaggle.com/c/data-science-bowl-2018/overview. Accessed 15 Oct 2019
KiTS2019 (2019) 2019 kidney and kidney tumor segmentation challenge. https://kits19.grand-challenge.org/. Accessed 15 Oct 2019
Isensee F, Petersen J, Kohl S A A, Jager PF, Maier-Hein KH (2019) Nnu-net: breaking the spell on successful medical image segmentation. arXiv:1904.08128
Kingma DP, Ba LJ (2015) Adam: a method for stochastic optimization. In: 3rd International conference on learning representations (ICLR)
Shi Y, Qi F, Xue Z, Chen L, Ito K, Matsuo H, Shen D (2008) Segmenting lung fields in serial chest radiographs using both population-based and patient-specific shape statistics. IEEE Trans Med Imaging 27:481–94
Li X, Luo S, Hu Q, Li J, Wang D, Chiong F (2016) Automatic lung field segmentation in X-ray radiographs using statistical shape and appearance models. J Med Imaging Health Inform 6:338–48
Dawoud A (2010) Fusing shape information in lung segmentation in chest radiographs. In: Image analysis and recognition—ICIAR 2010, volume 6112 of lecture notes in computer science. Springer, Berlin, Heidelberg, pp 70–78
Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Zhiyun X, Karargyris A, Antani S, Thoma G, McDonald CJ (2014) Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans Med Imaging 33:577–90
Ibragimov B, Likar B, Pernuš F, Vrtovec T (2012) A game-theorteic framework for landmark-based image segmentation. IEEE Trans. Med. Imaging 31:1761–76
Seghers D, Loeckx D, Maes F, Vandermeulen D, Suetens P (2007) Minimal shape and intensity cost path segmentation. IEEE Trans Med Imaging 26:1115–29
Wu G, Zhang X, Luo S, Hu Q (2015) Lung segmentation based on customized active shape model from digital radiography chest images. J Med Imaging Health Inform 5:184–91
Yang W, Liu Y, Lin L, Yun Z, Lu Z, Feng Q, Chen W (2018) Lung field segmentation in chest radiographs from boundary maps by a structured edge detector. IEEE J Biomed Health Inform 22:842–51
Ibragimov B, Likar B, Pernuš F, Vrtovec T (2016) Accurate landmark-based segmentation by incorporating landmark misdetections. In: 13th international symposium on biomedical imaging (ISBI). IEEE, pp 1072–1075
Chondro P, Yao C-Y, Ruan S-J, Chien L-C (2018) Low order adaptive region growing for lung segmentation on plain chest radiographs. Neurocomputing 275:1002–11
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of CVPR 2017
RSNA (2018) Kaggle pneumonia detection challenge. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge. Accessed 15 Oct 2019
Karargyris A, Siegelman J, Tzortzis D, Jaeger S, Candemir S, Xue Z, Santosh K, Vajda S, Antani S, Folio L, Thoma G (2016) Combination of texture and shape features to detect pulmonary abnormalities in digital chest X-rays. Int J Comput Assist Radiol Surg 11:99–106
Zheng G, Chu C, Belavy DL, Ibragimov B, Korez R, Vrtovec T, Hutt H, Everson R, Meakin J, Andrade IL, Glocker B, Chen H, Qi Dou Q, Heng PA, Wang C, Forsberg D, Neubert A, Fripp J, Urschler M, Stern D, Wimmer M, Novikov AA, Cheng H, Armbrecht G, Felsenberg D, Li S (2017) Evaluation and comparison of 3d intervertebral disc localization and segmentation methods for 3d t2mr data: a grand challenge. Med Image Anal 35:327–344
Cong J, Zheng Y, Xue W, Cao B, Li S (2019) Ma-shape: modality adaptation shape regression for left ventricle segmentation on mixed MR and CT images. IEEE Access 7:16584–16593
Kholiavchenko M (2019) https://github.com/dirtmaxim/ca-ml-organ-segmentation. Accessed 15 Oct 2019
Jeager S, Candemir S, Antani S, Wang Y, Lu P, G Thoma (2014) Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg 4:475–477
Acknowledgements
This work was supported by the Russian Science Foundation (Grant No. 18-71-10072), Russian Foundation for Basic Research (Grant No. 18-47-160015), and Slovenian Research Agency—ARRS (Grant No. P2-0232). Due to the requirements of the Russian Science Foundation, we explicitly state that the methodology development and implementation were solely supported by the Russian Science Foundation (Grant No. 18-71-10072).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study has not been supported by any industrial company and does not serve to promote any commercial product. Anonymized publicly available databases of CXR were used in the conducted experiments.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kholiavchenko, M., Sirazitdinov, I., Kubrak, K. et al. Contour-aware multi-label chest X-ray organ segmentation. Int J CARS 15, 425–436 (2020). https://doi.org/10.1007/s11548-019-02115-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-02115-9