Abstract
Purpose
Multi-organ segmentation from CT images is an essential step for computer-aided diagnosis and surgery planning. However, manual delineation of the organs by radiologists is tedious, time-consuming and poorly reproducible. Therefore, we propose a fully automatic method for the segmentation of multiple organs from three-dimensional abdominal CT images.
Methods
The proposed method employs deep fully convolutional neural networks (CNNs) for organ detection and segmentation, which is further refined by a time-implicit multi-phase evolution method. Firstly, a 3D CNN is trained to automatically localize and delineate the organs of interest with a probability prediction map. The learned probability map provides both subject-specific spatial priors and initialization for subsequent fine segmentation. Then, for the refinement of the multi-organ segmentation, image intensity models, probability priors as well as a disjoint region constraint are incorporated into an unified energy functional. Finally, a novel time-implicit multi-phase level-set algorithm is utilized to efficiently optimize the proposed energy functional model.
Results
Our method has been evaluated on 140 abdominal CT scans for the segmentation of four organs (liver, spleen and both kidneys). With respect to the ground truth, average Dice overlap ratios for the liver, spleen and both kidneys are 96.0, 94.2 and 95.4%, respectively, and average symmetric surface distance is less than 1.3 mm for all the segmented organs. The computation time for a CT volume is 125 s in average. The achieved accuracy compares well to state-of-the-art methods with much higher efficiency.
Conclusion
A fully automatic method for multi-organ segmentation from abdominal CT images was developed and evaluated. The results demonstrated its potential in clinical usage with high effectiveness, robustness and efficiency.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Abdominal organ segmentation from computed tomography (CT) images is a crucial preprocess for computer-aided diagnosis (CAD) systems, radiotherapy planning as well as cancer delineation and staging [34], where the estimation of the anatomical boundary/volume needs to be accurate. Since diagnostic and interventional imagery of abdomen often consists of 3D image analysis, accurate image segmentation enables comprehensive analysis and quantitative measures of shapes and volumes of the target organs, which can be indicators of disorders [17]. In traditional clinical practice, the delineation of organs is often performed manually by radiologists in an organ-by-organ and slice-by-slice way, which is tedious, time-consuming and prone to intra- and inter-observer variability. While it is prohibitive to manually label large-scale datasets, its full automation with high efficiency would enable systematic segmentation of CT images on the fly as soon as the image is acquired. Thus, developing automatic segmentation algorithms is very important from the viewpoints of diagnostic efficiency and feasibility.
Two major challenges exist in automatic abdominal multi-organ segmentation from CT images [33]. The first one is to automatically locate the anatomical structures in the target image. Since abdominal organs are mainly surrounded by soft tissues, large variations exist in the location, shape and size of organs among individuals. The other challenge is to delineate the fuzzy boundaries between adjacent organs and soft tissues. Abdominal organs, such as the liver, often share similar appearance and intensities with surrounding tissues in CT images, especially in noncontrast-enhanced data.
The potential benefits of automated multi-organ segmentation have encouraged an active field of research [9, 17, 21, 29, 31, 34, 35], which is currently dominated by statistical shape models (SSMs) or atlas-based methods. For these prior knowledge-based methods, shape models or probability atlases are built to capture anatomical knowledge for the target organs. However, with limited number of data, it is very difficult to build a model that is general enough to capture the large variability of the deformable organs, e.g., the liver and spleen. Although more recent works focused on generating subject-specific priors by utilizing region-wise local atlas selection strategy [34], dictionary learning technique [31] or inter-organ spatial relations [21], the training and testing process of these methods is highly dependent on initialization/registration of the prior models, which are computational intense tasks.
Some learning-based methods [6, 8, 13] are proposed to give more efficient localization of the organs. For instance, landmark-based methods [13] aimed to detect anatomically meaningful locations to posit the organs. However, it may be difficult to determine reliable anatomical landmarks for the deformable abdominal organs. Random forest regression models [6, 8] used training samples with associated features to locate regions of interest containing the organs. Although being fast, the used features, which are designed or selected manually, are critical to its result. The network-based method proposed in [30] that combined features and classifiers in a multi-level hierarchical way also needs building handcrafted features to perform classification task.
Instead of relying on handcrafted features and/or computational intense registrations, the end-to-end learning-based convolutional neural network (CNN) enabling to learn hierarchical features has been introduced for both natural and medical image segmentation [5, 18, 27, 28]. Typically, there are two widely used strategies. The first one obtains target delineation by using a sliding-window setup and conducting patch-wise image classification [5]. Specifically, by providing a local patch, the label of its central pixel/voxel is predicted through the CNN. In [5], this type of strategy was designed to segment neuronal membranes from electron microscopy images. Recently, Cha et al. [2] have also used this patch-wise training strategy for bladder segmentation from CT urography in a slice-by-slice manner. To distinguish between the inside and the outside of the bladder within user-predefined bounding boxes, over 160 thousand patches were extracted for training. While methods in this sliding-window strategy provided large margin improvement in performance [5], they have several limitations. For instance, using only local patch for prediction, limited local context information can be utilized and therefore it is prone to failure to segment multiple objects with similar intensities/appearances such as abdominal CT images. The densely extracted patches processed by a CNN also pose challenges on computational cost and running time.
In this work, we build on another recently proposed strategy, the so-called fully convolutional network [18], which can perform end-to-end learning and pixel-to-pixel dense predictions with a whole image as input. With the help of in-network up-sampling layers, pixel-wise likelihood map can be obtained for whole image at a time. Previously, this framework was designed for 2D natural image segmentation [18] and recently modified for biomedical image segmentation [27]. In this study, we design a 3D fully convolutional neural network to perform multi-organ segmentation in abdominal CT images, which is an extension of our fully CNN-based liver segmentation methods [12, 19]. It should be noted that very recently 3D CNN has been successfully employed in medical imaging community and solved several segmentation tasks, such as brain lesion [1] and abdominal organ segmentation [7, 12, 19]. For example, in our previous works [12, 19], we utilized 3D fully CNN for liver segmentation in CT images. As a drastically progressing area, several semantic segmentation methods based on 3D fully CNN have been made available since the completion of this work. For instance, an extension of the U-net [27] named 3D U-net [4] was introduced and tested for kidney segmentation. A V-net [20], a modified U-net, was employed for prostate segmentation. A 3D extension of the deeply supervised network [16], named 3D DSN [7], was introduced for liver segmentation and posted after our submission.
For learning-based methods [7, 31, 34] including CNN-based methods, the learned predictions are usually refined by traditional methods for further accuracy improvement. Take the CNN for example, in [19] the CRF and graph cut algorithm, which is computational intense for 3D volume, was used for the refinement of liver segmentation. In [3, 7], a fully connected CRF, which has advantage in capturing complicated shaped object such as those with holes or thin structures, was used for liver segmentation refinement. In [2], Cha et al. employed the traditional level-set method, the speed of which was limited by small time step in discretization.
In this paper, we propose to address the problem of multiple organ segmentation with a combination of a 3D fully CNN and an energy-based refinement model. Specifically, a ten-layer 3D CNN with volumetric convolutions is designed, which contains seven contracting layers and three expanding layers to capture high-level features and perform voxel-to-voxel likelihood prediction. To capture large variation in intensity and positions across subjects, we augmented the data by applying translation transform and intensity scaling. The refinement of the CNN results is formulated as a multi-phase segmentation problem, which is solved with a time-implicit multiple surface evolution method [26] by taking advantage of modern convex optimization techniques. Coupling the two techniques is to take advantages of both the detection/prediction capacity of CNNs and the fine-grained localization accuracy of classical multi-phase segmentation methods, producing accurate segmentation results and recovering organ boundaries at a level of detail. Instead of being dependent on the costly registration in training and testing stage as shape and atlas-based methods [21, 31, 34], no registration steps are required in our method. While most previous CNN-based methods [4, 7, 19] focused on single-target segmentation, this study is an early attempt for the more challenging multiple abdominal organ segmentation. With major components of the algorithm implemented on graphic processing units (GPUs), our method obtains high computational efficiency. A large dataset containing diverse qualities of images with multiple evaluation measures has been used for method evaluation.
Methods
In this section, we describe the imaging data used in this study and the proposed multi-organ segmentation framework. Figure 1 shows the proposed segmentation pipeline that consists of two main steps: (i) 3D CNN-based multi-organ localization and segmentation, and (ii) refinement of the multi-organ segmentation. Firstly, a CT volume is fed into the trained 3D fully CNN to acquire probability maps for all voxels, which localize the organs and act as spatial priors for subsequent fine segmentation step. A simple binarization follows to generate the initial segmentation for the organs of interest. Next, intensity models, i.e., probability density functions (PDFs), are automatically estimated with the initial segmentation. Finally, a time-implicit level-set-based multi-phase segmentation approach is introduced for multi-organ refinement, which incorporates the image intensity model, prior probability maps, disjoint region constraint, as well as gradient edge map.
Dataset
One hundred and forty 3D abdominal CT scans with or without contrast enhancement acquired from 132 patients were used for our experiments. Among the 140 CT scans, 20 are from the public dataset MICCAI-SLiver07 [11] training set and 120 are obtained from local hospitals. These data are in three types of contrast-enhancement patterns, i.e., portal venous phase (67 cases), (early) arterial phases (60 cases) and noncontrast phase (13 cases). The CT scans are measured as \(512\times 512\times (79\sim 304)\) voxels with a spacing of \((0.55\sim 1.00)\times (0.55\sim 1.00)\times (1.00\sim 2.50)\) mm/voxel. The liver, spleen, left kidney and right kidney are considered as target organs to be segmented. The ground truth was obtained by trained technicians with the semiautomatic segmentation tool [22, 23], and then, the results were approved and revised by experienced radiologist.
Automated localization and segmentation
For the automated localization and delineation of organs in 3D CT scans, we design a deep volumetric fully CNN, which conducts end-to-end learning and provides voxel-to-voxel prediction. Convolutional neural networks are neural network architectures that use extensive weight-sharing to reduce the degrees of freedom of models that are spatially correlated [15]. As the main blocks of the CNN, convolutional layers and pooling layers are applied alternatively on the raw input image. Each layer takes as input the output of the preceding layer and thus builds a hierarchy of increasingly complex features [38]. In contrast to patch-wise learning, by employing up-sampling layers, the fully CNN is able to predict segmentation for the whole volume at once.
As for the first convolutional layer, a CT volume as a whole is taken as the input. In subsequent layers, the input image blocks consist of the feature maps of the previous layer. Features are extracted via a set of filters convolved over the input image, followed by a nonlinear activity function. Denote \(x_r^{l-1}\) as the rth output feature map of (\(l-1\))th layer, and the sth output feature map of lth layer can be calculated as
where \(*\) is the three-dimensional convolutional operation, \(\omega _{sr}^l\) is the filter linking the rth input map and the sth output, and \(b_s^l\) is the according bias term. The function f denotes the element-wise activity function, which helps capture nonlinear transforms between the inputs and outputs. Here, we use the recently proposed nonlinearity Parametric Rectified Linear Unit (PReLU) [10] defined as
where \(a_s\) is also a trainable parameter that controls the slop of the negative part. With parameter \(a_s\), this activation can avoid zero gradients in comparison with ReLU. More importantly, we adaptively learn the PReLU parameters jointly with the whole model. As demonstrated in [10], the PReLU can improve model fitting with nearly zero extra computational cost and little overfitting risk.
Next, the convolved feature maps are sub-sampled via average-pooling layer, which introduces invariance to local deformations, such as translation, and reduces the computational cost. This operation takes the average values over sub-windows within each feature map. In our work, we use nonoverlapping sub-window of size \(2\times 2\times 2\).
In addition, some other layer types are used. Specifically, the local response normalization scheme [14] is applied after the first convolutional layer to enforce competitions between features at the same spatial location across different feature maps. In Doublesize layers [19], every 8 feature map channels are rearranged into \(2\times 2 \times 2\), i.e., doubling the size in each dimension and reducing the number of channels to 1/8.
Finally, an element-wise softmax nonlinearity normalizes the result of kernel convolutions into a multi-nominal distribution over the labels. Specifically, let \(\mathbf {b}\) be the vector of values of length n (the number of classification labels) at a given spatial position in the feature map, and it computes \(softmax(\mathbf {b})=\exp (\mathbf {b})/Z\), where \(Z=\sum _{i=1}^n \exp (b_i)\). The output values provide likelihood of a specific voxel belonging to the tissues. Thus, a probability map for each classification label is generated. Further, our approach performs a multi-class labeling by assigning each voxel the label with the largest probability, and this result is regarded as the initial segmentation of the organs.
The architecture
The detailed architecture is shown in Table 1. This network takes image blocks of size 496 \(\times \) 496 \(\times \) 279 as input and outputs four probability maps of size 248 \(\times \) 248 \(\times \) 256. The probability maps are then up-sampled to \(496\times 496\times 256\), and the values outside this block in the original image are set to 0. Thus every voxel in the target image is assigned the probability belonging to four tissue types (background, liver, spleen and kidneys). To make the training faster and satisfy the memory limit, the network is spread across four GPUs with parallelization scheme [14] in intermediate five layers (from \(Conv_3\) to \(Conv_7\)).
Network training
We train the network offline using CT scans with segmentations of the liver, spleen and both kidneys. Let \(\varTheta =\{W,B,A\}\) be the set of parameters to be estimated, where W, B, A correspond to the filters, biases and PReLU parameters, respectively. Given the predicted probability map of ith label \(L_i(I^{j};\varTheta )\) of jth training image \(I^j\) and the corresponding true label \(Y^j_i\), \(i=1,\ldots ,n, j=1,\ldots ,m\), where n is the number of classification labels and m is the number of training images, the parameters are optimized by minimizing the following cost function with respect to \(\varTheta \):
The first term computes the cross-entropy loss, and the second \(l_2\) regularization term is added to avoid overfitting. The cost function is minimized using the backpropagation algorithm [14].
Segmentation refinement via multiple surface evolution
The above CNN-based organ detection and segmentation generate spatial priors and give proper initial segmentations for the organs. Previous studies [31, 34] demonstrate performance improvement by combining the intensity information and spatial prior. Here, we combine the probabilistic priors and data-driven region and edge information in a multi-region segmentation model. In addition, a disjoint region constraint is introduced to enforce the exclusiveness between organs. Further, the model is efficiently solved in a time-implicit multi-phase level-set scheme in terms of convex optimization [26].
Denote the input 3D CT image \(I(x):\varOmega \rightarrow \mathcal {R}\), \(\varOmega \subseteq \mathcal {R}^3\) and consider n nonoverlapping regions \(\{\mathcal {C}_i\}_{i=1,\ldots ,n}\), such that \(\varOmega =\cup _{i=1}^n \mathcal {C}_i\), the multi-region segmentation model is defined as
which is a combination of the region-based data term
prior term
and edge-based regularization term
Here in the data term, \(F_i(I(x)):=p(I(x)|\mathcal {C}_i)\) is the probability density function estimated from the intensity distributions within region \(\mathcal {C}_i\) (\(i=1,\ldots ,n\)). In the prior term, we represent the prior probability map by negative log-likelihood to constrain the segmentation. The negative log-likelihood map describes the confidence that a certain voxel belongs to specific region. The lower the value of \(-\log L_i(x)\) is, the more likely the x belongs to region \(\mathcal C_i\), and vice versa. The last regularization term acts as a smooth term with the weight function defined as \(g(|\nabla I|)= 1/(1+\beta |\nabla I|^2)\), where \(\beta \) is a positive constant and fixed to 0.2 in our experiments. Note that the values of function g fall within the range [0,1] and g is an edge indicator that vanishes at object sharp boundaries. Spatially varying weights \(\lambda _{1,2}(\mathbf {x})\) are used to adaptively balance the three terms. Specifically, we set \(\lambda _{1,2}(\mathbf {x})=\alpha _{1,2}/(1+\beta |\nabla I|^2)\), where \(\alpha _{1,2}>0\) are positive constants. This makes the model selectively act as edge-based or region-based model in different regions.
Surface evolution using time-implicit level sets
For the multi-region segmentation problem (4), which can be viewed as the evolution of multiple surfaces with respect to a disjoint region constraint, many algorithms have been developed. A recent study [26] introduced a time-implicit multi-phase level-set scheme in terms of convex optimization with advantages in both implementation and computation, which is substantially distinct from the classical level-set approach [32]. Instead of one-by-one phase movement as in the classical approaches to multi-phase segmentation, the method used here propagates the surfaces of all phases simultaneously by means of minimizing costs w.r.t. region changes, which is a novel principle proposed in [26]. More importantly, large step sizes for surface propagation are allowed to ultimately improve efficiency. Each evolution step can be reformulated as a spatial continuous Potts problem [25], i.e., a continuous multi-region min-cut problem, which can be solved fast via a continuous max-flow algorithm [36]. Here, we describe the algorithm briefly and refer readers to [26] for details and proofs.
For n disjoint regions, \(\mathcal {C}_i, i=1,\ldots ,n\), the evolution of each region over the discrete time frame from t to \(t+1\) minimizes total cost of region changes. That is, given the current regions \(\mathcal {C}_i^t, i=1,\ldots ,n\), the new optimal surfaces \(\mathcal {C}_i^{t+1}, i=1,\ldots ,n\) minimize the energy:
subject to \(\varOmega =\cup _{i=1}^n\mathcal {C}_i, \mathcal {C}_k\cap \mathcal {C}_l=\emptyset , \forall k \ne l\). In the equation, two types of different regions are defined with respect to \(\mathcal {C}_i^{t+1}\).
-
1.
\(\mathcal {C}_i^+\) indicates expansion of \(\mathcal {C}_i^t\) w.r.t. \(\mathcal {C}_i^{t+1}\): \(x\in \mathcal {C}_i^+\), and it is outside \(\mathcal {C}_i^t\) at time t, but inside \(\mathcal {C}_i^{t+1}\) at \(t+1\); for such an expansion of x, with cost \(c_i^+(x)\).
-
2.
\(\mathcal {C}_i^-\) indicates shrinkage of \(\mathcal {C}_i^t\) w.r.t. \(\mathcal {C}_i^{t+1}\): \(x\in \mathcal {C}_i^-\), and it is inside \(\mathcal {C}_i^t\) at time t, but outside \(\mathcal {C}_i^{t+1}\) at \(t+1\); for such a shrinkage of x, with cost \(c_i^-(x)\).
By introducing the distance function \(dist(x,\partial \mathcal {C}_i^t)\) to the expansion and shrinkage cost, the surface is driven in a time-implicit manner [37] by both mean-curvature and region-based information that corresponds to the first two terms in (4), i.e., \(\lambda _1E_\mathrm{data}(\mathcal {C}) + \lambda _2E_\mathrm{prior}(\mathcal {C})\). The expansion and shrinkage cost functions are defined as
and
where \(dist(x,\partial \mathcal {C}_i^t)\) denotes the Euclidean distance of x to \(\partial \mathcal {C}_i\), and h is the surface evolution discrete time step.
Let \(u_i(x)\in \{0,1\},i=1,\ldots ,n\) be the indicator function of region \(\mathcal {C}_i\) and two cost functions \(D_i^s(x)\) and \(D_i^t(x)\) w.r.t. the current surfaces \(\mathcal C_i^t,i=1,\ldots ,n\) are defined:
The variational problem (8) can be expressed as the Potts problem
subject to \(\sum _{i=1}^n u_i(x)=1, \forall x\in \varOmega \). Detailed proof of the equivalence between (8) and (13) can be found in [26]. The global optimum of problem (13) can be approximated through convex relaxation and then be efficiently solved by the augmented Lagrangian method in a max-flow formulation [36]. In this way, the current surfaces propagate to the next position in each iteration and finally to the optimal position \(\mathcal {C}_i^*, i=1,\ldots ,n\) of model (4).
Experiments and results
We evaluated the proposed segmentation framework in the context of automated simultaneous segmentation of the liver, spleen and both kidneys. A fivefold cross-validation was conducted on 140 abdominal CT scans. In each cross, 112 CT scans were used for training the CNN and the rest 28 CT scans were for testing. The final error was averaged over the five crosses.
In order to perform quantitative evaluation, the segmentation results were compared with the ground truth. Volume-based and surface distance-based metrics, namely Dice similarity coefficient (DSC), Jaccard index (JI) and average symmetric surface distance (ASD), were computed. \(\hbox {DSC}=100\%\) and \(\hbox {JI}=100\%\) mean no under- or over-segmentation, and \(\hbox {ASD} = 0\) mm means perfect match between the automated segmentation and the ground truth surfaces.
Implementation details and parameter settings
Our implementation of the CNN was based on the cuda-convnet package.Footnote 1 For data preparation in both training and testing stage of the CNN, the input of the network was image blocks of size \(496\times 496\times 279\) that cropped from original CT volumes. The intensity range of input volumes was normalized to \([-2,2]\) firstly. Then, to capture large variation in intensity across subjects with different contrast-enhancement patterns, intensities were randomly permuted. Specifically, I(x) was translated by \(\nu \) and then multiplied by \(\sigma \), where \(\nu \in [-0.5,0.5]\) and \(\sigma \in [0.5,1.5]\) were two random values.
The hyperparameters of the CNN architecture described in Table 1 were selected by conducting a grid search on the training set. The filter weights were initialized randomly with Gaussian distribution \(N(0,1\times 10^{-2})\). During training, weights were updated by stochastic gradient descent algorithm with a momentum of 0.9. The biases and PReLU parameters in convolutional layers were initialized to 0 and 0.25, respectively. The number of epochs was fixed to 60 in all cross-validations. The regularization coefficient \(\mu \) in model (3) was set to \(5\times 10^{-4}\). The learning rate was initialized to \(2.5\times 10^{-8}\) and then decreased by a factor of \(\sqrt{2}\) every 8 epochs after the 40th epoch.
In the multi-organ refinement step, each slice of the input images and probability maps was downsampled to \(256\times 256\) to reduce the computation cost. Data were smoothed using anisotropic diffusion [24] as preprocessing. According to magnitude analysis and experience, the parameters in model (4) were set as \(\alpha _1=0.35\) and \(\alpha _2=0.30\). The discrete time step in surface evolution was set as \(h=100\).
Computational time
Computations were carried out on a Windows desktop with Intel Xeon E5-2680 CPU (2.70 GHz) and four graphics cards of NVIDIA GeForce GTX 980. The training of the CNN was implemented using parallel computing architecture with four graphics cards. In the multi-region segmentation step, the convex max-flow algorithm was based on the (GPU version) code of Yuan et al. [36]. Other computations were programmed in MATLAB 2014b. Training the CNN took approximately 12 h. The testing time of the CNN on an abdominal volume of size \(512\times 512\times 279\) took around 6 s. The surface refinement step took about 119 s. The average total time of testing a case in our dataset was about 125 s.
Segmentation accuracy
Quantitative results of our method on the segmentation of the liver, spleen and both kidneys from images of various contrast-enhancement patterns are summarized in Table 2. The dataset is separated into three parts, i.e., portal venous phase, artery phase and noncontrast phase, and two kinds of results that corresponds to the initial segmentation (I) and the final segmentation (F) are listed for each part. Figure 2 shows the box plots of results on 140 CT volumes with respect to the initial and final segmentations. The computed metrics in Table 2 indicate that the trained CNN can predict good initial segmentation with an accuracy over 89% for all the four organs (in terms of DSC). Also, the combination of the CNN and time-implicit level-set evolution achieved final segmentations with great agreement with the ground truth. The refinement step boosted the segmentation performance over the initial results significantly (\(p<0.001\) for all organs).
Figure 3 shows illustrative examples of organ localization by the CNN. The two rows correspond to good and bad results in terms of DSC. The good cases lead to DSCs of 96.7% for the liver, 95.3% for the spleen and 95.5% for the kidneys, while the bad cases gave DSCs of 82% for the liver, 62.3% for the spleen and 62.9% for the kidneys. As can be inferred from box plots of initial segmentations in Fig. 2, the CNN can locate the organs accurately and predict rough shapes of the organs for most cases in the dataset.
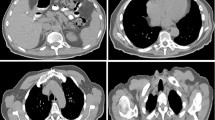
Figure 4 presents three typical examples of segmentation results for the liver, spleen and kidneys from images of different contrast patterns. The first three columns give axial projections of the CT volumes, and the fourth column provides 3D visualization of the automated segmentations. We can see that the automated segmentations agreed well with the ground truth.
Effect of the probabilistic prior on the proposed model
To better understand the role of each step, we compared the outcomes of the CNN, the surface evolution-based refinement without probabilistic prior and the complete proposed model. The results for one typical volume are shown in Fig. 5. As can be seen in the first row, the outcome of the CNN provided an accurate location for the liver, but there was under-segmentation in the left liver lobe. The outcome of the surface refinement without probabilistic prior leaked to the surrounding tissues due to low contrast at the borders. The complete proposed model evolved the initial surface closer to the ground truth. Similar behaviors can be observed in segmentation of the spleen (row two) and kidneys (row three).
Effect of the CNN parameters
Since the depth and width of the CNN are crucial to its performance, we have studied the effect of different CNN parameters on segmentation accuracy and the results are shown in Fig. 6. To determine the number of convolutional layers, layers from \(Conv_3\) to \(Conv_7\) of the CNN (see Table 1) are adjusted to devise five architectures with 6–10 convolutional layers. The accuracies of initial segmentations for the liver, spleen and kidneys are shown in Fig. 6a. It indicates that as convolutional layer number increases, the accuracy in terms of DSC arises. In addition, we test the effect of the convolution filter number, which decides the “width” of the network. We devised three architectures denoted as “CNN1,” “CNN2” and “CNN3,” which correspond to small, middle and big networks, respectively. The networks “CNN1” and “CNN2” contain {32, 128, 128, 128, 128, 128, 128, 128, 32, 4} and {64, 256, 256, 256, 256, 256, 256, 256, 128, 4} filters in the ten convolutional layers, respectively. The network “CNN3” is the architecture described in Table 1. Comparable results of the three architectures in Fig. 6b show that accuracy can be improved as the network becomes wider. Therefore, in our experiments we set the convolutional number as 10 and used the big network to achieve highest accuracy while making the computation memory satisfied on our machines.
Typical examples of segmentation results. From up to down, segmentation results for CT volumes of portal venous phase, artery phase and noncontrast, respectively. From 1st column to 3rd column are results for the liver, spleen and kidneys in 2D axial view, respectively. The automated segmentation is outlined in green, and the ground truth is shown in red. The 4th column provides 3D renderings of the segmentation results, where the liver is red, spleen is blue, and kidneys are yellow
Comparable results for the liver, spleen and kidneys of one typical CT volume (from top to down). From left to right are results of the CNN, the surface refinement without probabilistic prior and the proposed method, respectively. The automated segmentation is outlined in green, and the ground truth is colored in red. Note the regions of leakage indicated by red arrow
Effect of the size of training data
One central important factor of the CNN is the availability of large amount of labeled data, which is rather difficult for medical images. To evaluate the sensitivity of our method with respect to the size of training data, we tested its generalization ability in an experiment where the number of training data was decreased gradually. This experiment was tested on 28 data, with the number of training data varying from 28 to 112. Note in this experiment no training data were used for testing.
Figure 7 shows the initial and final segmentation results of our method with different number of training data. The accuracy of initial segmentation by the CNN arose as the number of training data increased, while the final segmentation result was relatively stable. It indicates that by enlarging the size of training dataset, the performance of our CNN-based organ detection and segmentation can be improved. When the number of training data is limited (e.g., 28 CT images), the initial segmentation shows severe under-/over-segmentation. This is because the complex CNN learning architecture with many parameters to learn can easily lead to overfitting on a small training set, where the diversity of data is very limited. Therefore, the low generalization ability and poor segmentation results can be expected. With more data for training, the performance of our CNN model is improved obviously. When the size of training data becomes large, the segmentation errors mainly exist in the precise border localization. Although deeper models with pooling and convolution can increase invariance and receptive fields thus capture high-level features, they also smooth responses and lower the boundary localization accuracy. Taking advantage of the segmentation refinement method, more details at the organ boundaries can be recovered. From Fig. 7, we can see that the results can achieve sound accuracy even with limited training subjects.
Comparison with state-of-the-art methods
It is always difficult to directly compare segmentation performance with different methods, as they usually use different datasets, different qualities of manual segmentations and different evaluation metrics. Here, our method was roughly compared with six state-of-the-art automatic multi-organ localization/segmentation methods [8, 9, 17, 21, 31, 34] based on the same metrics in Table 3. As for the initial segmentation, the proposed CNN can localize object organs more accurately than the random forest-based organ localization method [8] in terms of DSC and ASD. As for the final segmentation, the proposed method obtained higher DSCs for the liver, spleen and kidneys than four compared methods except for Linguraru et al. [17]. In terms of JI and ASD, the proposed method also achieved competitive performance. The work by Linguraru et al. [17] constructed a 4D graph model with shape constraints from probabilistic atlas to segment multiple organs from multi-phase CT images and achieved impressive results. However, since this approach was evaluated on a small dataset of high-resolution CT images and utilized a general probabilistic atlas, it might be challenged by more diverse datasets. Other works are based on probabilistic atlas [31, 34], SSMs [9] or combination of both [21]. The works of Tong et al. [31] and Wolz et al. [34] need registration process, which is computational intense.
Although the comparison is conducted on different datasets of different sizes, Table 3 still indicates our method’s advantage in runtime over other methods. The computational cost in different studies is not necessarily linear with the size of CT volumes. The runtime of Wolz et al. [34] is defined by the nonrigid registration step, and the overall time is around 3 h. The runtime of Tong et al. [31] increases approximately linearly with the number of selected atlases during training, which is significant to the segmentation accuracy. For image scans with slice sickness of 1 mm, the runtime of Linguraru et al. [17] is about 2 h per scan. In our method, the CNN-based organ detection spent around 6 s for each CT volume of size \(512\times 512\times 279\), comparable to that of Gauriau et al. [8]. The average runtime of our refinement method on our dataset is around 119 s, and the total time of our method is 125 s per volume, which is much lower than those of aforementioned methods.
Discussion and conclusions
In this paper, we developed a fast automatic multi-organ segmentation framework based on a deep 3D fully CNN and a multi-phase surface evolution-based refinement model. The proposed method can simultaneously localize and segment 4 organs, i.e., the liver, spleen and both kidneys, from CT images of various contrast-enhancement patterns with high accuracy and efficiency. To automatically detect the organs from complex backgrounds, a deep CNN was trained to learn subject-specific probability maps, which provided both spatial prior constraints and proper initialization for subsequent fine segmentation. Compared to SSMs/atlas-based methods [9, 17, 21, 31, 34], our organ localization needs no shape model/atlas construction/registration, complicated initial position searching or shape deformation. Compared to regression method [8], the CNN-based organ detection is advantageous to avoid complex feature extraction and selection process and can achieve higher accuracy. Compared to our previous work [19] that also used fully CNN for liver segmentation, this work considers the simultaneous segmentation of multiple organs, which is a much harder task due to the diverse variations of organ shapes, appearances and locations across subjects and highly similar organ intensity and appearances in each CT volume. To construct a more powerful network, wider hidden layers and an improved activation function PReLU [10] have been used. For the multi-organ segmentation problem, we formulated the CNN-guided refinement as a multi-phase segmentation problem, which is solved by a recently proposed convex optimization-based time-implicit multi-phase level-set algorithm [26]. Compared to multi-label graph cut method, this algorithm can successfully avoid metrification artifacts and can be easily implemented on GPUs to significantly improve computational efficiency with much lower memory cost.
A fivefold cross-validation on 140 CT scans showed that the proposed method yielded mean DSCs of \(96.0\pm 1.5\), \(94.2\pm {5.4}\) and \({95.4}\pm 3.2\%\) for the liver, spleen and kidneys, respectively. The mean JIs for the liver, spleen and kidneys were \(92.4\pm 2.8\), \({89.5}\pm 6.9\) and \(91.3\pm 5.3\%\), respectively. In addition, the mean ASDs were less than 1.3 mm for all organs. By implementing core components of the algorithm on GPUs, the average computational time on a CT volume was about 125 s, which was much less than most current methods in literature.
Although the proposed method performs well on a large and diverse dataset, the experiments and the method still have several limitations. First, the performance comparison has only be conducted using different datasets. A more thorough comparison of different methods on the same dataset will be valuable. Unfortunately, there is no public dataset available to evaluate multiple abdominal organ segmentation methods. Although the Visceral Anatomy3 Extended BenchmarkFootnote 2 is an excellent collection of data for this target, its online evaluation environment is currently only based on CPU computation, which makes it difficult to evaluate our GPU-implemented method on this benchmark. Second, although our method can perform well on most pathological data, such as livers with tumors and enlarged spleens, in current form it does not handle data with missing organs or organ of different topologies. Even if an organ is missing, the CNN-based object detection will give a localization prediction. In Fig. 8a we present an example of localization in a CT image with a missing right kidney. The kidney is located under the spleen and so does the final segmentation result even if the organ is not present. Another special case of the organ with abnormal topology is shown in Fig. 8b. It is a horseshoe kidney, i.e., the two kidneys are merged. However, the trained CNN still predicts two separate kidney regions and restricts the final segmentation. We can see that in the CNN training process, the spatial information and topology are encoded into the network. The inferred priors are dominated by the most training data of complete organs and regular topology. Third, the parameters of the refinement model are empirically set without optimal searching on a training set. In the future work, we will conduct parameter learning in a data-driven style. In addition, inspired by the work of Dou et al. [7], the refinement part may be further improved by utilizing the CRF method.
In conclusion, quantitative evaluation on a large and diverse dataset and comparisons with state-of-the-art automatic methods showed that the proposed automatic method can localize and segment the organs, i.e., liver, spleen and kidneys, accurately and efficiently. The high values of DSC and JI alone with low ASD suggest that our method could be potentially used for quantitative measurement of the organs in clinical application. In the future, we expect to adapt the proposed framework for more organs, such as pancreas and gallbladder.
References
Brosch T, Tang LY, Yoo Y, Li DK, Traboulsee A, Tam R (2016) Deep 3d convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35(5):1229–1239
Cha KH, Hadjiiski L, Samala RK, Chan HP, Caoili EM, Cohan RH (2016) Urinary bladder segmentation in ct urography using deep-learning convolutional neural network and level sets. Med Phys 43(4):1882–1896
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2015) Semantic image segmentation with deep convolutional nets and fully connected crfs. In: International conference on learning representations (ICLR)
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds) International conference on medical image computing and computer-assisted intervention, (MICCAI), Springer, pp 424–432
Ciresan D, Giusti A, Gambardella LM, Schmidhuber J (2012) Deep neural networks segment neuronal membranes in electron microscopy images. In: Advances in neural information processing systems, pp 2843–2851
Criminisi A, Robertson D, Konukoglu E, Shotton J, Pathak S, White S, Siddiqui K (2013) Regression forests for efficient anatomy detection and localization in computed tomography scans. Med Image Anal 17(8):1293–1303
Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng PA (2016) 3d deeply supervised network for automatic liver segmentation from ct volumes. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 149–157
Gauriau R, Cuingnet R, Lesage D, Bloch I (2015) Multi-organ localization with cascaded global-to-local regression and shape prior. Med Image Anal 23(1):70–83
He B, Huang C, Jia F (2015) Fully automatic multi-organ segmentation based on multi-boost learning and statistical shape model search. VISCERAL@ ISBI 2015 VISCERAL Anatomy3 Organ Segmentation Challenge, p 18
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision, pp 1026–1034
Heimann T, van Ginneken B, Styner M, Arzhaeva Y, Aurich V, Bauer C, Beck A, Becker C, Beichel R, Bekes G, Bello F, Binnig G, Bischof H, Bornik A, Cashman P, Chi Y, Cordova A, Dawant B, Fidrich M, Furst J, Furukawa D, Grenacher L, Hornegger J, Kainmuller D, Kitney R, Kobatake H, Lamecker H, Lange T, Lee J, Lennon B, Li R, Li S, Meinzer HP, Nemeth G, Raicu D, Rau AM, vanRikxoort E, Rousson M, Rusko L, Saddi K, Schmidt G, Seghers D, Shimizu A, Slagmolen P, Sorantin E, Soza G, Susomboon R, Waite J, Wimmer A, Wolf I (2009) Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imaging 28(8):1251–1265
Hu P, Wu F, Peng J, Liang P, Kong D (2016) Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol 61:8676–8698. doi:10.1088/1361-6560/61/24/8676
Jawarneh MS, Mandava R, Ramachandram D, Shuaib IL (2010) Automatic initialization of contour for level set algorithms guided by integration of multiple views to segment abdominal ct scans. In: 2010 Second international conference on computational intelligence, modelling and simulation (CIMSiM). IEEE, pp 315–320
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Lai M (2015) Deep learning for medical image segmentation. arXiv preprint arXiv:1505.02000
Lee CY, Xie S, Gallagher P, Zhang Z, Tu Z (2015) Deeply-supervised nets. In: AISTATS, vol 2, p 6
Linguraru MG, Pura JA, Pamulapati V, Summers RM (2012) Statistical 4d graphs for multi-organ abdominal segmentation from multiphase ct. Med Image Anal 16(4):904–914
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Lu F, Wu F, Hu P, Peng Z, Kong D. Automatic 3d liver location and segmentation via convolutional neural networks and graph cut. Int J Comput Assist Radiol Surg. doi:10.1007/s11548-016-1467-3
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. arXiv preprint arXiv:1606.04797
Okada T, Linguraru MG, Hori M, Summers RM, Tomiyama N, Sato Y (2015) Abdominal multi-organ segmentation from ct images using conditional shape-location and unsupervised intensity priors. Med Image Anal 26(1):1–18
Peng J, Dong F, Chen Y, Kong D (2014) A region-appearance-based adaptive variational model for 3d liver segmentation. Med Phys 41(4):043,502
Peng J, Hu P, Lu F, Peng Z, Kong D, Zhang H (2015) 3d liver segmentation using multiple region appearances and graph cuts. Med Phys 42(12):6840–6852
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12(7):629–639
Potts RB (1952) Some generalized order-disorder transformations. In: Mathematical proceedings of the Cambridge philosophical society, vol 48. Cambridge University Press, pp 106–109
Rajchl M, Baxter JS, Bae E, Tai XC, Fenster A, Peters TM, Yuan J (2015) Variational time-implicit multiphase level-sets. In: Energy minimization methods in computer vision and pattern recognition. Springer, pp 278–291
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Roth HR, Lu L, Farag A, Shin HC, Liu J, Turkbey EB, Summers RM (2015) Deeporgan: multi-level deep convolutional networks for automated pancreas segmentation. In: Medical image computing and computer-assisted intervention. Springer, pp 556–564
Saito A, Nawano S, Shimizu A (2016) Joint optimization of segmentation and shape prior from level-set-based statistical shape model, and its application to the automated segmentation of abdominal organs. Med Image Anal 28:46–65
Selver MA (2014) Segmentation of abdominal organs from ct using a multi-level, hierarchical neural network strategy. Comput Methods Programs Biomed 113(3):830–852
Tong T, Wolz R, Wang Z, Gao Q, Misawa K, Fujiwara M, Mori K, Hajnal JV, Rueckert D (2015) Discriminative dictionary learning for abdominal multi-organ segmentation. Med Image Anal 23(1):92–104
Vese LA, Chan TF (2002) A multiphase level set framework for image segmentation using the mumford and shah model. Int J Comput Vis 50(3):271–293
Wang C, Smedby O (2014) Automatic multi-organ segmentation using fast model based level set method and hierarchical shape priors. Proc VISCERAL Chall ISBI 1194:25–31
Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D (2013) Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans Med Imaging 32(9):1723–1730
Xu Z, Burke RP, Lee CP, Baucom RB, Poulose BK, Abramson RG, Landman BA (2015) Efficient multi-atlas abdominal segmentation on clinically acquired ct with simple context learning. Med Image Anal 24(1):18–27
Yuan J, Bae E, Tai XC, Boykov Y (2010) A continuous max-flow approach to potts model. In: European conference on computer vision. Springer, pp 379–392
Yuan J, Ukwatta E, Tai X, Fenster A, Schnoerr C (2012) A fast global optimization-based approach to evolving contours with generic shape prior. University of California, Los Angeles. Technical report CAM 12, vol 38
Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D (2015) Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 108:214–224
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 11271323, 91330105, 11401231), Zhejiang Provincial Natural Science Foundation of China (Grant No. LZ13A010002), Natural Science Foundation of Fujian Province (Grant No. 2015J01254) and Science Technology Foundation for Middle-aged and Young Teacher of Fujian Province (Grant No. JA14021). J. Peng was also supported by Promotion Program for Young and Middle-aged Teacher in Science and Technology Research of Huaqiao University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
This article does not contain any studies with human participants or animal performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Peijun Hu and Fa Wu have contributed equally in this work.
Rights and permissions
About this article
Cite this article
Hu, P., Wu, F., Peng, J. et al. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int J CARS 12, 399–411 (2017). https://doi.org/10.1007/s11548-016-1501-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-016-1501-5