Abstract
Purpose
Virtual digital resources and printed models have become indispensable tools for medical training and surgical planning. Nevertheless, printed models of soft tissue organs are still challenging to reproduce. This study adopts open source packages and a low-cost desktop 3D printer to convert multiple modalities of medical images to digital resources (volume rendering images and digital models) and lifelike printed models, which are useful to enhance our understanding of the geometric structure and complex spatial nature of anatomical organs.
Materials and methods
Neuroimaging technologies such as CT, CTA, MRI, and TOF-MRA collect serial medical images. The procedures for producing digital resources can be divided into volume rendering and medical image reconstruction. To verify the accuracy of reconstruction, this study presents qualitative and quantitative assessments. Subsequently, digital models are archived as stereolithography format files and imported to the bundled software of the 3D printer. The printed models are produced using polylactide filament materials.
Results
We have successfully converted multiple modalities of medical images to digital resources and printed models for both hard organs (cranial base and tooth) and soft tissue organs (brain, blood vessels of the brain, the heart chambers and vessel lumen, and pituitary tumor). Multiple digital resources and printed models were provided to illustrate the anatomical relationship between organs and complicated surrounding structures. Three-dimensional printing (3DP) is a powerful tool to produce lifelike and tangible models.
Conclusions
We present an available and cost-effective method for producing both digital resources and printed models. The choice of modality in medical images and the processing approach is important when reproducing soft tissue organs models. The accuracy of the printed model is determined by the quality of organ models and 3DP. With the ongoing improvement of printing techniques and the variety of materials available, 3DP will become an indispensable tool in medical training and surgical planning.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Human anatomical organs are the fundamental resource used by medical students to learn about the structures of the human body. During past centuries, donated bodies were the primary source of anatomical organs for medical schools. Medical trainees were afforded the opportunity to see and feel real organs in dissection classes [1, 2]. Due to the scarcity of donated bodies, the high cost of maintaining cadavers, and ethical concerns [3–5], fewer trainees now have the opportunity to touch real anatomical organs; new educational resources that are easier to obtain and use are therefore urgently needed.
Thanks to progress in neuroimaging technologies, computerized tomography (CT), and magnetic resonance imaging (MRI) have become widely used tools for producing medical images of living subjects. Medical images, volume rendering images, and digital models are then used to display the geometric structures and details on computer screens and tablets; the limitations of these are that they can only be viewed and not touched [6–8]. Previous studies have proved that lifelike printed models may be employed as a useful educational tool to enhance the understanding of the geometric structure and the complex spatial characteristics of anatomical organs [5, 9].
Since Levy et al. implemented their selective laser sintering technique to produce a physical model of the temporal bone [10], three-dimensional printing (3DP) has been promoted as one of the most important tools available. The principle of 3DP is to produce a physical model from a digital model using additive layer manufacturing. Because of the high accuracy and low cost of producing the replica of the digital model, 3DP has been widely applied in surgical planning [11–14], implant design [15, 16], education and training [4, 5, 17], and forensic science [18].
Although previous studies have already described the process of converting medical images to printed models [3, 4, 7, 18–20], most studies concentrate on producing models of the bones and only few soft tissue organs, because of the challenge of their segmentation and reconstruction. Here, we shall describe the method for converting multiple modalities of medical images to digital resources (volume rendering images and digital models) and printed models for both hard and soft tissue organs. By considering the quality of medical image reconstruction, we present both visual and subjective qualitative assessment along with numerical and objective quantitative assessment. We integrate open source packages and a low-cost desktop 3D printer to reproduce digital and printed models of the cranial base, tooth and dental pulp cavity, the brain, blood vessels of the brain, parts of the heart chambers and vessel lumen, and pituitary tumors. These multiple resources are an available and cost-effective way to enhance the understanding of anatomical organs.
Methods
Medical images acquisition
The acquisition of medical images is the first step in producing digital resources and printed models. In order to enhance the contrast between regions of interest (ROI) and surrounding structures and to reduce the complexity and time cost of ROI extraction, the appropriate modality should be chosen. We use computed tomography (CT) to construct the cranial base and cone beam computed tomography (CBCT) images to construct the tooth. Alternatively, we use magnetic resonance imaging (MRI) to construct the brain and pituitary tumors, time-of-flight magnetic resonance angiography (TOF-MRA) to construct the blood vessels of the brain, and computed tomography angiography (CTA) to construct the heart chambers and vessel lumen. Medical images are archived in standard digital imaging and communications in medicine (DICOM) format.
Visualization of medical images
Volume rendering is an available and effective tool to illustrate both the interior and boundary of the volume data with transparency and various colors [6]. Trainees translate, rotate, and zoom 3D medical images in order to view the details and anatomic relationship between various organs and their surrounding structures without segmentation and reconstruction. However, volume rendering results are only image projections produced by simulating the absorption of light along the ray path. The open source package Visualization Toolkit (VTK) was chosen to perform volume rendering in real time, integrating a number of volume rendering algorithms [21]. We provide 3D texture-based volume rendering and ray tracing tools to realize the volume rendering.
Digital and printed model production
The procedures for generating digital models from medical images essentially consist of three steps: medical image segmentation, reconstruction, and mesh processing. Image segmentation is actually an optional step, and digital models could be directly generated using iso-surface extraction. However, some irrelevant triangle meshes and small holes are generated during this procedure. To address this problem, mesh processing is an indispensable tool to enable us to obtain higher accuracy in the digital model. Subsequently, the digital model is archived as a stereolithography (STL) format file and imported to a low-cost desktop printer to produce a printed model. A typical sequence of these procedures is shown in Fig. 1. These steps are discussed in detail below.
Image segmentation
The goal of image segmentation is to extract the ROI from medical images. The low contrast between the ROI and surrounding structures presented a challenge. The choice of modality and various segmentation algorithms were investigated to extract the ROI. We utilized the open source package Medicine Insight Segmentation and Registration Toolkit (ITK) to develop segmentation tools that integrate threshold, level sets, watershed, region growing, etc. [21].
We provided several tools to realize ROI segmentation for various organs. The automatic threshold segmentation tool was a fast and effective tool to distinguish the ROI from high-contrast images by setting a range of values, e.g., the bone was easily distinguished from CT images. The ROI was extracted by comparing the density value of each pixel with a given threshold.
Automatic extraction of soft tissue organs has always been a challenge in medical image segmentation. Watershed was one of the classic region-based segmentation algorithms to partition images into several regions and basins by a local minimum of density value until the water level has reached the highest possible value. Then, morphological region growing was used to merge various basins following connectivity and density similarity [22]. Another available tool for soft tissue organ segmentation was the level set approach, a numerical technique for tracking the evolution of interfaces [23]. With this tool, the operator was able to coarsely draw the close curve, and the fine boundary of ROI was detected after several iterations.
Mesh reconstruction
Once ROI was extracted, mesh reconstruction was performed to generate 3D meshes. The marching cubes algorithm was one of the most common methods to convert voxel data to a series of triangle meshes by extracting iso-surfaces [24]. This algorithm calculated the triangle vertices using linear interpolation between adjacent images and determined the topology according to a lookup table and the density value of each voxel. The original volume data without segmentation can also be reconstructed using marching cubes, i.e., the pixel of each voxel was labeled by comparing the intensity value with the given range [25]. Alternatively, contour-based triangulation was another reconstruction method on the basis of boundaries of ROI extraction and determined the topology by connecting adjacent contours of contiguous images. VTK integrated both two methods and the reconstructed models can be archived in the STL format.
Digital resources and printed model of cranial base. a Axial view; b sagittal view; c coronal view; d bone segmentation; e digital skull model; f display digital model with medical images in the common coordinate system; g convert STL format file to series of layers using Makerware software. Each layer includes supporting structures and printed model; h printed model of cranial base after manually removing supporting structures
Mesh processing
Some small holes and self-intersection triangle meshes occurred during the course of reconstruction. In addition, some irrelevant triangle meshes were generated without accurate segmentation. To address this problem, interactive triangle meshes processing was an effective solution. The open source package MeshLab was easily used to provide mesh editing, hole filling, smoothing, and self-intersecting inspection [26]. An operator lacking extensive 3D modeling skills could also select and remove irrelevant meshes, fill gaps and holes, and ensure the high quality of digital models.
3D printing
There exist many types of 3DP techniques able to satisfy various requirements for printability [27]. Having considered the costs of the printing device and printing materials, the Makerbot Replicator 2 (Makerbot, New York, USA) and polylactide (PLA) filament materials were used. The Makerbot Replicator is a type of fused deposition modeling (FDM) printer that utilizes a high-temperature nozzle to heat the PLA filament to a semiliquid state and prints it one layer at a time until every layer has been printed.
Once a highly accurate digital model had been generated after mesh processing, this model was imported to the Makerware software, which is bundled with the Makerbot Replicator. Translation, rotation, and scaling transformation were carried out to ensure that the printable model was vertically located at the top of the build plate and not beyond the printing space. Makerware automatically slices the digital model into a series of layers, and supporting structures are generated on the build plate for construction since PLA should be deposited on a layer. The printing precision of the 3D printer needs to be set. The higher the printing precision is, the more time will be taken for printing. We must balance the time cost and the accuracy required for each printed model. The printing precision was set to 0.2 mm for the bones and 0.1 mm for soft tissue organs. In most situations, since supporting structures are tightly attached to both the printed models and build plate, we had to use pliers to remove supporting structures; caution should be exercised when removing supporting structures from the tiny printed models.
Digital resources and printed model of parts of heart chambers and vessel lumen. a Heart CTA image; b coarse digital model of heart chambers and vessel lumen using marching cubes without accurate segmentation by setting a given range; c Fine digital model after mesh processing, removing irrelevant triangle meshes and filling holes; d display digital model with medical images in the common coordinate system; e 3D printer and printed model; f printed model of part of heart chambers and vessel lumen after manually removing supporting structures; and g comparison of digital model and printed model
Digital resources and printed model of blood vessel. a TOF-MRI image; b blood vessel volume rendering; c digital model of blood vessel from automatic image segmentation; d align the digital model from segmentation (red color) and digital model from region grow (yellow color); e the spatial structure between digital models of blood vessel and head; f display digital model with medical images in the common coordinate system; and g printed model of blood vessel
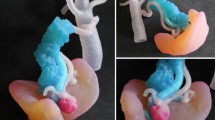
Digital resources and printed model of pituitary tumor. a MRI image; b interactive extraction of pituitary tumor; c manual segmentation result; d medical images; e volume rendering; f digital model of tumor; g display digital model with medical images in the common coordinate system; and h printed model of pituitary tumor
Accuracy evaluation
To assess the accuracy of medical image reconstruction, the present study took both qualitative and quantitative assessments. Qualitative assessment is accomplished by means of visual inspection and the subjective evaluation method. Medical images and the reconstructed model were aligned in the common coordinate system and users interactively watched and inspected these resources from different viewpoints. The objective is a quantitative assessment, and a numerical tool is used to visualize the geometric deviation. An optical scanner has enabled highly accurate digitization of real objects, and we utilized a Handyscan 700 laser scanner (CreaForm Company, Canada) to acquire the digital model. The quantitative comparison of dimensional errors was conducted by comparing the reconstructed model and corresponding scanned model using the open source package CloudCompare, version 2.6.2.
Results
Multiple digital resources (volume rendering images and digital models) and printed models were reproduced from multiple modalities of medical images of living subjects (Figs. 2, 3, 4, 5, 6, 7). Details of these cases, including the choice of modality data, voxel dimension, physical resolution and the number of vertices and triangle meshes in the digital model, are given in Table 1. We provide medical images (coronal plane, sagittal plane, and transverse plane), volume rendering images, digital models, and printed models to illustrate the geometric structure and anatomical relationship with surrounding structures.
CT and CBCT images were the best choices for bone segmentation (Figs. 2, 3). The bone was extracted using the threshold tool by setting a given value (Fig. 2d) and the digital model was generated using marching cubes (Fig. 2e). After the digital model was imported to the 3D printer, each layer of the printed model and corresponding supporting structures were generated by slicing (Fig. 2g). Supporting structures had to be removed manually by pliers as shown in Fig. 2h. Since a high level of accuracy was required for the tooth, CBCT images were an effective choice as a high-resolution form of imaging, which have widespread application in oral and maxillofacial radiology. Both the tooth and dental pulp cavity are shown in Fig. 3.
The production of digital and printed models of soft tissue organs has always been a challenge in medical training and in clinics due to the low contrast of the ROI and their surrounding structures. We have, however, succeeded in producing digital resources and printed models of soft tissue organs from multiple modalities of medical images (Figs. 4, 5, 6, 7).
The texture-based volume rendering algorithm was used to visualize the brain (Fig. 4c). Our previous study reported the use of the watershed and level set algorithm to extract brain tissues from MRI images [28]. The brain digital model was generated after segmentation (Fig. 4d). The medical images and digital model were located in the common coordinate system (Fig. 4e), and Fig. 4f shows the corresponding printed model.
CTA medical images were chosen (Fig. 5a) to reproduce the digital resources and printed model for parts of the heart chambers and vessel lumen, and the pixel of each voxel was labeled as 0 or 1 by comparing the density value of each pixel with the given range. Marching cubes was performed to generate digital models without accurate segmentation (Fig. 5b). The irrelevant triangle meshes were interactively removed using MeshLab and only the digital model of the heart chambers and vessel lumen was retained (Fig. 5c). Figure 5e, f, g shows the corresponding printed model.
Ray tracing was used to visualize the structure of the blood vessel from TOF-MRA images by setting the transfer function and color mapping (Fig. 6a, b). The digital model was first generated using an automatic segmentation method (Fig. 6c). However, some tiny structures were missing because of incomplete segmentation. To generate the complete model, another digital model was generated after region grow segmentation (Fig. 6d). We interactively marked seed points on the region of the blood vessel for segmentation, and the corresponding model was reconstructed. The irrelevant triangle meshes were removed using MeshLab. Subsequently, two digital models were aligned to generate a fine model (Fig. 6e). The spatial relationships between the medical images and digital model are presented in Fig. 6f. The printed model of the blood vessel is shown in Fig. 6g, and the supporting structures should be removed carefully as the tiny printed model was fragile.
Although manual segmentation is tedious work and requires the guidance of someone with anatomical knowledge, it is nevertheless the most accurate method to extract ROI from low-contrast brightness images. Before interactive segmentation, we adjusted the images’ brightness and contrast by setting the window level and width. The ROI was interactively selected by pressing and dragging the mouse (Fig. 7b). The pituitary tumor was extracted from MRI images (Fig. 7c, d). Volume rendering using the texture-based method, digital model, and printed model respectively are shown in Fig. 7e, h.
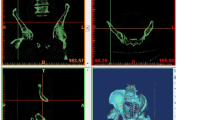
Qualitative and quantitative assessments have been carried out to determine the accuracy of converting medical images to digital models. As shown in Figs. 2f, 4e, 5d, 6f, and 7g, medical images and corresponding reconstructed models were located in the common coordinate system, and these discrepancies were empirically and subjectively viewed. To quantitatively assess the accuracy of reconstruction, the deviation between the reconstructed model and the scanned model was calculated. The skull of the cadaver was firstly reconstructed from CT images, which consisted of 950,426 vertices and 1,828,794 triangle meshes (Fig. 8a). The digital model produced by the laser scanner consisted of 1,551,041 vertices and 3,091,750 triangle meshes (Fig. 8b). As a result of pose inconsistencies between the two skull models (Fig. 8c), we aligned the two models in the common coordinate system (Fig. 8d). The geometric deviations are presented in Fig. 8e. Many of the highest deviations (blue and red regions) were located in the area around the tooth, zygoma, orbit, and lateral bulge of the chin mound.
Discussion
Medical schools have to cope with an ongoing shortage of donated bodies for education and research, so trainees have few opportunities to feel real organs. Multiple modalities of medical images are used to acquire data from living subjects, and then two-dimensional images and three-dimensional resources (volume rendering images and digital models) are provided to supplement training resources. Volume rendering is a useful approach to illustrate the geometrical structure and anatomic relationships between organs and their surrounding structures without image segmentation, but these can only be viewed and not printed. The procedures for constructing digital models are complicated due to the complexity of ROI segmentation, one advantage of them, however, is that can be printed as tangible models.
One limitation in the application of 3DP is the availability of digital models [29]. Since they are more easily distinguished, most previous studies have utilized digital resources and printed models of bones (such as the skull and vertebra) [5, 10, 18, 30–35]. Few soft tissue organs have been able to be used in clinical case studies or surgical training because of the challenge of soft tissue organ segmentation. This study describes the method for converting multiple modalities of medical images to digital resources. The choice of modality is very important, because resolution and contrast brightness affect the quality of reconstruction, i.e., bones are easily distinguished from CT images but difficult to extract from MRI images. Automatic segmentation approaches are used to reduce the complexity and time cost to realize image segmentation. However, some tiny structures and details may be overlooked as a result of incomplete segmentation. The most promising solution is to utilize multiple segmentation and reconstruction approaches to generate digital models, and a better model will be reproduced after alignment, avoiding the limitations of the single-choice approach.
During the past ten years, the application of 3DP has been invaluable in anatomical training and surgical education, because it both avoids certain ethical concerns and provides more realistic educational resources [3, 5, 9, 17, 18, 36, 37]. 3DP is low in cost, high in accuracy, and involves a short production time. Multiple copies of dissected specimens and clinical case studies can be produced to scale in any size. Previous studies have shown that 3D printed models are more intuitive compared with digital models and have significantly higher effectiveness in capturing complex 3D spatial relationships [5, 9, 29, 36]. Our studies provide multiple printed models for hard and soft tissue organs in order to better explain the spatial structure of anatomical organs.
The procedures for producing printed models involve the acquisition of images, image segmentation, reconstruction, mesh processing, and 3DP. The main challenge to the accuracy of printed models remains in producing a high-quality digital model and the high precision of 3DP [29]. Fasel et al. suggested that many structures of the printed models were incorrect due to the inaccuracy of the digital model and the limiting layer thickness of 3DP [32]. To assess the accuracy of the digital model, this study presents both qualitative and quantitative assessments. Assessors can easily utilize qualitative methodology to detect any discrepancies when we align digital models and medical images in the common ordinate system. The discrepancies between the pixels of a medical image and the corresponding vertex of a digital model can be empirically viewed when some structures are not reproduced from the medical images due to incomplete segmentation. Subjective visual inspection is important, but it is not sufficient.
Quantitative methodology is a powerful tool to numerically compute these kinds of deviation. A common numerical approach is to statically analyze dimension errors between a reconstructed model and a real object. Three-dimensional scanning is a highly accurate method for acquiring the geometrical structure of a complex object, and any errors in dimension can be accurately calculated by comparing the real object and the reconstructed model. Considering the working principle of the laser scanner and the geometrical structure of soft tissue organs are easily influenced by gravity, this quantitative method is preferred for the assessment of bones.
Several researchers have expressed concern about the accuracy of printed models using 3DP. Most studies reported that printed models of anatomical structures can be printed with sufficient accuracy and that there was no significant difference between printed models and real objects [3–5, 9, 17, 38]. Three-dimensional printing will not result in any loss of accuracy once the printing resolution of the 3D printer is higher than the thickness of medical images. Silva et al. asserted that the precision of 3DP was acceptable and may be useful in most maxillofacial surgeries [20]. Wu et al. reported that spinal printed models were accurate enough for surgeons [39].
There are still some limitations in the application of 3DP: (a) the printable area of the 3D printer is limited so that larger digital models (such as pelvis, femur, and spine) have to be cropped into several small components. The complete printed model is produced by pasting together the printed model of each component. (b) Low-cost FDM printers and PLA materials have been successfully used for education and training, but more lifelike printed models of soft tissue organs with full colors and elasticity are necessary to satisfy increasing educational demands. However, these printers and printing materials are still expensive.
Conclusion
This study presents the method for converting medical images to digital resources and printed models using open source packages and a low-cost desktop 3D printer, well within the budget of medical schools and hospitals. The effectiveness of this method has been demonstrated by producing models of the cranial base, tooth, the brain, blood vessels of the brain, parts of the heart chambers and vessel lumen, and pituitary tumors. These various techniques, including volume rendering, reconstruction, and 3DP, are feasible and enable trainees to better understand the spatial structure of anatomical organs.
References
Winkelmann A (2007) Anatomical dissection as a teaching method in medical school: a review of the evidence. Med Educ 41(1):15–22. doi:10.1111/j.1365-2929.2006.02625.x
Sugand K, Abrahams P, Khurana A (2010) The anatomy of anatomy: a review for its modernization. Anat Sci Educ 3(2):83–93. doi:10.1002/ase.139
McMenamin PG, Quayle MR, McHenry CR, Adams JW (2014) The production of anatomical teaching resources using three-dimensional (3D) printing technology. Anat Sci Educ 7(6):479–486. doi:10.1002/ase.1475
AbouHashem Y, Dayal M, Savanah S, Štrkalj G (2015) The application of 3D printing in anatomy education. Med Educ Online 20:29847. doi:10.3402/meo.v20.29847
Li Z, Li Z, Xu R, Li M, Li J, Liu Y, Sui D, Zhang W, Chen Z (2015) Three-dimensional printing models improve understanding of spinal fracture–a randomized controlled study in China. Sci Rep 5:11570. doi:10.1038/srep11570
Drebin RA, Carpenter L, Hanrahan P (1988) Volume rendering. SIGGRAPH Comput Graph 22(4):65–74. doi:10.1145/54852.378484
Rengier F, Mehndiratta A, von Tengg-Kobligk H, Zechmann CM, Unterhinninghofen R, Kauczor HU, Giesel FL (2010) 3D printing based on imaging data: review of medical applications. Int J CARS 5(4):335–341. doi:10.1007/s11548-010-0476-x
Noguera JM, Jiménez JJ, Osuna-Pérez MC (2013) Development and evaluation of a 3D mobile application for learning manual therapy in the physiotherapy laboratory. Comput Educ 69:96–108. doi:10.1016/j.compedu.2013.07.007
Preece D, Williams SB, Lam R, Weller R (2013) “Let’s get physical”: advantages of a physical model over 3D computer models and textbooks in learning imaging anatomy. Anat Sci Educ 6(4):216–224. doi:10.1002/ase.1345
Levy RA, Guduri S, Crawford RH (1994) Preliminary experience with selective laser sintering models of the human temporal bone. Am J Neuroradiol 15(3):473–477
Wagner JD, Baack B, Brown GA, Kelly J (2004) Rapid 3-dimensional prototyping for surgical repair of maxillofacial fractures: a technical note. J Oral Maxillofac Surg 62(7):898–901. doi:10.1016/j.joms.2003.10.011
Malik HH, Darwood AR, Shaunak S, Kulatilake P, El-Hilly AA, Mulki O, Baskaradas A (2015) Three-dimensional printing in surgery: a review of current surgical applications. J Surg Res 199(2):512–522. doi:10.1016/j.jss.2015.06.051
Darwood A, Collier J, Joshi N, Grant WE, Sauret-Jackson V, Richards R, Dawood A, Kirkpatrick N (2015) Re-thinking 3D printing: a novel approach to guided facial contouring. J Craniomaxillofac Surg 3(7):1256–1260. doi:10.1016/j.jcms.2015.06.001
Waran V, Devaraj P, Hari Chandran T, Muthusamy KA, Rathinam AK, Balakrishnan YK, Tung TS, Raman R, Rahman ZA (2012) Three-dimensional anatomical accuracy of cranial models created by rapid prototyping techniques validated using a neuronavigation station. J Clin Neurosci 19(4):574–577. doi:10.1016/j.jocn.2011.07.031
Lee MY, Chang CC, Ku YC (2008) New layer-based imaging and rapid prototyping techniques for computer-aided design and manufacture of custom dental restoration. J Med Eng Technol 1:83–90. doi:10.1080/03091900600836642
Giannatsis J, Dedoussis V (2007) Additive fabrication technologies applied to medicine and health care: a review. Int J Adv Manuf Technol 40(1–2):116–127. doi:10.1007/s00170-007-1308-1
Vaccarezza M, Papa V (2015) 3D printing: a valuable resource in human anatomy education. Anat Sci Int 90(1):64–65. doi:10.1007/s12565-014-0257-7
Ebert LC, Thali MJ, Ross S (2011) Getting in touch-3D printing in forensic imaging. Forensic Sci Int 211(1–3):e1–6. doi:10.1016/j.forsciint.2011.04.022
Marro A, Bandukwala T, Mak W (2016) Three-dimensional printing and medical imaging: a review of the methods and applications. Curr Probl Diagn Radiol 45(1):2–9. doi:10.1067/j.cpradiol.2015.07.009
Silva DN, Gerhardt de Oliveira M, Meurer E, Meurer MI, Lopes da Silva JV, Santa-Barbara A (2008) Dimensional error in selective laser sintering and 3D-printing of models for craniomaxillary anatomy reconstruction. J Craniomaxillofac Surg 36(8):443–449. doi:10.1016/j.jcms.2008.10.008
Schroeder WJ, Ibáñez L, Martin KM (2004) Software process: the key to developing robust, reusable and maintainable open-source software. IEEE Int Symp Biomed Imaging (1):648–651. doi:10.1109/ISBI.2004.1398621
Vincent L, Soille P (1991) Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE T Pattern Anal 13(6):583–598. doi:10.1109/34.87344
Osher S, Fedkiw RP (2001) Level set methods: an overview and some recent results. J Comput Phys 169(2):463–502. doi:10.1006/jcph.2000.6636
Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. SIGGRAPH Comput Graph 21(4):163–169. doi:10.1145/37402.37422
He HG, Tian J, Zhao MC, Yang H (2002) A 3D medical imaging surface reconstruction scheme based on segmentation. J Softw 13(2):219–226
Cignoni P, Callieri M, Corsini M, Dellepiane M, Ganovelli F, Ranzuglia G (2008) Meshlab: an open-source mesh processing tool. Eurographics Ital Chapt Conf 2008:129–136
Zhang L, Dong H, Saddik AE (2015) From 3D sensing to printing: a survey. ACM T Multim Comput 12(2):27. doi:10.1145/2818710
Shui WY, Zhou MQ, Geng GH (2009) Automatic brain tissue extraction approach of magnetic resonance head images. J Softw 20(5):1139–1145
Fredieu JR, Kerbo J, Herron M, Klatte R, Cooke M (2015) Anatomical models: a digital revolution. Med Sci Educ 25(2):183–194. doi:10.1007/s40670-015-0115-9
McGurk M, Amis A, Potamianos P, Goodger N (1997) Rapid prototyping techniques for anatomical modelling in medicine. Ann R Coll Surg Engl 79(3):169–174
Salmi M, Paloheimo KS, Tuomi J, Wolff J, Makitie A (2013) Accuracy of medical models made by additive manufacturing (rapid manufacturing). J Craniomaxillofac Surg 41(7):603–609. doi:10.1016/j.jcms.2012.11.041
Fasel JH, Beinemann J, Schaller K, Gailloud P (2013) A critical inventory of preoperative skull replicas. Ann R Coll Surg Engl 95(6):401–404. doi:10.1308/003588413X13629960046994
Huotilainen E, Jaanimets R, Valasek J, Marcian P, Salmi M, Tuomi J, Makitie A, Wolff J (2014) Inaccuracies in additive manufactured medical skull models caused by the DICOM to STL conversion process. J Craniomaxillofac Surg 42(5):e259–265. doi:10.1016/j.jcms.2013.10.001
Ma B, Kunz M, Gammon B, Ellis RE, Pichora DR (2014) A laboratory comparison of computer navigation and individualized guides for distal radius osteotomy. Int J CARS 9(4):713–724. doi:10.1007/s11548-013-0966-8
Lambrecht JT, Berndt DC, Schumacher R, Zehnder M (2009) Generation of three-dimensional prototype models based on cone beam computed tomography. Int J CARS 4(2):175–180. doi:10.1007/s11548-008-0275-9
Weiss TL, Zieselman A, Hill DP, Diamond SG, Shen L, Saykin AJ, Moore JH (2015) The role of visualization and 3-D printing in biological data mining. BioData Min 8(1):1–7. doi:10.1186/s13040-015-0056-2
Watson RA (2014) A low-cost surgical application of additive fabrication. J Surg Educ 71(1):14–17. doi:10.1016/j.jsurg.2013.10.012
Naftulin JS, Kimchi EY, Cash SS (2015) Streamlined, inexpensive 3D printing of the brain and skull. PloS One 10(8):e0136198. doi:10.1371/journal.pone.0136198
Wu A-M, Shao Z-X, Wang J-S, Yang X-D, Weng W-Q, Wang X-Y, Xu H-Z, Chi Y-L, Lin Z-K (2015) The accuracy of a method for printing three-dimensional spinal models. PloS One 10(4):e0124291. doi:10.1371/journal.pone.0124291
Acknowledgments
The authors would like to thank the anonymous reviewers. This work is supported by the National Science Foundation of China (61402042) and Beijing Natural Science Foundation (4152028).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors, Wuyang Shui, Mingquan Zhou, Shi Chen, Zhouxian Pan, Qingqiong Deng, Yong Yao, Hui Pan, Taiping He, Xingce Wang, have declared that no competing interests exist.
Ethical approval
The local ethics committee considered that this study had been carried out in accordance with the Declaration of Helsinki.
Informed consent
Informed consent was obtained from all patients for being included in the study.
Rights and permissions
About this article
Cite this article
Shui, W., Zhou, M., Chen, S. et al. The production of digital and printed resources from multiple modalities using visualization and three-dimensional printing techniques. Int J CARS 12, 13–23 (2017). https://doi.org/10.1007/s11548-016-1461-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-016-1461-9