Abstract
Purpose Differentiation of glioblastomas from metastases is clinical important, but may be difficult even for expert observers. To investigate the contribution of machine learning algorithms in the differentiation of glioblastomas multiforme (GB) from metastases, we developed and tested a pattern recognition system based on 3T magnetic resonance (MR) data.
Materials and Methods Single and multi-voxel proton magnetic resonance spectroscopy (1H-MRS) and dynamic susceptibility contrast (DSC) MRI scans were performed on 49 patients with solitary brain tumors (35 glioblastoma multiforme and 14 metastases). Metabolic (NAA/Cr, Cho/Cr, (Lip \(+\) Lac)/Cr) and perfusion (rCBV) parameters were measured in both intratumoral and peritumoral regions. The statistical significance of these parameters was evaluated. For the classification procedure, three datasets were created to find the optimum combination of parameters that provides maximum differentiation. Three machine learning methods were utilized: Naïve-Bayes, Support Vector Machine (SVM) and \(k\)-nearest neighbor (KNN). The discrimination ability of each classifier was evaluated with quantitative performance metrics.
Results Glioblastoma and metastases were differentiable only in the peritumoral region of these lesions (\(p<0.05\)). SVM achieved the highest overall performance (accuracy 98 %) for both the intratumoral and peritumoral areas. Naïve-Bayes and KNN presented greater variations in performance. The proper selection of datasets plays a very significant role as they are closely correlated to the underlying pathophysiology.
Conclusion The application of pattern recognition techniques using 3T MR-based perfusion and metabolic features may provide incremental diagnostic value in the differentiation of common intraaxial brain tumors, such as glioblastoma versus metastasis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Primary glioblastomas multiforme (GB) and intracranial metastases are the most commonly identified brain tumors in the adult population. Conventional MR imaging of glioblastomas and solitary intracranial metastatic lesions may be indistinguishable, often displaying lack of differentiation between these two entities, as their imaging characteristics and contrast-enhancement patterns may be similar in many cases. Preoperative differentiation between these lesions may contribute to a more efficient treatment planning and follow-up, especially in the cases when brain metastases are detected before the primary cancer. In these cases, if glioblastoma has been withdrawn from the differential diagnosis, accurate identification of metastases would be important for early search of the primary tumor, a safer location to biopsy for histo-pathological diagnosis and the possible further treatment with neoadjuvant therapy [1].
Advanced MR imaging techniques, such as proton magnetic resonance spectroscopy (\(^{1}\)HMRS) and Dynamic Susceptibility Contrast Enhanced (DSC) MR imaging, provide information regarding the physiological and metabolic characterization of brain tissue. In vivo \(^{1}\)HMRS provides a metabolic profile of brain tumors, measuring specific aminoacids-as N-acetylaspartate (NAA), Choline (Cho), Creatine (Cr), lipids (Lip) and lactate (Lac) and their relative ratios [1], whereas DSC-MRI enables non-invasive qualitative and quantitative measurements of tumor vascularity via the relative cerebral blood volume (rCBV) parameter [1, 2].
The previous studies have sought to distinguish glioblastomas from intracranial metastatic brain tumors using the aforementioned MRI methods [1–8]. Regarding the intratumoral region, the majority of previous researchers have reported that both \(^{1}\)HMRS and DSC MRI do not contribute significantly in the differentiation of these tumor groups, due to the similarities of the metabolite ratios and the increased vascularity between these tumors [1, 2, 5–7]. Consequently, the interest of investigators included the peritumoral regions due to the different patho-physiological mechanisms involved, regarding the infiltrating or non-infiltrating nature of the lesions [1, 2, 5–7, 9–12].
However, all the previous reports did not investigate the nonlinear relationships between the various MR parameters extracted from these techniques. The analysis of these large amounts of data with extremely significant diagnostic value may be a time-consuming process, requires specific expertise and may not be feasible during the clinical routine, especially because these data are mainly numeric.
At this point, pattern recognition techniques can be applied in order to investigate the complex intra-variable relationships and potentially aid the differential diagnosis of common intraaxial brain tumors, such as GBs and metastatic lesions. The Machine Learning discipline provides the mathematical and computational mechanisms to take advance of the available biological knowledge and data gathered from the problem domain [13].
Recently, a great effort has been made to develop intelligent systems for brain tumor diagnosis, automatic processing, classification, evaluation and representation of spectroscopic data [14–18]. Nevertheless, the previous studies used only a single MR sequence and did not investigate the simultaneous contribution of multiple MR imaging parameters. Only a few researchers achieved to combine multi-parametric data provided by conventional MRI and either 1H-MRS or perfusion MRI [19–25].
Li et al. [19] trained SVM classifier using signal intensity on the T1-weighted and T2-weighted images or blood supply for glioma grading, achieving accuracies between 83.21 and 88.33 %. Zacharaki et al. [21] applied nonlinear SVMs for the discrimination of glioblastomas from metastases scoring 81 % accuracy and concluded that the parameters extracted from the rCBV maps proved to be particularly important, since they were top-ranked in most classification pairs investigated. Devos et al. [22] compared three classification techniques for automated brain tumor diagnosis. The authors observed that nonlinear LS-SVMs reached a significantly better performance than the linear techniques, proving that several diagnostic problems have a nonlinear behavior. A multi-project and multicenter evaluation of automatic brain tumor classification has been also reported [23]. The authors applied several classifiers using as features short TE \(^{1}\)HMRS signals. The accuracy scored for the differentiation of glioblastomas and intracranial metastases was not higher than 78 %.
However, to the best of our knowledge, none of the aforementioned studies have used a combination of quantitative features extracted from \(^{1}\)HMRS and DSC-MR imaging in a classification scheme. Therefore, our study was concentrated on the utilization of parameters which may be easily extracted by the user in every clinical center and may not require further post-processing before being used as input in a pattern recognition procedure.
Hence, the purpose of this study was to evaluate the contribution of pattern recognition techniques using \(^{1}\)HMRS and DSC-MRI data as classification features in the differentiation of glioblastomas from cerebral metastases and to detect the optimum set of metabolic and perfusion parameters in terms of potential diagnostic value. The overall workflow of our study is presented in Fig. 1.
A schematic diagram of the workflow of the present study. Additional to conventional MR imaging, proton MR spectroscopy, dynamic susceptibility contrast MRI and biopsy was performed in all clinical cases. The biopsy outcome determined the supervised classification task, while the metabolic and perfusion data (NAA/Cr, Cho/Cr, (Lip\( \,+\, \)Lac)/Cr and rCBV) were used as features in the pattern recognition techniques
Methods and materials
Patients
Our prospective clinical study was approved by the Hospital Institutional Review Board committee. Patients with a solitary brain tumor with conventional MR imaging characteristics compatible with a glioblastoma or a metastatic lesion participated in our study. Our inclusion criteria were adult, cooperative patients with a solitary, inhomogeneous, contrast-enhancing brain lesion. Exclusion criteria were children, multiple lesions, prior surgery, and chemotherapy or radiation therapy. Written informed consent was obtained from all patients included in our study, after being approved by the hospital’s ethics committee, according to the Declaration of Helsinki.
\(^{1}\)HMRS and DSC-MRI were performed on 49 patients (aged 32–73 years) with a solitary brain tumor (35 glioblastomas multiforme and 14 metastases). Particularly the 14 metastatic lesions consisted of 12 lung, and 2 breast primary tumors. The occurrence of a single brain metastasis is relatively rare compared to multiple metastatic lesions and glioblastomas. This fact determined the number of metastatic tumors in this study. This may be considered as a potential limitation of such studies. Hence, it follows that the inclusion of a larger cohort of patients is expected to improve our preliminary results. All clinical cases were evaluated by two radiologists, and the diagnosis was suggested before surgery. It has to be mentioned that the differential diagnosis of gliomas may include other lesions such as abscesses or lymphomas; however, these lesions may be easily differentiated from GBs as they characteristically present restricted diffusion, while gliomas and metastatic lesions do not. All patients underwent gross total or partial surgical resection of their lesions, and the surgical procedures were performed within a month from the neuroimaging analysis. A histo-pathological diagnosis (biopsy) was obtained in all cases and was considered as the gold standard.
Conventional MR imaging, \(^{1}\)HMRS and DSC-MRI examination protocols
The study was performed on a 3-Tesla MRI whole-body scanner (GE, Healthcare, Signa®HDx) applying a standardized MRI, \(^{1}\)HMRS and DSC-MRI examination patient protocol, using a 4-channel birdcage phased-array head coil.
\(^{1}\)HMRS data acquisitions were performed using PROton Brain Exam (PROBE) Single-Voxel (SV) spectroscopy and two-dimensional-MRSI (2D-MRSI) before contrast administration in order to avoid signal disturbance. Data were acquired using Point-RESolved Spectroscopy (PRESS) pulse sequence with automatic shimming and Gaussian water suppression. Measurement parameters used in SV scans were 1,500/35 msec (TR/TE), 128 signal acquisitions (Nacq), and voxel size was chosen not to be less than 3.375 cm\(^{3}\) for adequate SNR. Measurement parameters used in 2D-MRSI were 1,000/144 msec (TR/TE), 16\(\times \)16 phase encoding steps, 10 mm section thickness, and the field of view (FOV) size was adjusted to each patient’s brain anatomy. In every region of interest, 1 to 4 voxels (depending on the phase/frequency encoding used and tumor size) were used during the study from the 2D-MRSI technique, in order to produce the final spectrum, as illustrated in Fig. 2. The positioning of the acquisition voxel was carefully done on one of the most central slices of the tumor in order to ensure accurate placement. In cases of small or odd shaped tumors, we acquired a quick axial T2 image of a few very thin slices (3 mm) positioned over the tumor to guide the placement of the voxel.
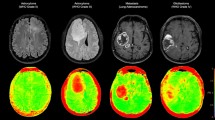
A 55-year-old man presenting a glioblastoma multiforme (first column) and a 60-year-old man presenting a metastatic lung tumor (second column). Upper row Axial T1-weighted images after contrast agent administration, CBV maps and signal intensity-time curves (a, b). Lower row Single-Voxel 1H-MRS. Voxel-localization and indicative spectra from the intratumoral and the peritumoral area respectively (c, d)
The DSC-MR images were acquired with a single shot gradient-echo echo planar imaging sequence (TR/TE \(=\) 2,000/20.7 msec, flip angle\( \,=\, \)60, FOV\( \,=\, \)24, thickness\( \,= \)5 mm with gap\( \,=\, \)0 mm, NEX\( \,=\, \)1) during the first pass of bolus of contrast material at a dose of \(>\)0.4 mmol/kg body weight. The section thickness and location of the perfusion-weighted MR data set were determined by using axial T1-weighted images after contrast injection to locate the lesion and axial T2-weighted images to locate the peritumoral T2 signal abnormality.
Data post-processing
\(^{1}\)H MR Spectroscopy: DSC-MR imaging
The delineation of the tumors was conducted by two separate radiologists in order to reduce inter-observer variability. In cases where the delineated areas presented controversies between the two radiologists, an average demarcation was taken into account. The intratumoral region of the lesions was defined as the area presenting a hyperintese signal on T2-weighted images in combination with cystic and necrotic portions, and a heterogenous or a ring-shaped contrast enhancement in T1-post-contrast imaging. The peritumoral area of both tumor types presented a hyperintese signal on T2-FSE and T2-FLAIR images, and an iso/hypointense T1 signal after contrast administration. The area exactly outside the margin of the solid part of the tumor and its surrounding was defined as the peritumoral area. However, especially in some glioblastoma cases, due to the peritumoral infiltration, this delineation was not always directly feasible, therefore an area extending 1 cm away from the presumed tumoral margin was considered as the peritumoral region.
Spectra for each patient were acquired from the intratumoral and peritumoral regions (Fig. 2). Spectroscopic data analysis and calculation of metabolite ratios were performed on an Advantage Linux workstation using the Functool software (General Electric Healthcare). The metabolic ratios of NAA/Cr and Cho/Cr were calculated for two different echo times (35 and 144 msec), while (Lip \(+\) Lac)/Cr ratio was measured using a TE of 144 msec [26]. For both spectroscopic techniques, a rectangular ROI was localized by using the transverse T2-weighted FLAIR or T2-weighted FSE, sagittal T1-weighted FSE and coronal T2-weighted FSE imaging sequences. Post-processing of the raw spectral data included baseline correction, frequency inversion and phase shift. Gaussian curves were fitted to NAA, Cho, Cr, Lipid and Lactate peaks for peak area determination.
For DSC-MRI, the data were processed on the GE workstation using the Functool software. T2*-weighted images were firstly corrected for motion artifacts with BrainStat software. The CBV map (approximated by using the negative enhancement integral) was then overlaid on T2-weighted or T1 post-contrast images. In the intratumoral region, five to ten ROIs, ranging in size from 25 to 62 mm\(^{2}\) each, were placed in the areas presenting increased perfusion, as seen on the CBV colored overlay maps (Fig. 2), and the maximum of all values was recorded. This is the so-called “hot-spot” analysis. However, it has to be mentioned that this analysis might be susceptible to user-dependent errors and may lead to a slight under- or overestimation of the rCBV measurements. Emblem et al. [27] suggested “Histogram analysis” as an alternative method for perfusion quantification since it reduces the user-dependency and allows the reproducibility of the results. The ROI placement within the tumor was carefully performed. For this purpose, combined information from post-contrast T1-weighted images, T2-weighted FSE images, and T2*-weighted images was used. The peritumoral region was defined to be within 1 cm outside the enhancing tumor margin presenting the highest CBV value, where three to six ROIs were placed along the peritumoral area to measure CBV. The CBV value of the contralateral normal side was measured as well. Finally, the rCBV ratio was calculated by dividing the CBV value either from the intratumoral or peritumoral area defined above, with the CBV value from the contralateral normal side.
ROI placement was performed without knowledge of the histological information. The CBV measurements were performed by two separate readers.
It should be noted that in the intratumoral measurements, obvious necrosis, cysts, hemorrhage, edema, calcification and normal appearing brain tissue were excluded from the ROI whenever possible, in order to avoid false lesion estimation. Regarding the peritumoral area, for lesions that were located close to major vascular structures, the signal intensity-time curve was carefully inspected, as it may clearly indicate vessels that produce very large signal intensity changes [28]. Furthermore, we applied a gamma-variate function to the first pass bolus curve in order to correct for contrast agent extravasation. However, it has to be mentioned that this method generates lower CBV map SNR than numerical integration [29, 30].
Statistical analysis
We grouped the patients according to tumor type (glioblastoma/metastasis). Statistical analysis was performed using the SPSS (v17) statistical software package. Parameter values were expressed as mean \(\pm \) SD. The Mann–Whitney test was employed to compare metabolic and perfusion values between glioblastomas and metastases, and logistic regression analysis was applied in order to investigate the multiparametric relations. Nevertheless, these relationships may often be nonlinear and complex; hence, classical statistical analysis may not be sufficient to reveal these limitations.
Classification methods
In this present study, the classification procedure was based on three classification algorithms: support vector machine (SVM), Naïve Bayes and KNN.
The SVMs first map the attribute vectors into a feature space either linearly or nonlinearly, according to the selected kernel function. Then, within this feature space, an optimal hyperplane is constructed, which separates all the data points of two classes. The best hyperplane for an SVM is the one with the largest margin between classes [31, 32]. The Naïve Bayesian Classifier is a probabilistic classifier which assumes that features are independent.
If the observed feature values of an instance and the prior probabilities of classes are given, then the probability that an instance belongs to a specific class can be estimated. The class with the highest estimated probability is the class prediction [33, 34].
The KNN algorithm compares the test sample with the available training samples and finds the ones that are more similar (“nearest”) to it. When the \(k\)-nearest training samples are found, the class label in majority is assigned to the new sample [20, 35, 36].
Datasets Specification
By applying supervised classification methods, machine learning classifiers can be used to provide binary outcomes in order to distinguish metastases from GBs. Thus, we sought to identify the optimum combination of parameters for tumor classification which might accent the underlying pathophysiology. We started by evaluating the two basic parameters (NAA/Cr as a marker of neuronal viability, and rCBV as an index of tumor neovascularization) which have been validated as significant indices in GB versus Metastasis differentiation according to the literature [1, 10–12]. Then, we continued by successively including the additional parameters (Cho/Cr as a marker of tumor aggressiveness and (Lip \(+\) Lac)/Cr as a marker of necrosis) in order to investigate the potential improvement of the classification results.
Therefore, three datasets were created and evaluated. The first dataset (DS1) consisted of the NAA/ Cr ratio and rCBV. The second dataset (DS2) consisted of DS1 \(+\) Cho/Cr ratio, and the third dataset (DS3) consisted of DS2 \(+\) (Lip \(+\) Lac)/Cr ratio. The aforementioned datasets were applied for each region of interest in order to train and test each classifier. All the created datasets are summarized in Table 1.
Classification procedure
In our study, the classes of GB and metastasis overlapped so we had to deal with a nonlinear classification task, where the patterns could not linearly separate. Regarding the SVM method in order to solve the binary classification problem, we trained the classifier utilizing the most commonly used kernel functions (linear, polynomial, radial basis function). The classification results showed that the highest classification performance was achieved when the RBF kernel function was applied. Critical parameters were the parameter \(\sigma \) of the kernel function and the regularization parameter \(C\), which determines the trade-off between minimizing the training errors, as well as the model complexity [37]. The optimization was accomplished by a grid search execution in order to identify a good pair of (\(C\), \(\sigma \)), so that the classifier can accurately predict unknown data. Various pairs of (\(C\), \(\sigma \)) values were tested and the one with the best cross-validation accuracy was chosen. We tested growing sequences of \(C\) and \(\sigma \) (\(C\) \(=\) 0.01, 0.05, 0.1, 0.5, 1, 2, 10, 11, 100, 1,000, \(\sigma \) \(=\) 1000, 100, 11, 10, 2, 1, 0.5, 0.1, 0.05, 0.01).
In \(k\)-nearest algorithm, one of the most important key issues which affect the performance of a classifier is the choice of \(k\). According to Qi [38], the values of the \(k\) parameter should normally be odd numbers and less than the square root of the number of samples in the data set. Li et al. [39] 2011 set \(k\) to the square root of the total number of variables of the input dataset. It must be noted that if \(k\) is too small, then the result can be sensitive to noise points. On the other hand, if \(k\) is too large, then the neighborhood may include too many points from other classes [40]. In our study, the number of elements was 49, so we tried the odd numbers which were less and equal to the square root of 49 (\(k\) \(=\) 1, 3, 5, 7) per cross-validation fold. Finally, the Euclidean distance function was used in the KNN classifier.
Eighteen binary classifiers were created in total, six (3 intratumoral, 3 peritumoral) for each of the three classification methods applied. During the classification procedure, we performed the 10-fold cross-validation method in order to evaluate each classifier. This procedure was repeated 200 times, each time splitting randomly every dataset, in order to avoid bias possibly introduced by the selection of a specific training and test set. The performance of each classifier was measured in terms of: test accuracy (percentage of correctly classified cases), sensitivity (proportion of actual positives which are correctly identified), specificity (proportion of negatives which are correctly identified), and F1 (provides a more balanced evaluation of a classifier’s performance by averaging precision and recall:
Precision is the number of correct results divided by the number of all returned results and Recall is the number of correct results divided by the number of results that should have been returned.
Furthermore, in order to test the performance of the classifiers even further, we calculated the Geometric Mean of Recalls:
as well as Balance Error Rate (BER) and Error Rate (ERR):
where \(n_{A}\) is the number of cases of class A, \(e_{A}\) the number of misclassified cases, \(n_{B}\) the number of cases of class B and \(e_{B}\) is the number of misclassified cases. BER is useful when one class is underrepresented compared to the other class as observed in our study [23].
This large number of performance metrics was calculated in order to have the most accurate overall evaluation as possible. Lastly confusion matrices were computed for over 200 runs of stratified random sampling (Table 3).
Finally, in order to validate the performance of the trained classifiers, we used an independent test set from our center, processed in the same conditions as described in the imaging protocols previously. The test set consisted of 20 patients with a histopathological diagnosis (14 glioblastoma and 6 lung metastases).
Results
Statistical analysis
The initial preoperative radiologists’ diagnosis of these tumors resulted in 73.4 % accuracy (36 out of 49 cases correctly diagnosed) for the first radiologist (R1), and 79.6 % accuracy (39 out of 49 cases correctly diagnosed) for the second radiologist (R2). It has to be stressed here that we excluded the cases where the radiologists included both lesion types in their differential diagnosis. Hence, the sensitivity and specificity for the two blind evaluators were respectively R1: 0.78 and 0.71 and R2: 0.71 and 0.82.
Regarding statistical analysis, the results of the Mann–Whitney test for the metabolic and perfusion parameter values (mean \(\pm \) SD) and the results of the logistic regression analysis for each dataset for both regions of interest are summarized in Table 2.
The differences in the metabolic ratios between glioblastomas and metastases did not reach statistical significance in the intratumoral region, revealing the lack of classical statistical tests to differentiate between these lesions using spectroscopic data. Nevertheless, a trend of intratumoral Cho/Cr and (Lip \(+\) Lac)/Cr ratios toward higher values for metastases was observed, when compared to that of glioblastomas. However, due to the wide corresponding standard deviations, these tendencies were not statistically confirmed. Similar results were observed in the comparison of mean rCBV ratios between the two tumor groups, as their difference did not reach statistical significance.
Comparing the metabolic and perfusion parameters in the peritumoral area between the two tumor groups, rCBV, NAA/Cr, and long TE Cho/Cr ratios were significantly different (\(p<0.05\)), reflecting the difference in the pathophysiological properties in the periphery of the two lesions (Table 2). The findings of Logistic Regression analysis showed that the two tumor groups could not be differentiated by any of the three datasets in the intratumoral region. On the contrary, regarding the peritumoral region, all three datasets significantly differentiated glioblastomas from metastases, and it was observed that the additional inclusion of parameters in the datasets does not always lead to an improved statistical outcome.
Classification
The performance of SVM, Naïve Bayes and KNN classifiers for both regions of interest is shown in Table 1. For the intratumoral area, the classifier that reached the highest overall performance was the SVM for DS3 (Acc. 0.97), although it also showed very high performance for the other two datasets (Acc. 0.95 and 0.92 respectively). Naïve Bayes and KNN classifiers presented lower performances compared to SVM; however, their highest performance was also observed for DS3.
In the peritumoral region, SVM presented again the highest discrimination ability (Acc. 0.98) between the two tumor groups, but this time using the first data set (DS1), followed by Naïve Bayes also using DS1. The KNN classifier presented the lowest differentiation ability for all feature combinations.
The confusion matrices per classifier are presented in Table 3, where the superiority of SVM is verified. The Metastasis class was considered as the true-po sitive class and the GB class as the true negative. In the intratumoral area, SVM presented the highest performance using DS3 where all GB cases were correctly classified, whereas only one metastatic case was misclassified for the same dataset. Similarly, in the peritumoral area, the highest performance in GB and metastasis classification was achieved by SVM using DS1, with all clinical cases being correctly classified. Naïve-Bayes proved to be second best in both regions of interest, whereas KNN presented the highest percentage of misclassified clinical cases for all datasets.
Regarding the validation procedure, the prediction results of the independent test set were in accordance with the results derived from the evaluation procedure of the classifiers’ performance.
Discussion
In the present study, we sought to investigate the contribution of pattern recognition techniques in the differentiation of a common differential diagnostic problem in the clinical routine, that of glioblastomas versus intracranial metastases. We evaluated the performance of different classifiers in the discrimination of the two types of tumors using combinations of metabolic and perfusion parameters. Moreover, we sought to identify the optimum set of features in terms of potential diagnostic value.
Intratumoral area
All classifiers differentiated significantly GBs from metastases (Table 1), in contrast to statistical analysis which despite the observed tendency of Cho/Cr and (Lip \(+\) Lac)/Cr for higher values in metastatic lesions, could not reach significant values, most probably due to the large standard deviations observed (Table 2). Regarding the overall classification performance, SVM reached the highest value among all classifiers for all three datasets as this is shown in Table 1. Naïve-Bayes and KNN presented greater variations in their performance depending on the dataset used, hence proved to be more sensitive to feature selection. Moreover, the analysis of the different datasets revealed the significant role that the underlying pathophysiology may play in the classification outcome, since all classifiers presented the highest performance when the features of DS3 were used. This might be attributed to the fact that the destruction of neurons as well as the increased vasculature (features of DS1) are common characteristics of the two tumor entities, while the inclusion of additional tumor features such as aggressiveness (as represented by Cho in DS2) and necrosis (as represented by Lip \(+\) Lac in DS3) optimize further the classification outcome. Although glioblastomas and metastases share common lipid and Choline profiles intratumorally, it must be noted that their lipid signal arises from different origins, such as pure tumor necrosis for infiltrative tumor and less necrosis combined with lipid membrane structure for migratory tumor cells [41].
Peritumoral area
For the peritumoral area, all classifiers differentiated significantly GBs from metastases (Table 1). In this case, statistical analysis also revealed significant differences between the two tumor entities (Table 2). This fact obviously posits the hypothesis that there should be a distinct differentiation between infiltrative and non-infiltrative nature outside the tumor. Indeed, the NAA/Cr and the rCBV ratios proved significantly different (\(p=0.01\) and \( < \)0.01 respectively) verifying the destruction of neurons and increased angiogenesis in the peritumoral area of an infiltrating lesion such as a GB, while around a metastasis, one should expect a normal metabolic profile.
Evaluating the overall classification performance, again SVM reached the highest value among all classifiers for all three datasets as this is shown in Table 1, followed by Naïve-Bayes, which presented equally strong discrimination ability. KNN presented an overall low performance for all datasets. A very interesting finding of this study is that the highest performance was reached using the features of DS1, contrary to the observations of the intratumoral case. One would expect that the inclusion of additional features would obviously lead to a better classification outcome. Nevertheless, this is strongly correlated again to the underlying pathophysiology. Indeed, we established that the successive inclusion of the Cho/Cr and (Lip \( + \) Lac)/Cr ratio as additional features degraded the performance of all classifiers in the peritumoral area. As it has been previously reported, peritumoral lipids and lactate do not aid in tumor characterization, due to the absence of necrosis in this area [10]. Moreover, the presence of Cho signal although helpful has not been reported to be an exclusive characteristic regarding infiltration. Nevertheless, it has been reported that the Cho signal may significantly contribute in the differentiation of these lesions [1, 3]. This means that infiltration of GBs may be present in the peritumoral region although not detectable yet in terms of increased Cho signal [12]. More importantly, the previous finding emphasizes the ability of the classifiers to identify nonlinear intra-variable relationships, which accent the underlying pathophysiology irrespective of the number of features used.
Classifier performance
Evaluating the overall classification performance which is depicted in Table 1, it is clear that the SVM classifier has the best discrimination ability for both regions of interest, followed by Naive Bayes especially in the case of the peritumoral region. KNN presented the lowest performance in both tumor regions.
SVMs superiority may be attributed to the concept of margin. Under this concept, SVM realize the principle of data-dependent structure risk minimization, using the relation between the target function and the data set, without depending on the of data set’s dimension. At the same time, SVM minimize the structure risk of both complexity and minimizing loss and can tolerate the noise and the fuzzy value in the data set [19]. Furthermore, SVM superiority may be due to the fact that they belong to the general category of kernel methods. This has the advantage of generating nonlinear decision boundaries, despite the small data size, using methods designed for linear classifiers and allow the user to apply a classifier to data that have no obvious fixed-dimensional vector space representation [37, 42].
Naïve Bayes owes its good performance to the zero-one loss function used in classification. This function defines the error as the number of incorrect predictions. Unlike other loss functions, such as the squared error, it has the key property that it does not penalize inaccurate probability estimates-as long as the greatest probability is assigned to the correct class [43–45]. Furthermore, Naive Bayes computes adistinct kernel estimation for each feature of every class, creating multiple (Gaussian) distributions which is generally more effective than using a single distribution [46]. A potential limitation of Naïve Bayes classifier is the assumption of independence between attributes, which is difficult to accomplish in datasets that consist of medical data. Although in practice this assumption is not fully satisfied, studies have shown that Naïve Bayes is effective in medical applications [34].
On the contrary, the KNN technique is a conceptually and computationally quite simple method, as it calculates distances between nearest neighbors in the feature space. Therefore, due to the methods simplicity on very difficult classification tasks, such as the differentiation of ambiguous brain lesions (GB and Metastasis), KNN may be outperformed by more complex techniques, such as SVM and Naïve-Bayes algorithms, as observed in this study. Additionally, KNN is sensitive to irrelevant or redundant features degrading the overall classification performance [47].
Our initial results in the case of this particular differential diagnostic problem of GB versus Metastasis are encouraging and indicate that pattern recognition techniques may provide incremental diagnostic value over the radiologists’ analysis, as well as over simple statistical analysis of the imaging data.
On the other hand, the analysis of these large amounts of data with extremely significant diagnostic value may be a time-consuming process; it requires specific expertise and may not be feasible during the clinical routine especially because these data are mainly numeric. Hence, classification algorithms such as SVM can be applied in the clinical environment to the benefit of patient treatment.
Lastly, it has to be mentioned that in our classification procedure, we used quantitative features extracted from 1H-MRS and DSC-MRI. Nonetheless, the combination of these features with additional features such as texture analysis and genotypic information of the tumor may improve the overall accuracy of the classification procedure which can then be extended to other differential diagnostic problems.
Conclusion
The simultaneous analysis and evaluation of multiple numerical parameters provided by advanced MR imaging techniques, such as 1H-MR spectroscopy and dynamic susceptibility contrast enhanced MR Imaging, may be challenging in a clinical environment. In the present study, we investigated the contribution of pattern recognition techniques in the differentiation of glioblastomas and metastatic tumors, when metabolic and perfusion data are used as classification features. Our results indicate that these techniques may provide incremental diagnostic value in the differentiation of these common intraaxial brain tumors. The SVM algorithm demonstrated the highest classification performance for both intra and peritumoral regions.
Hence, the complex and nonlinear relationships between many MR variables can be simplified by multivariate classification methods, and differences related to intra-variable correlations may be further accented between tumor types. Consequently, pattern recognition techniques may constitute an important supplementary tool and substantially aid in the differential diagnosis.
References
Chiang IC et al (2004) Distinction between high-grade gliomas and solitary metastases using peritumoral 3-T magnetic resonance spectroscopy, diffusion, and perfusion imaging. Neuroradiology 46(8):619–627
Liu X et al (2011) MR diffusion tensor and perfusion-weighted imaging in preoperative grading of supratentorial nonenhancing gliomas. Neurol Oncol 13(4):447–455
Cha S (2009) Neuroimaging in neuro-oncology. Neurotherapeutics 6(3):465–477
Toh CH et al (2008) Primary cerebral lymphoma and glioblastoma multiforme: differences in diffusion characteristics evaluated with diffusion tensor imaging. Am J Neuroradiol 29(3):471–475
Law M et al (2002) High-grade gliomas and solitary metastases: differentiation by using perfusion and proton spectroscopic MR imaging. Radiology 222(3):715–721
Howe FA et al (2003) Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med 49(2):223–232
Fan G, Sun B, Wu Z, Guo Q, Guo Y (2004) In vivo single-voxel proton MR spectroscopy in the differentiation of high-grade gliomas and solitary metastases. Clin Radiol 59(1):77–85
Al-Okaili RN et al (2007) Intraaxial brainmasses MR imaging- based diagnostic strategy-initial experience. Radiology 243(2): 539–550
Weber MA et al (2006) Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology 66(12):1899.S–1906.S
Chawla et al (2010) Proton magnetic resonance spectroscopy in differentiating glioblastomas from primary cerebral lymphomas and brain metastases. J Comput Assist Tomogr 34(6):836–841
Lee EJ et al (2011) Diagnostic value of peritumoral minimum apparent diffusion coefficient for differentiation of glioblastoma multiforme from solitary metastatic lesions. Am J Roentgenol 196(1):71–76
Tsougos I et al (2012) Differentiation of glioblastoma multiforme from metastatic brain tumor using proton magnetic resonance spectroscopy, diffusion and perfusion metrics at 3 T. Cancer Imaging 12:1–14. doi:10.1102/1470-7330.2012.0038
García-Gómez JM (2011) Brain tumor classification using magnetic resonance spectroscopy. In: Tumors of the central nervous. System, vol 3. Springer, pp 5–19
INTERPRET Consortium, “INTERPRET”. Web site, 1999–2001. IST-1999-10310, EC. http://gabrmn.uab.es/interpret/
Tate AR et al (2006) Development of a decision support system for diagnosis and grading of brain tumours using in vivo magnetic resonance single voxel spectra. Nucl Magn Reson Biomed 19(4):411–434
eTUMOUR Consortium, “eTumour: Web accessible MR Decision support system for brain tumour diagnosis and prognosis, incorporating in vivo and ex vivo genomic andmetabolomic data”.Web site. FP6-2002-LIFESCIHEALTH 503094, VI framework programme, EC. http://cordis.europa.eu/search/index.cfm?fuseaction=proj.document&PJ_RCN=7921577. Accessed 6 Oct 2012
Gonzalez Velez H et al (2009) HealthAgents: distributed multi-agent brain tumor diagnosis and prognosis. Appl Intell 30(3): 191–202
Arús C et al (2006) On the design of a web-based decision support system for brain tumour diagnosis using distributed agents. In: IAT Workshops, pp 208–211
Li G, Yang J, Ye C, Geng D (2006) Degree prediction of malignancy in brain glioma using support vector machines. Comput Biol Med 36(3):313–325
Zacharaki EI, Kanas VG, Davatzikos C (2011) Investigating machine learning techniques for MRI-based classification of brain neoplasms. Int J Comput Assist Radiol Surg 6(6):821–828
Zacharaki EI et al (2009) Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 62(6):1609–1618
Devos A et al (2005) The use of multivariate MR imaging intensities versus metabolic data from MR spectroscopic imaging for brain tumour classification. J Magn Reson 173(2):218–228
Garcia-Gomez JM et al (2009) Multiproject-multicenter evaluation of automatic brain tumor classification by magnetic resonance spectroscopy. MAGMA 22(1):5–18
Blanchet L et al (2011) Discrimination between metastasis and glioblastoma multiforme based on morphometric analysis of MR images. Am J Neuroradiol 32(1):67–73
Dimou I et al (2011) Brain lesion classification using 3T MRS spectra and paired SVM kernels. Biomed Signal Process Control 6(3):314–320
Kousi E et al (2012) Spectroscopic evaluation of Glioma grading at 3T: the combined role of short and long TE. Sci World J 2012:546171
Emblem KE et al (2008) Glioma grading by cerebral blood volume maps. Radiology 247(3):808–817
Zhang H (2008) Perfusion MR imaging for differentiation of benign and malignant meningiomas. Neuroradiology 50(6):525–530
Boxerman JL, Schmainda KM, Weisskoff RM (2006) Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with Glioma tumor grade, whereas uncorrected maps do not. Am J Neuroradiol 27:859–867
Knopp EA, Cha S, Johnson G, Mazumdar A, Golfinos JG, Zagzag D et al (1999) Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology 211(3):791–798
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Chang CC, Lin CJ (2001) LIBSVM: a library for support vector machines. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm
John GH, Langley P (1995) Estimating continuous distributions in Bayesian classifiers. In: (UAI’95) Philippe B, Steve H (eds) Proceedings of the eleventh conference on uncertainty in artificial intelligence. Morgan Kaufmann Publishers Inc., San Francisco, pp 338–345
Kazmierska J, Malicki J (2008) Application of the Naïve Bayesian Classifier to optimize treatment decisions. Radiother Oncol 86(2):211–216
Cover TM, Hart PE (1967) Nearest neighbor pattern classification. Inst Electr Electron Eng Trans Inf Theory 13:21–27
Wang J, Neskovic P, Cooper LN (2007) Improving nearest neighbor rule with a simple adaptive distance measure. Pattern Recognit Lett 28(2):207–213
Lukas L et al (2004) Brain tumor classification based on long echo proton MRS signal. Artif Intell Med 31(1):73–89
Qi H (2002) Feature selection and kNN fusion in molecular classification of multiple tumor types. In: Proceedings of the mathematics and engineering techniques in medicine and biological sciences. Las Vegas, Nevada
Li S, Harner EJ, Adjeroh DA (2011) Random KNN feature selection-a fast and stable alternative to random forest. BMC Bioinform 12:450
Wu X et al (2008) Top 10 algorithms in data mining. Knowl Inf Syst 14(1):1–37
Opstad KS et al (2004) Differentiation of metastases from high-grade gliomas using short echo time 1H spectroscopy. J Magn Reson Imaging 20(2):187–192
Ben-Hur A, Weston J (2010) A user’s guide to support vector machines. In: Methods in molecular biology. Data Mining Techniques for the Life Sciences, vol 609. Springer, Berlin, pp 223–239
Domingos P, Pazzani M (1997) On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn 29(2–3): 103–130
Friedman JH, Fayyad U (1997) On bias, variance, 0/1-loss, and the curse-of-dimensionality. Data Min Knowl Discov 1(1):55–77
Frank E, Trigg L, Holmes G, Witten IH (2000) Technical note: Naive Bayes for regression. Mach Learn 41(1):5–25
Williams N, Zander S, Armitage G (2006) A preliminary performance comparison of five machine learning algorithms for practical IP traffic flow classification. ACM SIGCOMM Comput Commun Rev 36(5)
Cunningham P, Delany SJ (2007) k-Nearest Neighbour Classifiers. Technical Report UCD-CSI-2007-4
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Evangelia Tsolaki and Patricia Svolos contributed equally to this work.
Rights and permissions
About this article
Cite this article
Tsolaki, E., Svolos, P., Kousi, E. et al. Automated differentiation of glioblastomas from intracranial metastases using 3T MR spectroscopic and perfusion data. Int J CARS 8, 751–761 (2013). https://doi.org/10.1007/s11548-012-0808-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-012-0808-0