Abstract
In this paper, a novel hierarchical multi-class SVM (H-MSVM) with extreme learning machine (ELM) as kernel is proposed to classify electroencephalogram (EEG) signals for epileptic seizure detection. A clinical EEG benchmark dataset having five classes, obtained from Department of Epileptology, Medical Center, University of Bonn, Germany, is considered in this work for validating the clinical utilities. Wavelet transform-based features such as statistical values, largest Lyapunov exponent, and approximate entropy are extracted and considered as input to the classifier. In general, SVM provides better classification accuracy, but takes more time for classification and also there is scope for a new multi-classification scheme. In order to mitigate the problem of SVM, a novel multi-classification scheme based on hierarchical approach, with ELM kernel, is proposed. Experiments have been conducted using holdout and cross-validation methods on the entire dataset. Metrics namely classification accuracy, sensitivity, specificity, and execution time are computed to analyze the performance of the proposed work. The results show that the proposed H-MSVM with ELM kernel is efficient in terms of better classification accuracy at a lesser execution time when compared to ANN, various multi-class SVMs, and other research works which use the same clinical dataset.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
EEG is a noninvasive testing method, which contains very useful information related to different physiological states of the brain, and thus, it is an effective tool for understanding the complex dynamical behavior of the brain. Since EEG is noninvasive, it can be recorded over a long time span which is very important for monitoring neurological disorders like epileptic seizures which are ephemeral. Epilepsy is a disorder of the normal brain function, by which approximately 1 % of the world’s population suffers. These EEG recordings are visually inspected by highly trained professionals for detecting epileptic seizures. This information is then used for clinical diagnosis and possible treatment plans. The process is time-consuming and expensive [15].
Research on seizure detection began in the 1970 s and various methods addressing this problem have been presented. Liu et al. proposed the time-domain method which searches for periodic, rhythmic patterns in EEG similar to the ones occurred during seizure activity. The authors analyzed the autocorrelation of EEG to provide a measure for rhythmicity [30]. Event-related EEG changes over the primary motor cortex are then analyzed off-line from the EEG recordings [37]. In the frequency domain, seizure detection relies on the differences in the frequency-domain characteristics of the normal and epileptic EEG [10]. Since the EEG is nonstationary in general, it is most appropriate to use time–frequency-domain methods like wavelet transforms (WT) [2, 26, 54] which do not impose the quasi-stationarity assumption on the data like the time- and frequency-domain methods. WT provides both time and frequency views of a signal simultaneously which makes it possible to accurately capture and locate transient features in the data like the epileptic spikes. He et al. [16] proposed a method for removing ocular artifacts based on adaptive filtering. Nonlinear measures like correlation dimension (CD), largest Lyapunov exponent (LLE), and approximate entropy (ApEn) quantify the degree of complexity in a time series. These measures help understand EEG dynamics and underlying chaos in the brain signals [25]. ApEn is a statistical parameter, widely used in the analysis of physiological signals, such as estimation of regularity in epileptic seizure time series data [39, 40]. Diambra et al. [6] have shown that the value of the ApEn drops abruptly due to the synchronous discharge of large groups of neurons during an epileptic activity. Thus, it is a suitable feature to characterize the EEG signals. Andrzejak et al. [3, 4] used CD to characterize the interictal EEG for seizure prediction and found that the CD values calculated from interictal EEG recordings are significantly lower for the epileptogenic zone as compared to other areas of the brain.
Artificial neural networks (ANN) have been widely applied to classify EEG signals over the last two decades [8, 27, 41]. A variety of different ANN-based approaches were reported in the literature for epileptic seizure detection [28, 38, 44]. Kalayci et al. [23] used wavelet transform to capture characteristic features of the EEG signals and then combined with ANN to get satisfying classification result. Auto regressive coefficients are extracted as feature vectors from EEG segment, and then a neural network classifier is used to classify each EEG segment into different sleep stages. The BioSleep package produces reasonable results with the comparison of human scoring in the third-part evaluation [22]. Nigam et al. [35] described a method for automated detection of epileptic seizures from EEG signals using a multistage nonlinear preprocessing filter for extracting two features: relative spike amplitude and spike occurrence frequency. These features were fed to a diagnostic artificial neural network. Mohseni et al. [33] applied short-time Fourier transform analysis of EEG signals and extracted features based on the pseudo-Wigner–Ville and the smoothed pseudo-Wigner–Ville distribution and used these features as inputs to an ANN for classification. Jahankhani et al. [21] decomposed EEG signal with WT into different sub-bands and extracted statistical information from the wavelet coefficients. He utilized radial basis function network (RBF) and multi-layer perceptron network (MLP) as classifiers. Erfanian et al. [7] presented an adaptive noise canceller (ANC) filter using an artificial neural network for real-time removal of electro-oculogram interference from electroencephalogram (EEG) signals. Subasi [43, 46] decomposed the EEG signal into time–frequency representations using DWT. Some features such as the mean of the absolute value, average power, standard deviation, variance, and ratio of the absolute mean value derived from the wavelet coefficients are calculated and applied to different classifiers, such as feed-forward error back-propagation artificial neural network (FEBANN), dynamic wavelet network (DWN), dynamic fuzzy neural network (DFNN), and mixture of expert system (ME), for epileptic EEG classification. Success of variance in seizure detection is well established [31]. In the work of Srinivasan et al. [42], features from the time domain and frequency domain are employed individually or jointly for classifying EEG signals.
The high classification results showed that the Elman recurrent neural network combination feature exhibited excellent discrimination performance. In [12], Lyapunov exponents were extracted from EEG signals with Jacobi matrices and then applied as inputs to recurrent neural networks (RNNs) to obtain good classification results. Ubeyli [49, 50] classified the EEG signals by combining Lyapunov exponents and fuzzy similarity index. Several entropy measures were investigated for discriminating EEG signals [24]. Connectivity techniques can be used to show real-time changes in the brain state in response to stimuli [19]. This can allow researchers’ insight into the effects of gaming on brain in real time. The classification ability of the entropy measures was tested through an adaptive neuro-fuzzy inference system [45]. Guo et al. [13] decompose original EEG signal first into several sub-bands through four-level multi-wavelet transform with repeated row preprocessing for each sub-band signal, and then calculated ApEn feature to classify the EEG signals using three-layer multi-layer perceptron neural network with Bayesian regularization back-propagation training algorithms. Neural network is an information processing system, and it has been the choice of many researchers for the classification due to its special characteristics such as self-learning, adaptability, robustness, and massive parallelism. In ANNs, knowledge about the problem is distributed through the connection weights of links between neurons. The neural network has to be trained to adjust the connection weights and biases in order to produce the desired mapping. ANNs are widely used in the biomedical area for modeling, data analysis, regression, and classification.
Nicolaou et al. [34] proposed approximate entropy drops which occurred during seizure intervals and employed this as a feature for automatic seizure detections using SVM. Ubeyli [51] carried out a study for classification of EEG signals by combining the model-based methods and least-square support vector machine (SVM). Iscan et al. [20] proposed to combine the time- and frequency-feature approach for the classification of healthy and epileptic EEG signals using different classifiers including SVM. Acharya et al. [1] extracted four entropy-based nonlinear features from EEG data and trained seven classifiers. Hsu et al. [17] developed a method using the SVM classifier with nonlinear features for automatic seizure detection in EEG signals. Varun Joshi et al. [52] presented a new method for electroencephalogram (EEG) signal classification based on fractional-order calculus. Generally to train a SVM classifier, the user must determine a suitable kernel function, optimum hyper parameters, and proper regularization parameter. This goal is usually accomplished by cross-validation techniques. The cross-validation technique can be used to select parameters. The performance of SVM largely depends on the kernels. But selecting the appropriate kernel functions, which are well suited to the specific problem such as seizure detection, is very difficult. Speed and size is another problem of SVM both in training and testing. In terms of running time, SVMs are slower than other machine learning techniques, but provides better performance with respect to classification accuracy. Basically, SVM is binary classifier, and there are variations of SVM for multi-classification such as one versus one and one versus rest, and DAG MSVMs are available, but they require N(N − 1) SVMs for an N class problem which takes much computation time. So, the work for multi-class SVM classifiers and also to customize the kernel function for seizure detection is a scope for further research. So, the present work contributes the following.
-
i.
A new kernel called ELM kernel for SVM
-
ii.
A new multi-classification scheme called hierarchical MSVM
The proposed scheme is tested using the complete five classes of benchmark clinical EEG dataset recorded from five healthy subjects and five epileptic patients during both ictal and interictal periods. Since the dataset is hierarchical in nature, the proposed hierarchical approach is much suitable. It is shown that the new scheme is able to detect epileptic seizures with very high classification accuracy at a lesser execution time. The paper is organized as follows. Section 2 describes the benchmark dataset and proposed methods such as wavelet transform-based feature extraction and a novel hierarchical multi-class SVM classifier with ELM kernel in this work. Section 3 presents the various experiments carried out and results. In Sect. 4, the evaluation procedure and the experimental results are discussed. Concluding remarks on the effectiveness of the present study and hints for the future researcher are furnished in Sect. 5.
2 Methods
2.1 Dataset description
The benchmark EEG data [3] used in this work are obtained from University of Bonn, Germany. The data are available in public domain that consists of five different sets {A, B, C, D, E}. Each dataset consists of 100 single-channel EEG epochs of 23.6-s duration. The data were recorded with 128-channel amplifier system and digitized at 173.61 Hz sampling rate and 12-bit A/D resolution. The description of the dataset is summarized in Table 1. The experimental setup followed in this paper on this benchmark dataset is also adopted by number of researchers [3, 5, 13, 14, 29, 36, 47, 48, 51].
The dataset is hierarchical in nature. The dataset can be classified as normal and seizure in first level. Then from the normal subset they can be further classified as normal-eye-opened and normal-eye-closed in the second level. As well as in the same level, the seizure subset can be classified as during-seizure and seizure-free. And in the last level, the seizure-free subset can be further classified as hippocampal and epileptogenic. So the hierarchical multi-class SVM approach is very much suitable for this particular benchmark dataset.
2.2 Proposed methodologies
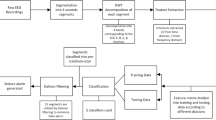
The EEG signal classification for epileptic seizure detection consists of main modules such as a feature extractor that generates wavelet-based features from the EEG signals and a feature classifier (H-MSVM) that outputs the class. The block diagram of the proposed approach is illustrated in Fig. 1.
2.3 Wavelet transform-based feature extraction
Transforming the input data into a set of features which reduces dimensionality is called feature extraction. WT has several advantages, which can simultaneously possess compact support, orthogonality, symmetry, short support, and higher-order approximation. WT is widely applied in biomedical engineering areas for solving a variety of real-life problems. WT provides a more flexible way of time–frequency representation of a signal by allowing the use of variable-sized windows. In WT, long time windows are used to get a fine low-frequency resolution, and short time windows are used to get high-frequency information. Thus, WT gives precise frequency information at low frequencies and precise time information at high frequencies. This makes the WT suitable for the analysis of irregular data patterns, such as impulses occurring at various time instances. So, WT is an effective tool for classification and analysis of nonstationary signal, such as EEG signals. Wavelet decomposition of a source EEG signal has been done up to fifth level using Daubechies wavelet of order 2. As DB2 has asymmetric properties, orthogonality and its smoothing feature made it more suitable to analyze and detect changes of nonstationary signal such as EEG [11]. A rectangular window, which was formed by 256 discrete data, has been selected so that the EEG signal is considered to be stationary in that interval. Wavelet transformation employs two sets of functions called scaling functions and wavelet functions, which are related to low-pass and high-pass filters, respectively. The decomposition of the source EEG signal into the different frequency bands is obtained by consecutive high-pass and low-pass filtering of the time-domain signal. The procedure of multi-resolution decomposition of a signal x[n] is schematically shown in Fig. 2. The multi-resolution analysis, using five levels of decomposition, yields six separate EEG sub-bands. Table 2 summarizes wavelet sub-bands, frequency ranges, and features of the proposed work.
In the present work, the dimensionality reduction is carried out from wavelet coefficients of the source EEG data which is discussed as follows. After wavelet decomposition, the source EEG signal is transformed into 4108 wavelet coefficients and decomposed into six sub-bands such as D1, D2, D3, D4, D5 and A5, and considering all the features for classification increases the computation time [11]. In order to further decrease the dimensionality of the extracted features and reducing computation time, six features have been extracted from each sub-band, and so totally 36 features are used to characterize the EEG signals for classification. The following wavelet-based nonlinear features such as (i) approximate entropy, (ii) largest Lyapunov exponent, and linear features such as (iii) minimum, (iv) maximum, (v) mean, and (vi) standard deviation have been extracted from each sub-band. The statistical features have the advantages of familiarity and efficiency and also have advantages when making inferences. In this work, in addition to linear statistical features such as minimum, maximum, mean and standard deviation, nonlinear features such as ApEn and LLE are used to characterize the signal variance. The nonlinear features have a great advantage in reflecting the chaotic behavior and serve as useful features in classifying the EEG signals [11]. Table 2 summarizes wavelet sub-bands, frequency ranges, and features of the proposed work.
2.3.1 Approximtate entropy (ApEn)
The approximate entropy measures the predictability of the current amplitude values of a physiological signal based on the previous amplitude values. This measure can quantify the complexity or irregularity of the system.
-
i.
Let the values containing N wavelet coefficients in each sub-band be X = [x(1), x(2), x(3), …, x(N)].
-
ii.
Let x(i) be a subsequence of X such that x(i) = [x(i), x(i + 1), x(i + 2), …, x(i + m − 1)] for 1 ≤ i ≤ N − m, where m represents the number of samples used for the prediction.
-
iii.
Let r represent the noise filter level that is defined as
$$r = k \times {\text{SD}}\quad \;{\text{for}} \; k = 0,0.1,0.2,0.3, \ldots ,0.9$$(1)where SD is the standard deviation of the data sequence X.
-
iv.
Let {x(j)} represent a set of subsequences obtained from x(j) by varying j from 1 to N. Each sequence x(j) in the set of {x(j)} is compared with x(i), and in this process, two parameters, namely Cim(r) and Cim + 1(r), are defined as follows:
$$C_{i}^{m} \left( r \right) = \frac{{\mathop \sum \nolimits_{j = 1}^{N - m} k_{j} }}{N - m}$$(2)where
$$k = \left\{ {\begin{array}{*{20}l} {1,} \hfill & {{\text{if}}\; |x\left( i \right) - x\left( j \right)\quad {\text{for}}\quad 1 \le j \le N - m} \hfill \\ {0,} \hfill & {\text{otherwise}} \hfill \\ \end{array} } \right.$$and
$$C_{i}^{m + 1} \left( r \right) = \frac{{\mathop \sum \nolimits_{j = 1}^{N - m} k_{j} }}{N - m}$$(3) -
v.
ApEn is calculated using Cim(r) and Cim + 1(r) as follows:
$${\text{ApEn}} = \, \frac{1}{N - m}\left[ {\sum\limits_{i = 1}^{N - m} {\ln \left( {\frac{{C_{i}^{m} (r)}}{{C_{i}^{m + 1} (r)}}} \right)} } \right]$$(4)
2.3.2 Largaest Lyapunov exponent (LLE)
Largest Lyapunov exponents are computed from each sub-band. The Lyapunov exponent quantifies the nonlinear chaotic dynamics of the signal and measures how fast nearby trajectories in the dynamic system diverge. The general formula of Lyapunov exponent is given as follows:
where \(\varDelta x_{ij} \left( 0 \right) \, = x\left( {t_{i} } \right) - x\left( {t_{j} } \right)\) is the displacement vector at the time point t i , that is the perturbation of the fiducial orbit observed at t j with respect to t i , while \(\varDelta x_{ij} (\varDelta t) \, = x(t_{i} + \varDelta t) - x(t_{j} - \varDelta t)\) is the same vector after time Δt. The vector x(t i ) is the point in the fiducial trajectory for t = t i and x(t j ) is a properly chosen vector adjacent to x(t i ) in the phase space and N is the number of data points.
2.3.3 Maximum value
Maximum Value = Largest value among the wavelet coefficients in each sub-band.
2.3.4 Minimum value
Minimum Value = Smallest value among the wavelet coefficients in each sub-band.
2.3.5 Mean
Mean of the wavelet coefficients is computed in each sub-band.
where x is the wavelet coefficients in each sub-band, N is the length of the wavelet coefficients in each sub-band, i varies from 1 to N, and j varies from 1 to 6 (sub-bands)
2.3.6 Standard deviation
Standard deviation of the wavelet coefficients is computed in each sub-band.
where x is the wavelet coefficients in each sub-band, N is the length of the wavelet coefficients in each sub-band, i varies from 1 to N, and j varies from 1 to 6 (sub-bands)
2.4 Classification using hierarchical multi-class SVM with ELM kernel
2.4.1 Proposed hierarchical multi-class SVM
In this paper, a new scheme called hierarchical multi-class SVM with an ELM kernel is proposed for the classification of EEG signals. The SVM is a binary classifier, which can be extended by fusing several of its kind into a multi-class classifier. The binary SVM is fused into multi-class SVM by hierarchical approach, since this particular dataset is hierarchical in nature this approach is very much suitable. The dataset is partitioned into two nonoverlapping data subsets at different levels. These two subsets are used as positive and negative samples to train a SVM classifier. Each classifier divides the data into two sets, N − 1 such classifiers are needed to solve an N class classification problem. This scheme is in tree structure, where each node of the tree represents a SVM classifier. The proposed H-SVM structures are composed at different levels; each level consists of a finite number of SVM classifiers. In every node of the tree one, SVM one vs. rest problem is computed. The ways to build the hierarchical multi-class SVM classifier (SVM tree) in the training phase is described, and the means to use the SVM tree to classify new input patterns during the test phase are illustrated. The training for the hierarchical SVM tree classifier starts from the training dataset. Figure 3 illustrates the schematic diagram of the proposed hierarchical multi-class SVM classifier. The first two sets include surface EEG recordings that are collected from five healthy subjects using a standardized electrode placement scheme. The subjects were awake and relaxed with their eyes open and closed, respectively. The data for the last three sets are obtained from five epileptic patients undergoing presurgical evaluations. The third and the fourth datasets consist of intracranial EEG recordings during seizure-free intervals (interictal periods) from within the epileptogenic zone and opposite the epileptogenic zone of the brain, respectively. The data in the last set were recorded during seizure activity (ictal periods) using depth electrodes placed within the epileptogenic zone. The dataset contains five classes. After training, SVM tree classifier contains four-node SVM classifiers. At the top level, the dataset {ABCDE} is divided into to set {AB} and {CDE} by SVM1. At the second level, dataset {AB} is divided into {A} and {B}, respectively, by SVM2; dataset {CDE} is divided into {CD} and {E} by SVM3. Finally, the dataset {CD} is divided into {C} and {D} by SVM4. Both the training and testing phases of the classifier are carried out in a top-down manner.
2.4.2 Extreme learning machine (ELM)
Extreme learning machine [18] is a currently popular neural network architecture based on random projections. It has one hidden layer with random weights, and an output layer whose weights are determined analytically. Both training and prediction are fast when compared with many other nonlinear methods. This work points out that ELM, although introduced as a fast method for training a neural network, is in some sense closer to a kernel method in its operation. A fully trained neural network has learned a mapping such that the weights contain information about the training data.
The following, including algorithm, is an abridged and slightly modified version of ELM introduction in [32]. The ELM algorithm was originally proposed in [18], and it makes use of the single-layer feed-forward neural network (SLFN). The main concept behind the ELM lies in the random choice of the SLFN hidden layer weights and biases. The output weights are determined analytically, thus the network is obtained with a very few steps and with low computational cost. Consider a set of N distinct samples (x i , y i ) with x i ∈ R d1 and y i ∈ R d2; then, a SLFN with H hidden units is modeled as the following sum
With f being the activation function, \(w_{i}\) the input weights, \(b_{i}\) the biases and \(\beta_{i}\) the output weights.
In the case where the SLFN perfectly approximates the data, the errors between the estimated
Outputs y i and the actual outputs y i are zero, and the relation is
which writes compactly as \(H\beta = Y,\) with
and \(\beta = \left( {\beta_{1}^{\text{T}} \ldots \beta_{\text{H}}^{\text{T}} } \right)\) and \(Y = \left( {y_{1}^{\text{T}} \ldots y_{\text{H}}^{\text{T}} } \right)^{\text{T}}\).
Theorem states that with randomly initialized input weights and biases for the SLFN, and under the condition that the activation function is infinitely differentiable, then the hidden layer output matrix can be determined and will provide an approximation of the target values which is as good as expected. (nonzero training error). The way to calculate the output weights b from the knowledge of the hidden layer output matrix H and target values, is proposed with the use of a Moore–Penrose generalized inverse of the matrix H, denoted as H†. Overall, the ELM algorithm is summarized below.
2.4.3 ELM algorithm
Given a training set (x i , y i ) with x i ∈ ℜd1 and y i ∈ ℜd2, an activation function f: ℜ → ℜ and the number of hidden nodes H.
-
1.
Randomly assign input weights \(w_{i}\) and biases \(b_{i}\), i ∈ [1, H];
-
2.
Calculate the hidden layer output matrix H;
-
3.
Calculate output weights matrix b = H†Y.
Number of hidden units is an important parameter for ELM and should be chosen with care. The selection can be done for example by cross-validation, information criteria, or starting with a large number and pruning off the network.
2.4.4 Analysis of ELM
Essential property of a fully trained neural network is its ability to learn features on data. Features extracted by the network should be good for predicting the target variable of a classification/regression task. In a network with one hidden and one output layer, the hidden layer learns the features, while the output layer learns a linear mapping. This could be considered as the first nonlinear mapping of data into a feature space and then performing a linear regression/classification in that space. ELM has no feature learning ability. It projects the input data into whatever feature space, the randomly chosen weights happen to specify, and learns a linear mapping in that space. Parameters affecting the feature space representation of a data point are type and number of neurons, and the variance of hidden layer weights. Training data can affect these parameters through model selection, but not directly through any training procedure. This is similar to what a support vector machine does. A feature space representation for a data point is derived using a kernel function with a few parameters, which are typically chosen by some model selection outline. Features are not learned from data, but dictated by the kernel. Weights for linear classification or regression are then learned in the feature space. The biggest difference is that ELM explicitly generates the feature space vectors but in SVM or other kernel method only similarities between feature space vectors are used.
2.4.5 Kernel functions
The idea of the kernel function is to enable operations to be performed in the input space rather than the potentially high-dimensional feature space. Hence the inner product does not need to be evaluated in the feature space. The function is expected to perform mapping of the attributes of the input space to the feature space. The kernel function plays a critical role in SVM and its performance. It is based upon reproducing kernel Hilbert Spaces.
If K is a symmetric positive definite function, which satisfies Mercer’s Conditions,
Then the kernel represents a legitimate inner product in feature space. The training set is not linearly separable in an input space. The training set is linearly separable in the feature space. This is called the “kernel trick.”
The different kernel functions are listed below.
-
1.
Lineal kernel:
$$K\left( {x_{i} , y_{i} } \right) = x_{i}^{\text{T}} y_{i}$$(13) -
2.
Multi-layer perceptron kernel:
$$K\left( {x_{i} , y_{i} } \right) = \tanh \left( {sx_{i}^{\text{T}} y_{i} + t^{2} } \right)$$(14)where s is scale parameter and t is the bias.
-
3.
Polynomial kernel:
$$K\left( {x_{i} , y_{i} } \right) = x_{i}^{\text{T}} y_{i} + t)^{d}$$(15)where t is the intercept and d is the degree of the polynomial.
-
4.
Radial basis function:
Gaussian radial basis function: radial basis functions most commonly with a Gaussian form
$$K\left( {x_{i} , y_{i} } \right) = \exp \left( { - \frac{{x_{i} - y_{i}^{2} }}{{2\sigma^{2} }}} \right)$$(16)Exponential radial basis function: a radial basis function produces a piecewise linear solution which can be attractive when discontinuities are acceptable.
$$K\left( {x_{i} , y_{i} } \right) = \exp \left( { - \frac{{x_{i} - y_{i} }}{{2\sigma^{2} }}} \right)$$(17)where \(\sigma^{2}\) is the variance of the Gaussian kernel.
There are many more including Fourier, splines, B-splines, additive kernels, and tensor products.
2.4.6 ELM kernel
The architecture of SVM with new ELM kernel is illustrated in Fig. 4. ELM uses a fixed mapping from data to feature space. In derivation of the neural network kernel, which has infinite number of hidden units, and when the weights are integrated out, the resulting function is parameterized in terms of weight variance [53]. We interpret ELM as an approximation to this infinite neural network. This idea has been suggested for support vector machine in [9], which has been the main inspiration for our work. An attempt based on the same idea is done in Gaussian process classification. Authors of [9] proposed using ELM hidden layer to form a kernel to be used in SVM classification. The ELM kernel function is defined as
that is, the data are fed through the ELM hidden layer to obtain the feature space vectors, and their covariance is then computed and scaled by the number of hidden units.
A proper kernel function for a certain problem is dependent on the specific data. Here ELM has been used as a new kernel for SVM for the classification of EEG signals. ELM is fast, but it does not search for maximum-margin hyperplanes. Instead, they minimize a sum of squared errors between the class labels and the multi-layer perceptron output. This kind of criterion is not really suitable for classification. The study proposes to merge both the SVM and ELM approaches in order to obtain models which (i) are fast to train and (ii) are maximum-margin classifiers.
3 Results
A five-class EEG signal classification problem is dealt with, which is the assignment of subjects to one of five predetermined classes. The proposed technique for classification of the EEG signals was implemented by using MATLAB (R2013a) software package running in an Intel Core 2 Duo processor with 2.8 GHz. For classification of EEG signals, 500 signals are used (Dataset A-E each contains 100 signals). From these by cross-fold selection method, 50 % of the nonoverlapped data are used for training and remaining 50 % of the nonoverlapped data for testing.
The classification and misclassification results of various SVMs used in the H-MSVM for the classification of the EEG signals are given in Table 3. The challenge is set A is confused with set B and set C with set D. From these matrices, the number of EEG signals which are correctly classified and misclassified could be identified. The classification accuracy of the proposed approach has been compared with other existing classifiers such as multi-class SVMs (one vs. one, one vs. rest, DAG), artificial neural network (ANN). It can be seen from Table 4 that the proposed H-MSVM achieves highest classification accuracy over other methods. Table 5 presents the values of the statistical parameters such as sensitivity, specificity and classification accuracy of the proposed H-MSVM classifier for various EEG Dataset {A, B, C, D, E}. The proposed classifier achieves an overall classification accuracy of 94 %.
Stringent experiments have been conducted using hold-out and cross-validation methods on the entire dataset [3]. (i) to study statistical relevance of the dataset; (ii) to study the generalization ability of the proposed method to an independent dataset; (iii) and to study the variability to changes in the training/testing data. The results are presented in Table 6. From this table, it is evident that the proposed method, when subjected to cross-validations, yields consistent classification accuracy with less variations across different runs. This table also demonstrates the generalization ability of the proposed method and statistical relevance of the data using hold-out validations. It is observed from the table that the classification accuracy is monotonically increasing, when more samples are used for training than testing. The proposed method stabilizes at 94 %, in which 50 % samples are used for training and remaining 50 % samples are used for testing. The improvements in terms of classification accuracy at 60:40 ratio are 1 %, which is marginal. The classification accuracy of 97 % is achieved, when it is overtrained using 80:20 ratio.
Table 7 presents a comparison between the approach proposed and other existing research works which use the same benchmark EEG dataset. Complete five-class EEG dataset {A, B, C, D, E} which are more challenging to classify are used. Most of the existing researchers have used only two-class or three-class problems. Only a few researchers used the complete five-class dataset. The new hierarchical multi-class SVM with ELM kernel and the features of wavelet transform-based statistical coefficients, approximate entropy, and largest Lyapunov exponents were used in the study to classify the EEG signals indicating higher performance than that of the other existing research works.
4 Discussion
Figure 5 compares between-class distance and within-class distance for various hierarchical classes of datasets based on the features. From the figure, it is observed that within-class distance was minimum and between-class distance was maximum. So the extracted features are well suited for discriminating various classes.
Table 8 presents the comparison of classification accuracies and number of SVMs required for various multi-class SVMs. Table 9 summarizes classification accuracy and execution time of various SVM kernels. It is proved that the computation time for ELM kernel is much lesser than other kernels with comparable classification accuracy. Using RBF kernel, the accuracy increases, reaches its maximum, and then decreases. In contrast, the accuracy with ELM kernel quickly stabilizes for each dataset. Experiments have been conducted using SVM as classifier employing RBF kernel and ELM kernel. It is observed that the classification accuracy of the RBF kernel does not stabilize quickly, whereas ELM kernel quickly gets stabilized. Also it is observed that the classification accuracy of RBF kernel-based classifier, after reaching its maximum, started decreasing, and this is due to the fact that the RBF Kernel is not immune to over-fitting. This work considered as a complete five-class dataset {ABCDE} of EEG for the classification. In the proposed hierarchical multi-class SVM classifier, the computational complexity is lesser when compared with other multi-class SVM classifiers that is N − 1 where N is the number of classes, since other multi-class SVM classifiers require number of SVMs which is same as the number of classes. For the example application, other multi-class SVM classifiers require N(N − 1) SVMs. Here in the proposed classifier, only four SVMs were used in three levels (hierarchical tree). The smallest computation of classifying the test pattern is just one SVM evaluation when the decision could be made at the top node. The worst case is N − 1 SVM evaluations when four SVM nodes classifiers have to be traversed before the classification decision is arrived at. The test phase for one pattern by one-against-one, one-against-rest, and DAG SVM approaches require N SVM evaluations. Compared with those approaches, the proposed SVM tree classifier is more efficient in the test phase. The efficiency gained in testing phase is very important for many practical applications. The classification stage in application such as epileptic seizure detection in real time requires fast response. Additional experiments have been carried out using clinical EEG data, acquired from 20 epileptic patients who had been under the evaluation and treatment in the Neurology Department of Sri Ramakrishna Hospital, Coimbatore, India, for detecting epileptic seizure. The proposed method achieves 98 % classification accuracy and suits for real-time clinical utilities.
5 Conclusions
The proposed approach has successfully classified complete range of EEG datasets (multi-classes A–E) with the emphasis on epileptic seizure detection. When compared to other classification schemes, the proposed method is efficient in terms of classification accuracy and computation complexity. Moreover, the hierarchical structure generated in the approach indicates the interclass relationships among different classes and dataset. The proposed approach achieves 94 % classification accuracy, which positively proved that this method is successful. This paper also proposes an approach merging both the SVM and ELM framework. Experiments show that the accuracy of SVM classifiers with ELM kernel is better, when compared with the standard RBF kernels. The results from this work can be expanded to include a more complete range of pathologies. Possible directions for further work include optimizing the features and kernel parameters using particle swarm optimization and developing real-time epileptic seizure detection and monitoring system.
References
Acharya UR, Molinari F, Sree SV, Chattopadhyay S (2012) Automatic diagnosis of epileptic EEG using entropies. Biomed Signal Process Control 7(4):401–408
Adeli H, Zhou Z, Dadmehr N (2003) Analysis of EEG records in an epileptic patient using wavelet transform. J Neurosci Methods 123:69–87
Andrzejak RG, Lehnertz K, Rieke C (2001) Indications of nonlinear deterministic and finite dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys Rev E 64(6):061907
Andrzejak RG, Widman G, Lehnertz K (2001) The epileptic process as nonlinear deterministic dynamics in a stochastic environment: an evaluation on mesial temporal lobe epilepsy. Epilepsy Res 44:129–140
Chandaka S, Chatterjee A, Munshi S (2009) Cross-correlation aided support vector machine classifier for classification of EEG signals. Expert Syst Appl 36(2):1329–1336
Diambra L, Figueiredo J, Malta C (1999) Epileptic activity recognition in EEG recording. Phys A Stat Mech Appl 273(3–4):495–505
Erfanian A, Mahmoudi B (2005) Real-time ocular artifact suppression using recurrent neural network for electro-encephalogram based brain–computer interface. Med Biol Eng Comput 43:296–305
Foo SY, Stuart G, Harvey B, Meyer Baese A (2002) Neural network based EEG pattern recognition. Eng Appl Artif Intell 15:253–260
Frenay B Verleysen M (2010) Using SVMs with randomised feature spaces: an extreme learning approach. In: Proceedings of the ESANN, pp 315–320
Gotman J, Flanagah D, Zhan J, Rosenblat B (1997) Automatic seizure detection in the newborn: methods and initial evaluation. Electroencephalogr Clin Neurophysiol 103:356–362
Guler I, Ubeyli (2009) Multiclass support vector machines for EEG-signals classification. IEEE Trans Inf Technol Biomed 11(2):117–126
Guler N, Ubeyli E, Guler I (2005) Recurrent neural networks employing Lyapunov exponents for EEG signals classification. Expert Syst Appl 29(3):506–514
Guo L, Riveero D, Pazaos A (2010) Epileptic seizure detection using multiwavelet transform based approximate entropy and artificial neural networks. J Neurosci Methods 193:156–163
Guo L, Rivero D, Dorado J, Rabunal JR, Pazos A (2010) Automatic epileptic Seizure detection in EEG based on line length feature and artificial neural network. J Neurosci Methods 191:101–109
Hasan O (2009) Automatic detection of epileptic seizures in EEG using discrete wavelet transform and approximate entropy. Expert Syst Appl 36:52027–52036
He P, Wilson G, Russel C (2004) Removal of ocular artifacts from electro-encephalogram by adaptive filtering. Med Biol Eng Comput 42(3):407–412
Hsu KC, Yu SN (2010) Detection of seizures in EEG using subband nonlinear parameters and genetic algorithm. Comput Biol Med 40:823–830
Huang GB, Zhu QY, Siew CK (2006) Extreme learning machine: theory and applications. Neurocomputing 70:489–501
Hwang H-J, Kim K-H, Jung Y-J, Kim D-W, Lee Y-H, Im, C-H (2011) An EEG-based real-time cortical functional connectivity imaging system. Med Biol Eng Comput 49(9):985–995
Iscan Z, Dokur Z, Demiralap T (2011) Classification of electroencephalogram signals with combined time and frequency features. Expert Syst Appl 38:10499–10505
Jahankhani P, Kodogiannis V, Revett K (2006) EEG signal classification using wavelet feature extraction and neural networks. In: IEEE John Vincent Atanasoff 2006 international symposium on modern computing, pp 52–57
Jennifer C, John GG, Phil HJ, Griffiths Clive J, Drinnan Michael J (2006) Comparison of manual sleep staging with automated neural network-based analysis in clinical practice. Med Biol Eng Comput 44(1/2):105–110
Kalayci T, Ozdamar O (1995) Wavelet preprocessing for automated neural network detection of EEG spikes. IEEE Eng Med Biol Mag 14(2):160–166
Kannathal N, Choo ML, Acharya UR, Sadasivan PK (2005) Entropies for detection of epilepsy in EEG. Comput Methods Programs Biomed 80:187–194
Kannathal N, Choo M, Acharya U, Sadasivan P (2005) Entropies for detection of epilepsy in EEG. Comput Methods Programs Biomed 80(3):187–194
Khan YU, Gotman J (2003) Wavelet based automatic seizure detection in intracerebral electroencephalogram. Clin Neurophysiol 114:898–908
Kiymik MK, Akin M, Subasi A (2004) Automatic recognition of alertness level by using wavelet transform and artificial neural network. J Neurosci Methods 139:231–240
Kiymik MK, Subasi A, Ozcalik HR (2004) Neural networks with periodogram and autoregressive spectral analysis methods in detection of epileptic seizure. J Med Syst 28(6):511–522
Liang SF, Wang HC, Chang WL (2010) Combination of EEG complexity and spectral analysis for epilepsy diagnosis and seizure detection. In: EURASIP journal on advances in signal processing, p 853434
Liu A, Hahn JS, Heldt GP, Coen RW (1992) Detection of neonatal seizures through computerized EEG analysis. Electroencephalogr Clin Neurophysiol 82:30–37
McSharry PE, He T, Smith LA, Tarassenko L (2002) Linear and non-linear methods for automatic seizure detection in scalp electro-encephalogram recordings. Med Biol Eng Comput 40:447–461
Miche Y, Sorjamaa A, Bas P, Simula O, Jutten C, Lendasse A (2010) OP-ELM: optimally pruned extreme learning machine. IEEE Trans Neural Netw 21(1):158–162
Mohseni H, Maghsoudi A, Kadbi M, Hashemi J, Ashourvan A (2006) Automatic detection of epileptic seizure using time–frequency distributions. In: IET 3rd international conference on advances in medical, signal and information processing, vol 14
Nicolaou N, Georgiou J (2012) Detection of epileptic electroencephalogram based on permutation entropy and support vector machine. Expert Syst Appl 39:202–209
Nigam V, Graupe D (2004) A neural-network-based detection of epilepsy. Neurol Res 26(1):55–60
Ocak H (2009) Automatic detection of epileptic seizures in EEG using discrete wavelet transform and approximate entropy. Expert Syst Appl 36(2):2027–2036
Peltoranta M, Pfurtscheller G (1994) Neural network based classification of non-averaged event-related EEG responses. Med Biol Eng Comput 32:189–196
Petrosian A, Prokhorov D, Homan R, Dashei R, Wunsch D (2000) Recurrent neural network based prediction of epileptic seizures in intra and extracranial EEG. Neurocomputing 30:201–218
Pincus SM (1991) Approximate entropy as a measure of system complexity. Proc Natl Acad Sci USA 88:2297–2301
Radhakrishnan N, Gangadhar B (1998) Estimating regularity in epileptic seizure time-series data: a complexity-measure approach. IEEE Eng Med Biol 17(3):89–94
Schaltenbrand N, Lengelle R, Toussaint M (1996) Sleep stage scoring using the neural network model: comparison between visual and automatic analysis in normal subjects and patients. Sleep 19(1):26–35
Srinivasan V, Eswaran C, Sriraam N (2005) Artificial neural network based epileptic detection using time-domain and frequency-domain features. J Med Syst 29(6):647–660
Subasi A (2005) Epileptic seizure detection using dynamic wavelet network. Expert Syst Appl 29(2):343–355
Subasi A (2006) Automatic detection of epileptic seizure using dynamic fuzzy neural networks. Expert Syst Appl 31:320–328
Subasi A (2007) Application of adaptive neuro-fuzzy inference system for epileptic seizure detection using wavelet feature extraction. Comput Biol Med 37(2):227–244
Subasi A (2007) EEG signal classification using wavelet feature extraction and a mixture of expert model. Expert Syst Appl 32(4):1084–1093
Subasi A, Gursoy MI (2010) EEG signal classification using PCA, ICA, LDA and support vector machine. Expert Syst Appl 37:8659–8666
Tzallas A, Tsipouras M, Fotiadis D (2007) Automatic seizure detection based on time–frequency analysis and artificial neural networks. Comput Intell Neurosci 13, Article ID 80510
Ubeyli E (2006) Analysis of EEG signals using Lyapunov exponents. Neural Netw World 16(3):257–273
Ubeyli E (2006) Fuzzy similarity index employing Lyapunov exponents for discrimination of EEG signals. Neural Netw World 16(5):421–431
Ubeyli ED (2010) Least square support vector machine employing model-based methods coefficients for analysis of EEG signals. Expert Syst Appl 37:233–239
Varun J, Ram Bilas P, Antony V (2014) Classification of ictal and seizure-free EEG signals using fractional linear prediction. Biomed Signal Process Control 9:1–5
Williams CKI (1998) Computation with infinite neural networks. Neural Comput 10:1203–1216
Zarjam P, Mesbah M, Boashash B (2003) Detection of newborns EEG seizure using optimal features based on discrete wavelet transform. Proc IEEE Int Conf Acoust Speech Signal Process 2:265–268
Acknowledgments
The authors wish to thank Andrzejak et al. [3], for the EEG dataset available: (http://www.meb.unibonn.de/epileptologie/science/physik/eegdata.html) and Neurology Department of Sri Ramakrishna Hospital, Coimbatore, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Murugavel, A.S.M., Ramakrishnan, S. Hierarchical multi-class SVM with ELM kernel for epileptic EEG signal classification. Med Biol Eng Comput 54, 149–161 (2016). https://doi.org/10.1007/s11517-015-1351-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-015-1351-2