Abstract
This work involves retinal image classification and a novel analysis system was developed. From the compressed domain, the proposed scheme extracts textural features from wavelet coefficients, which describe the relative homogeneity of localized areas of the retinal images. Since the discrete wavelet transform (DWT) is shift-variant, a shift-invariant DWT was explored to ensure that a robust feature set was extracted. To combat the small database size, linear discriminant analysis classification was used with the leave one out method. 38 normal and 48 abnormal (exudates, large drusens, fine drusens, choroidal neovascularization, central vein and artery occlusion, histoplasmosis, arteriosclerotic retinopathy, hemi-central retinal vein occlusion and more) were used and a specificity of 79% and sensitivity of 85.4% were achieved (the average classification rate is 82.2%). The success of the system can be accounted to the highly robust feature set which included translation, scale and semi-rotational, features. Additionally, this technique is database independent since the features were specifically tuned to the pathologies of the human eye.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Ophthalmologists use digital fundus cameras [37] to non-invasively view the optic nerve, fovea, surrounding vessels and the retinal layer [18]. Often, this type of imaging technique is referred to as retinal imaging and the ophthalmologist may search the images for signs of various diseases. Since retinal imaging is non-invasive, there is a rapid increase in the number of images which are being collected. Diagnosing these large volumes of images is expensive, time consuming and may be prone to human error. To aid the doctors with this diagnostic task, a computer-aided diagnosis (CAD) scheme could offer an objective, secondary opinion of the images. Additionally, the same feature set could be used in a content-based image retrieval (CBIR) application, which would avoid the need for text annotations.
In the past, a lot of research has been dedicated to vessel and retinal segmentation as well as the registration of retinal images [16, 31, 33, 39, 48]. As of recently, automated eye disease detection has become more important and has received some attention from the research community. Brandon et al. developed a system to automatically classify different types of drusens [8] using pixel level, region level, area level and image level classification using feature thresholds and neural networks. The achieved results are promising (87% classification rates on 119 images), but feature thresholds were created experimentally and would be highly database dependant. Sinthanayothin et al. [37] created an automatic screening system which aims to detect diabetic retinopathy [exudates, microaneurysms and haemorrhages (HMA)]. First, the optic disc and blood vessels were located and removed. To detect exudates, recursive region growing was used based on a relative intensity measure. To detect HMA, image enhancement was performed followed by a threshold which classified HMA and non-HMA regions. There were no specific details on how the threshold was determined, so this threshold may be database dependant. Furthermore, since regions were grown according to intensity, this technique would not be robust to illumination changes (i.e. other retinal databases/imaging conditions). On 771 images, overall classification rates of 75.43% were achieved. Wang et al. [45] created a CAD system which detects exudates in retinal images and achieved classification rates of 85%. To increase performance, contrast enhancement was performed using an exponential mapping function. The parameters of the mapping function were found empirically and therefore are database dependant. Additionally, initial training was performed with manually segmented regions, which requires intervention from the trained professional. Zhou et al. [49] used several features to discriminate between hypertension and diabetic-related diseases of the eye. Twenty-six images were used which were acquired from 16 patients. Since data was collected from only a few patients, the classification rates may be skewed.
Although the discussed techniques achieve promising results, thresholds and parameters were specifically developed for the database used. Additionally, one technique was not fully automated since it required manual region segmentation. The proposed work aims to overcome these limitations by designing a system which is fully automated and database independent (i.e. features are not dependent on illumination conditions or the imaging system used). Furthermore, the previous works listed only detect a specific type of abnormality and lack the ability to diagnose various types of pathologies. To combat this, the proposed work utilizes a highly robust and descriptive feature set to diagnose a variety of pathologies, such as exudates, drusens, choroidal neovascularization, central vein and artery occlusion, histoplasmosis, hemi-central retinal vein occlusion, arteriosclerotic retinopathy and more. The retinal images are stored as lossy JPEG images, so feature extraction is completed in the compressed domain. Feature extraction in the compressed domain has become an important topic recently [2, 10, 11, 43, 46], since the prevalence of images stored in lossy formats far supersedes the number of images stored in their raw format. The rest of the paper is structured as follows: Section 2 contains the methodology used for designing features and the classification scheme used. Sections 3, 4, 5 contain the results, discussions and conclusions, respectively.
2 Methods
There are several challenges associated with the development of an automated classification scheme for retinal imagery: pathologies come in different shapes, forms, sizes and can occur in many different regions of the eye. The following subsections will detail the methods used to design a highly robust feature set which can account for all these scenarios.
2.1 Feature extraction problem formulation
In a feature extraction problem, important structures or events within the data are quantified with discriminatory descriptors. The extracted features are then fed into a classifier, which arrives at a decision. For retinal imagery, this decision is related to the diagnosis of the patient. Let \({{\mathcal{X}}} \in {{\mathbf{R}}}^n\) represent the signal space which contains all retinal images with the dimensions of n = N × N. Since the images contained within \({{\mathcal{X}}}\) can be expected to have a very high dimensionality n, using all these samples to arrive at a classification result would be prohibitive [13]. Furthermore, the original image space \({{\mathcal{X}}}\) is also redundant, which means that all the image samples are not necessary for classification. Therefore, to gain a more useful representation, a feature extraction operator f may map the subspace \({{\mathcal{X}}}\) into a feature space \({{\mathcal{F}}}\)
where \({{\mathcal{F}}} \in {{\mathbf{R}}}^k,\quad k\leq n\) and a particular sample in the feature space may be written as a feature vector: F = {F 1, F 2, F 2,..., F k }. In the case of highly discriminatory features, image classes would map to non-intersecting clusters in the feature space \({{\mathcal{F}}}.\) However, the spatial domain representation of the retinal images may not carry enough discriminating characteristics to result in high classification results. Therefore, prior to feature extraction, the retinal images may be transformed to another domain to gain a more descriptive representation.
Although it is important to choose features which provide maximum discrimination between image classes, it is also important that these features are robust. A feature is robust if it provides consistent results across the entire application domain [40]. To ensure robustness, the numerical descriptors should be rotation, scale and translation (RST) invariant. In other words, if the image is rotated, scaled or translated, the extracted features should be insensitive to these changes, or it should be a rotated, scaled or translated version of the original features, but not modified [28]. This is useful for classifying unknown image samples since they will not have structures with the same orientation and size as the images in the training set [22].
If a feature is extracted from a transform domain, it is also important to investigate the invariance properties of the transform since any invariance in this domain also translates to an invariance in the features. For instance, the 1-D Fourier spectrum is translation-invariant [38] since any translation in the time domain representation of the signal, does not change the magnitude spectrum in the Fourier domain.
2.2 Multiresolution transformation for retinal images
In general, biomedical signals contain a combination of information which is localized spatially (i.e. transients, edges) as well as structures which are more diffuse (i.e. small oscillations, texture) [41]. As shown by Fig. 1, the retinal images are also of this nature (nonstationary), since they contain both coarse or fine texture patterns (high frequency), as well as regions with slowly varying or constant pixel values (low frequency). In order to adequately represent these localized texture patterns, the proposed work utilizes a transformation which can provide a good description of these features. To achieve such a description, multiresolutional analysis (MRA) techniques will be utilized since they can represent localized image features with good space-frequency resolution. For discrete implementations, MRA can be realized with the discrete wavelet transform (DWT) [27, 28, 42, 44]. The DWT offers a multiresolutional representation by dyadically changing the size of the analysis window. Consequently, the basis functions are tuned to events which have high frequency components in a small analysis window (scale) or low frequency events with a large scale [9]. Therefore, in one set of basis functions, it is possible to decompose the image with various space-frequency resolutions. In this work, the 2-D DWT is realized using the lifting/filterbank technique [9, 28]. The wavelets and scaling basis functions are related to a set of 1-D lowpass (h o (n)) and highpass (h 1(n)) filter coefficients, where each filter is applied separably to the image. The 5/3 Le Gull wavelet is used since filter lengths are small and can warrant an efficient implementation [30, 47].
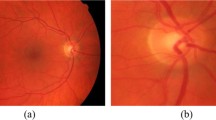
Retinal images (700 × 605) supplied by the STare public database [18]. a Normal retinal image, b normal retinal image, c retinal image with background diabetic retinopathy, d retinal image with central retinal vein occlusion
A 2-D DWT scheme is made up of basis functions which can decompose various scaled versions of the input image. In other words, the DWT is scale-invariant, i.e. any scaled version of the input image will be matched to a scaling function with the same scale in the basis family. As stated earlier, any transformation which is applied prior to feature extraction must also have invariant properties; thus the DWT can give way to scale-invariant features. Consequently, a single transformation can be used to capture various-sized pathologies (which is beneficial for retinal image classification since pathologies do not come in a predefined size).
Although the DWT provides good space-frequency localization and is scale-invariant, it is a well known fact that the DWT is shift-variant [9, 28]. The shift-variant property of the DWT is a direct result of the rate change operators [25]. For different translations of the input image, a different set of DWT coefficients would be generated. In fact, since decimation is carried out separately along the rows and columns of an image, the 2-D DWT would produce four different distributions of coefficients, which are in response to shifts of the input by: (0,0), (0,1), (1,0), (1,1), where the first index corresponds to the row shift and the second index corresponds to the column shift. The coefficients for all other shifts of the input can be obtained by circularly translating one of the sets of DWT coefficients created by one of the four fundamental shifts of the input [(0,0), (0,1), (1,0), (1,1)]. For instance, all other translations of the input by {0, 2, 4, 6, ...} rows and {0, 2, 4, 6, ...} columns would result in DWT representations which are space translated versions of the DWT coefficients computed from the input when it is shifted by (0,0). Similar results apply for the other input shifts (0,1), (1,0), (1,1), see [32]. Since different shifts of the input image results in a completely different set of amplitude values for the coefficients, the extraction of a consistent feature set is difficult [28, 41]. In order to extract shift-invariant features from the wavelet domain, shift-invariant algorithms must be investigated so that a consistent set of coefficients are chosen, regardless of the input image’s space translation. To achieve this, a shift-invariant discrete wavelet transform (SIDWT) may be performed on the input image f(x,y)
where \({\widetilde{F}}(k_1,k_2,j)\) are the wavelet coefficients. This representation would be considered shift-invariant if a shift of the input image (Δx, Δy) ∈ Z results in output coefficients which are exactly the same as \({\widetilde{F}}(k_1,k_2,j),\) or a spatially shifted version of it. This may be shown by
where k ′1 = k 1 + b 1· Δx and k ′2 = k 2 + b 2· Δy for some (b 1, b 2) ∈ Z. If the coefficients are exactly the same: b 1 = b 2 = 0.
2.3 Shift-invariant discrete wavelet transform (SIDWT)
The shift-variant property of the DWT is widely known and several solutions have been proposed. To achieve a shift-invariant representation, Mallat et al. use an overcomplete, redundant dictionary, which corresponds to filtering without decimation [6, 28]. From the filtered and fully sampled version of the image, local extrema are used for translation invariance since a shift in the input image results in a corresponding shift of the extrema [23, 28]. Since there is no decimation, each level of decomposition contains as many samples as the input image, thus making the algorithm computationally complex and memory intensive.
Simoncelli et al. [36] propose an approximate shift-invariant DWT algorithm by relaxing the critical sampling requirements of the DWT. This algorithm is known as the power-shiftable DWT since the power in each subband remains constant. As explained in [6], the shift-variant property is also related to aliasing caused by the DWT filters. The power shiftable transform tries to remedy this problem by reducing the aliasing of the mother wavelet in the frequency domain. The modifications to the mother wavelet result in a loss of orthogonality [26].
The Matching Pursuit (MP) algorithm can also achieve a shift-invariant representation, when the decomposition dictionary contains a large amount of redundant wavelet basis functions [29]. However, the MP algorithm is extremely computationally complex and arriving at a transformed representation causes significant delays [12]. Bradley combines features of the DWT pyramidal decomposition with the à trous algorithm [28], which provides a trade off between sparsity of the representation and time-invariance [6]. Critical sampling is only carried out for a certain number of subbands and the rest are all fully sampled. This representation only achieves an approximate shift-invariant DWT [6].
The algorithms discussed either try to minimize the aliasing error by relaxing critical subsampling and/or add redundancy into the wavelet basis set. However, these algorithms either suffer from lack of orthogonality (which is not always an issue for feature extraction), achieve an approximate shift-invariant representation, are computationally complex or require significant memory resources. To combat these downfalls, it is possible to use the SIDWT algorithm proposed by Beylkin, which computes the DWT for all circular shifts, in a computationally efficient manner [5]. Furthermore, the transformation utilizes orthogonal wavelets, thereby resulting in less redundancy in the representation [23]. Belkyn’s work has also been extended to 2-D signals by Liang et al. [23, 24, 26] and its performance in a biomedical image feature extraction application will be investigated.
2.4 SIDWT algorithm for retinal imagery
For different shifts of the input image, it was shown that the DWT can produce one of four possible coefficient distributions (after one level of decomposition). These four DWT coefficient sets (cosets) are not translated versions of one another and each coset is the DWT output response to one of four shifts of the input: (0,0), (0,1), (1,0), (1,1). All other shifts of the input (at this decomposition level) will result in coefficients which are shifted versions of one of these four cosets. Therefore, to account for all possible representations, these four cosets may be computed for each level of decomposition. This requires the LL band from each level to be shifted by the four translates {(0,0), (0,1), (1,0), (1,1)} and each of these new images can be separately decomposed to account for all representations. To compute the coefficients at the jth decomposition level, for the input shift of (0,0), the subbands LLj, LHj, HLj, HHj may be found by filtering the previous levels coefficients LLj+1, as shown below:
The subband expressions listed in Eqs. 2 through 5 contain the coefficients which would appear the same if LLj+1 is circularly shifted by {0, 2, 4, 6,..., s} rows and {0, 2, 4, 6,..., s} columns, where s is the number of row and column coefficients in each of the subbands for the level j + 1.
The subband coefficients which are the response to a shift of (0,1) in the previous level’s coefficients may be computed by
which contain all the coefficients for {0, 2, 4, 6,..., s} row shifts and {1, 3, 5, 7,..., s − 1} column shifts of LL j+1. Similarly, for a shift of (1,0) in the input, the DWT coefficients may be found by
which contain all the coefficients if the previous levels’ coefficients LL j+1 are shifted by {1, 3, 5, 7,..., s − 1} rows and {0, 2, 4, 6,..., s} columns. For an input shift of (1,1), the subbands may be computed by
Similarly, these subband coefficients account for all DWT representations corresponding to {1, 3, 5, 7,..., s − 1} row shifts and {1, 3, 5, 7,..., s − 1} column shifts of the input subband LL j+1.
Performing a full decomposition will result in a tree which contains the DWT coefficients for all N 2 circular translates of an N × N image. The number of coefficients in each node (per decomposition level) remains constant at 3N 2, and a complete decomposition tree will have N 2(3log2 N + 1) elements [23]. For a detailed review on how to address the coefficients of the SIDWT decomposition tree, see [23, 24]. A proper addressing scheme will help to find the wavelet transform for a particular translate (m, n), where m is the row shift and n is the column translate of the input image.
This new representation computes the 2-D DWT for all circular translates in a computationally efficient manner. To achieve shift-invariance, a method to select a consistent set of wavelet coefficients which are independent of the input translation must be determined. Such a method would choose the same basis set (which corresponds to a particular translate of the original image), regardless of the shift in the input image. Coifmen and Wickenhauser’s best basis selection technique [14] is utilized. See [21] for more details on the algorithm’s full implementation (including the best basis algorithm and the coefficient addressing scheme).
2.5 Multiscale texture analysis for retinal images
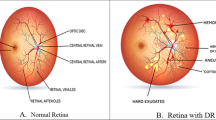
Normal retinal images are easily characterized by their overall homogeneous appearance. Other anatomical properties of a normal eye (such as the retina, fovea or veins) are embedded into this homogeneous texture. This is easily seen in Fig. 2a–c.
Retinal images which exhibit textural characteristics. a–c Normal, homogeneous retinal images, d background diabetic retinopathy (dense, homogeneous yellow clusters), e macular degeneration (large, radiolucent drusens with heterogeneous texture properties), f central retinal vein occlusion (oriented, radiating texture)
Unlike normal eyes, eyes which contain disease do not possess uniform texture qualities. Three cases of abnormal retinal images are shown in Fig. 2d–f. Diabetic retinopathy is characterized by exudates or lesions which are typically found in random whitish/yellow patches of varying sizes, locations [45] with relatively defined margins [37], as shown in Fig. 2a. The exudates occur in very dense, semi-homogeneous patches, which possess directional textural elements that can be differentiated from the homogeneous background. Furthermore, blood vessels rapidly grow in the retina to compensate for reduced blood supply caused by the exudates [4]. This may be characterized by oriented texture which is composed of a high concentration of blood vessels. Another clinical sign of diabetic retinopathy are microaneurysms and haemorrhages. Microaneurysms appear as small, red, round dots and haemorrhages can have “dot”, “blot” or “frame” configurations [37]. These two types of pathologies are the first clinically detectable signs of diabetic retinopathy [37] and may be detected with a texture analysis tool since they disrupt the homogeneity of the background.
Another type of eye disease is known as macular degeneration which can cause blindness if it goes untreated. Macular degeneration may be characterized by drusens, which appear as yellowish, cloudy blobs, which exhibit no specific size or shape and can appear with a variety of characteristics [8]. This is shown in Fig. 2e by the yellow “blobs” which are scattered throughout the eye. These structures are heterogenous and not as dense (radiopaque) as exudates. Other diseases include central retinal vein and/or artery occlusion and an example is shown in Fig. 2f. It is easy to notice from this image that an oriented texture pattern radiates from a central location.
Since normal and abnormal retinal images possess unique texture qualities which allow visual discrimination between the two classes, a texture analysis scheme which interprets the images in accordance to human texture perception should be adopted to differentiate between the retinal images (in fact, texture has been one of the most important image characteristics used to classify images [1, 19]). When a textured image is viewed, the human visual system can easily discriminate between textured regions. To try and understand how the human visual system can easily differentiate between textures, Julesz defined textons, which are elementary units of texture [20]. Various textured regions can be decomposed using these textons, which include elongated blobs, lines, terminators and more. It was also found that the frequency content, scale, orientation and periodicity of these textons can provide important clues on how to differentiate between two or more textured areas [20, 28]. These fundamental points have opened up a wide-variety of computer vision applications [1, 15, 38], which aim to mimic the properties of the human visual system when discriminating texture. Since many applications and algorithms are being built to assist humans with their everyday tasks, for interpretation of medical imagery it is only logical that these computing devices understand images the same way humans do.
To extract texture-based features, normalized graylevel co-occurrence matrices (GCMs) are used. Let each entry of the normalized GCM be represented as a probability distribution p(l 1, l 2, d, θ), where l 1 and l 2 are two graylevel values at a distance d and angle θ. Using normalized GCMs, statistical quantities which measure the uniformity of the images will be used, since relative homogeneity differentiates between normal and abnormal retinal images. The features which will be used are homogeneity (h), which describes how uniform the texture is and entropy (e), which is a measure of nonuniformity or the complexity of the texture.
If the region is homogeneous, there will be only a few rapidly changing graylevel values. As a result, only a small number of high probability values will be found in the GCM matrix, translating into a large h value. Conversely, if the texture is nonuniform, there will be lots of varying pixel values which would each carry a small probability value. This would cause the value of h to be small. For complex, random texture structures, the entropy will be large while a completely uniform texture will result in e = 0.
Traditionally, features based on the GCM have been extracted from the spatial domain [1, 19]. A weakness of this technique is that it doesn’t consider other important qualities which aid in texture discrimination. As Julesz has mentioned in [20], texture discrimination is also dependant on the perception of texture events at different scales, frequencies and orientations. Consequently, to gain a robust representation which is in accordance to human texture perception, textural features are computed from the wavelet domain. Extracting features from the wavelet domain will result in a localized texture description, since the DWT has excellent space-localization properties. To account for oriented texture, the GCMs are computed at various angles in the wavelet domain at d = 1 to account for fine texture. Typically, the DWT is not used for texture analysis due to its shift-variant property. However, using the SIDWT algorithm described will allow for the extraction of a consistent feature set, thus allowing for multiscale texture analysis.
2.5.1 Multiscale texture features
In the wavelet domain, GCMs are computed from each scale j at several angles θ. Each subband isolates different frequency components—the HL band isolates horizontal edge components, the LH subband isolates horizontal edges, the HH band captures the diagonal high frequency components and LL band contains the lowpass filtered version of the original. To demonstrate this localized frequency concept, please refer to Fig. 3 for one level of decomposition of a synthetic image. For a retinal image decomposition, see Fig. 4a, b. Note how the pathology in Fig. 4b is very localized and heterogeneous in all the subbands.
One level of DWT decomposition for retinal images. Contrast enhancement was performed in the higher frequency bands (HH, LH, HL) for visualization purposes. a Normal image decomposition showing an over the a homogeneous appearance of the wavelet coefficients in the HH, HL and LH bands, b decomposition of a retinal image with diabetic retinopathy, see Fig. 2c for original image. Note that in each of the higher frequency subbands the retinopathy appears as heterogeneous blobs (high-valued wavelet coefficients) in the center of the subband (which spatially corresponds to the center of the original image)
To capture these oriented texture components, the GCM is computed at 0° in the HL band, 90° in the LH subband, 45° and 135° in the HH band and 0°, 45°, 90° and 135° in the LL band to account for any directional elements which may still may be present in the low frequency subband. From each of these GCMs, homogeneity h and entropy e are computed for each decomposition level using Eqs. 18 and 19. For each decomposition level j, more than one directional feature is generated for the HH and LL subbands. The features in these subbands are averaged so that: features are not biased to a particular orientation of texture and the representation will offer some rotational invariance. The features generated in these subbands (HH and LL) are shown below. Note that the quantity in parenthesis is the angle at which the GCM was computed.
As a result, for each decomposition level j, two feature sets are generated:
where \({\widetilde{h}}^{j}_{\rm HH}, {\widetilde{h}}^{j}_{\rm LL}, {\widetilde{e}}^{j}_{\rm HH}\) and \({\widetilde{e}}^{j}_{\rm LL}\) are the averaged texture descriptions from the HH and LL band previously described and h j HL(0°), e j HL(0°), h j LH(90°) and e j LH(90°) are homogeneity and entropy texture measures extracted from the HL and LH bands. Since directional GCMs are used to compute the features in each subband, the final feature representation is not biased for a particular orientation of texture and may provide a semi-rotational invariant representation.
2.6 Pattern analysis/classification
Using the SIDWT and feature extraction operator f, the image space \({\mathcal{X}}\) has been mapped to the feature space \({{\mathcal{F}}},\) where each image is described by a feature vector F = {F 1 e , F 1 h ,..., F j e , F j h }, where j is the number of decomposition levels. After extracting features, it is necessary to classify this data into groups (i.e. classify each image as normal or abnormal). For each image and corresponding feature set F i , let \({{\mathcal{Y}}}\in\{1, 2, \ldots, C\}\) be the class label or category for image i and C is the maximum number of classes. For this particular application, C = 2; one class label is ‘normal’ and the other class label is “abnormal”. Therefore, each image may be classified into one of two groups: \({{\mathcal{Y}}}\in\{1, 2\}.\) To arrive at a classification result, it is necessary to define a classifier which maps the feature space into the decision regions \(d:{{\mathcal{F}}}\rightarrow {{\mathcal{Y}}}.\)
Prior to choosing a classifier, a series of factors must be considered which are related to the feature set and database size. Firstly, a large number of test samples are required to evaluate a classifier with low error (misclassification) rates [35] since a small database will cause the parameters of the classifiers to be estimated with low accuracy [34]. This requires the biomedical image database to be large, which may not always be the case since acquiring the images is always not easy and also the number of pathologies may be limited (i.e. data collection will have to continue for a number of years to get all the desired cases). If the extracted features are strong (i.e. the features are mapped into nonoverlapping clusters in the feature space) the use of a simple classification scheme will be sufficient in discriminating between classes. Therefore, linear discriminant analysis (LDA) [3, 13] will be the classification scheme of interest.
To enhance the reliability of LDA, the leave one out method (LOOM) is also used. Instead of dividing the N input samples into two equal sized sets of training and testing data, one sample is removed from the whole set and the discriminant functions are derived from the remaining N − 1 data samples. Then, using these discriminant scores, the left out sample is classified. This procedure is completed for all N samples. Since most biomedical image databases are expected to contain a small number of images, using LOOM will allow the classifier parameters to be estimated with least bias [17]. This is all completed at a cost of increased computational complexity, since the discriminant functions must be recalculated N times [7].
3 Results
The objective of the proposed system is to automatically classify various pathologies from normal retinal images. The retinal images used are 700 × 605, 24 bpp and lossy (.jpeg). Thirty-eight normal and 48 abnormal images were used (ground truth data is supplied with database). Pathologies of the abnormal images included: exudates, large drusens, fine drusens, choroidal neovascularization, central vein and artery occlusion, histoplasmosis, arteriosclerotic retinopathy, hemi-central retinal vein occlusion and more. Since the DWT requires the dimensions of the input image to be a factor of two, zero padding was completed prior to any analysis. The images were converted to grayscale prior to any processing to examine the feature set in this domain. Features were extracted for the first five levels of decomposition. Further decomposition levels will result subbands of 8 × 8 or smaller, which will result in skewed probability distribution (GCM) estimates and thus were not included in the analysis. Therefore, the extracted features are F j e and F j h for j = {1, 2, 3, 4, 5}. The block diagram of the proposed system is shown in Fig. 5.
In order to find the optimal sub-feature set, an exhaustive search was performed (i.e. all possible feature combinations were tested using the proposed multiscale texture classification scheme). The optimal classification performance was achieved by combining homogeneity features from the fourth decomposition level with entropy from the first, second and fourth decomposition levels. These four feature sets are shown below:
Using the optimal feature set shown above, the classification results for the retinal images are shown as a confusion matrix in Table 1 (specificity of 79% and sensitivity of 85.4% were achieved). From the misclassified cases, it was noticed that normal retinal images which possessed several, thick veins, which appeared in an oriented manner, were the cases that were misclassified. Since the texture analysis scheme searches for such texture qualities (i.e. heterogeneous oriented texture) for abnormal images, it is easy to understand why these types of images were misclassified. See Fig. 6 for such a case. It would be possible to obtain better performance if PCA (principal component analysis) was used to provide a greater separation between clusters in the feature space for cases which are “close calls”. Furthermore, two of the misclassified normal images had several imaging artifacts (i.e. some of the eye was missed in the image), which undoubtedly caused them to also be misclassified.
In terms of the abnormal images which were misclassified as normal, it was noted upon examination of the misclassified images that the pathologies were very difficult to detect, even with the human eye. See for example Fig. 6 which contains an example of an abnormal image which was classified as normal. As can easily be seen by this image, the pathology (fine drusens) are very unnoticeable and blend into the background, thus causing them to be classified as homogeneous, or normal.
To verify the optimality of this multiscale textural feature set, the classification performance of Table 1 is compared to homogeneity and entropy features extracted from the spatial domain. Again, d = 1 was used and GCMs were computed at four directions (0°, 45°, 90° and 135°) and both h and e were computed from these GCMs using Eqs. 18 and 19, respectively. The classification results for the three possible spatial feature combinations (i.e. only h, only e, h and e) are shown in Tables 2, 3, 4. As shown by these tables, the performance of the spatial features does not compare to the performance achieved by the multiresolution analysis scheme. Therefore, localization of the pathology in both scale and frequency is important for retinal image texture discrimination.
4 Discussions
The results show that the classification rates are quite high (average classification rate of 82.2%), indicating that the proposed system can differentiate between normal and abnormal retinal images with a high success rate even though a variety of pathologies were present. The choice of wavelet-based statistical texture measures (entropy and homogeneity) was critical since they were able to efficiently describe the localized texture properties of the images. This was further confirmed by computing the same features in the spatial domain, which resulted in poor classification performance due to lack of localization. The SIDWT allowed for the extraction of consistent (i.e. shift-invariant) features. The scale-invariant basis functions of the DWT captured pathologies of varying sizes within one transformation (i.e. scale-invariant). Furthermore many of the abnormal retinal images possess oriented texture elements (central vein/artery occlusion) and the overall success of the system can also be accounted to the fact that the oriented texture was properly characterized by (1) extracting features from the 2-D DWT, which isolates localized directional texture elements, (2) computing features for different angles to characterize the oriented textural properties and (3) averaging the features to obtain a non-biased, semi-rotational invariant representation.
As shown by other works in the area of retinal imaging (Sect. 1), the authors’ schemes were tuned to only detect a specific type of abnormality (i.e. drusens OR retinopathy). Therefore, a major success of this work is the development of a system which can differentiate between normal and several abnormalities of the human eye with high classification rates. Since multiple pathologies were classified within one framework, comparison with other works is not possible. Additionally, each of the related works used a number of processing steps prior to feature extraction which may vary from database to database. In the proposed work, no prior preprocessing is performed (aside from grayscale conversion) and the features are database independent. These results could be applied on other databases to get similar results since a highly discriminatory and robust (scale, translation and semi-rotational invariant) feature set was used.
Although the classification results are high, any misclassification can be accounted to cases where there is a lack of statistical differentiation between the texture uniformity of the abnormal and normal retinal images. This phenomena was shown in Fig. 6a, b. As stated, in future works, PCA could be used to decorrelate or separate the clusters even further in the feature space to increase classification performance.
Another important consideration about the classification results arise from the sizes of the databases. Since only a modest number of images were used, misclassification could result due to the lack of proper estimation of the classifiers parameters (although the scheme tried to combat this with LOOM). Additionally, finding the right trade off between number of features and database size is an ongoing research topic and has yet to be perfectly defined [17].
A last point for discussion is the fact that features were successfully extracted from the compressed domain of the retinal images. Since many forms of multi-media are being stored in lossy formats (.mp3, .mpeg, .jpeg), it is important that classification systems may also be successful when utilized in the compressed domain.
Overall, the system achieved good results in the presence of several pathologies and required no intervention from the ophthalmologist. Consequently, such a system could be employed in a clinical setting, as a computer-aided diagnosis system or content-based image retrieval system. Since the number of images being acquired in ophthalmology clinics around the world are rapidly increasing due to its non-invasive nature, such a scheme would help reduce the labourious task of interpretation or image archiving for the physician.
5 Conclusions
A unified feature extraction and classification scheme was developed using the discrete wavelet transform for retinal images. Textural features were extracted from the wavelet domain in order to obtain localized numerical descriptors of the relative homogeneity of the retinal images. To ensure the DWT representation was suitable for consistent extraction of features, a shift-invariant discrete wavelet transform (SIDWT) was computed. To combat the small database size, LDA classification was used with LOOM to try and gain a true approximation of the classifier’s parameters. Unlike other works, this work classifies multiple types of pathologies using one algorithm, thus not permitting fair comparison.
Eighty-six abnormal and normal retinal images were correctly classified at an average rate of 82.2%. The success of the system can be accounted to semi-rotational features and scale-invariance of the DWT, which permitted the extraction of consistent features for various sizes, forms and types of pathologies. Due to the success of the proposed work, it may be used in a CAD scheme or a CBIR application, to assist the ophthalmologists to diagnose and retrieve the images. Additionally, since the retinal images were stored in a lossy format and high classification rates were achieved, the system successfully extracted features from the compressed domain.
References
Aksoy S, Haralick R (1998) Textural features for image database retrieval. In: Proceedings of IEEE workshop on content-based access of image and video libraries, pp 45–49
Armstrong A, Jiang J (2001) An efficient image indexing algorithm in JPEG compressed domain. In: International conference on image processing, pp 350–351
Balakrishnama S, Ganapathiraju A, Picone J (1999) Linear discriminant analysis for signal processing problems. In: Proceedings of IEEE Southeastcon, pp 78–81
Barrett S, Rylander HG, III AW (1994) Automated lesion data base building for the treatment of retinal disorders. In: Proceedings IEEE international conference image processing, pp 426–430
Beylkin G (1992) On the representation of operators in bases of compactly supported wavelets. SIAM J Numer Anal 29:1716–1740
Bradley A (2003) Shift-invariance in the discrete wavelet transform. Digital image computing: techniques and applications, pp 29–38
Braga-Neto U, Dougherty E (2004) Is cross-validation valid for small-sample microarray classification? Bioinformatics 20(3):374–380
Brandon L, Hoover A (2003) Drusen detection in a retinal image using multi-level analysis. MICCAI 618–625
Burrus C, Gopinath R, Guo H (1998) Introduction to wavelets and wavelet transforms: a primer. Prentice Hall International, Houston
Chang S (1995) Compressed-domain techniques for image/video indexing and manipulation. In: International conference on image processing, pp 314–317
Chiu C, Wong H, Ip HS (2004) Compressed domain feature transformation using evolutionary strategies for image classification. In: International conference on image processing, pp 429–432
Cohen I, Raz S, Malah D (1997) Orthonormal shift-invariant wavelet packet decomposition and representation. Signal Process 57:251–270
Coifman R, Saito N (1995) Local discriminant bases and their applications. J Math Imaging Vis 5:337–358
Coifman R, Wickerhauser M (1992) Entropy-based algorithms for best basis selection. IEEE Trans Inf Theory 38:713–718
Falzon F, Mallat S (1998) Analysis of low bit rate image transform coding. IEEE Trans Signal Process 46(4):1027–1042
Foracchia M, Grisan E, Ruggeri A (2004) Detection of optic disc in retinal images by means of a geometrical model of vessel structure. IEEE Trans Med Imaging 23(10):1189–1195
Fukunaga K, Hayes R (1989) Effects of sample size in classifier design. IEEE Trans Pattern Anal Mach Intell 11(8):873–885
Goldbaum M (2002) STARE: structured analysis of the Retina. World Wide Web. http://www.parl.clemson.edu/stare/
Haralick R, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 3(6):610–621
Julesz B (1981) Textons, the elements of texture perception, and their interactions. Nature 290(5802):91–97
Khademi A (2006) Multiresolutional analysis for classification and compression of medical images. Master’s thesis. Ryerson University, Canada
Leung M, Peterson A (1992) Scale and rotation invariant texture classification. In: Conference record of the twenty-sixth asilomar conference on signals, systems and computers, pp 461–465
Liang J, Parks T (1994) A two-dimensional translation invariant wavelet representation and its applications. In: IEEE international conference on image processing, pp 66–70
Liang J, Parks T (1996) Translation invariant wavelet transforms with symmetric extensions. In: IEEE digital signal processing workshop, pp 69–72
Liang J, Parks T (1996) A translation-invariant wavelet representation algorithm with applications. IEEE Trans Signal Process 44(2):225–232
Liang J, Parks T (1998) Image coding using translation invariant wavelet transforms with symmetric extensions. IEEE Trans Image Process 7:762–769
Mallat S (1989) A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans Pattern Anal Mach Intell 11(7):674–693
Mallat S (1998) Wavelet tour of signal processing. Academic, USA
Mallat S, Zhang Z (1993) Matching pursuits with time–frequency dictionaries. IEEE Trans Signal Process 41:3397–3415
Marcellin M, Bilgin A, Gormish M, Boliek M (2000) An overview of JPEG-2000. In: Proceedings of the IEEE data compression conference. IEEE Computer Society, p 523
Matsopoulos G, Mouravliansky N, Delibasis K, Nikita K (1999) Automatic retinal image registration scheme using global optimization techniques. IEEE Trans Inf Technol Biomed 3(1):47–60
Pun C, Lee M (2004) Extraction of shift invariant wavelet features for classification of images with different sizes. IEEE Trans Pattern Anal Mach Intell 26(9):1228–1233
Raman B, Wilson M, Benche I, Soliz P (2003) A JAVA-based system for segmentation and analysis of retinal images. In: Proceedings of the IEEE symposium computer-based medical systems, pp 336–339
Raudys S, Jain A (1990) Small sample size effects in statistical pattern recognition: recommendations for practitioners and open problems. In: Proceedings of the 10th international conference on pattern recognition, pp 417–423
Raudys S, Jain A (1991) Small sample size effects in statistical pattern recognition: recommendations for practitioners. IEEE Trans Pattern Anal Mach Intell 13(3):252–264
Simoncelli E, Freeman W, Adelson E, Heeger D (1992) Shiftable multiscale transforms. IEEE Trans Inf Theory 38:587–607
Sinthanayothin C, Kongbunkiat V, Phoojaruenchanachai S, Singalavanija A (2003) Automated screening system for diabetic retinopathy. In: Proceedings of the 3rd international symposium on image and signal processing and analysis, pp 915–920
Tan T (1994) Scale and rotation invariant texture classification. In: IEEE colloquium on texture classification: theory and applications, pp 3/1–3/3
Teng T, Lefley M, Claremont D (2002) Progress towards automated diabetic ocular screening: a review of image analysis and intelligent systems for diabetic retinopathy. Med Biol Eng Comput 40(1):2–13
Umbaugh S, Wei Y, Zuke M (1997) Feature extraction in image analysis: a program for facilitating data reduction in medical image classification. IEEE Eng Med Biol Mag 16:62–73
Unser M, Aldroubi A (1996) A review of wavelets in biomedical applications. Proc IEEE 84(4):626–638
Vetterli M, Herley C (1992) Wavelets and filter banks: theory and design. IEEE Trans Signal Process 40(9):2207–2232
Voulgaris G, Jiang J (2001) Texture-based image retrieval in wavelets compressed domain. In: International conference on image processing, pp 125–128
Wang T, Karayiannis N (1998) Detection of microcalcifications in digital mammograms using wavelets. IEEE Trans Med Imaging 17:498–509
Wang H, Hsu W, Goh K, Lee M (2000) An effective approach to detect lesions in color retinal images. In: Proceedings of IEEE conference on computer vision and pattern recognition, pp 181–186
Xiong Z, Huang T (2002) Wavelet-based texture features can be extracted efficiently from compressed-domain for JPEG2000 coded images. In: International conference on image processing, pp 481–484
Zhang H, Fritts J (2004) An overview of JPEG-2000. In: Proceedings of SPIE: visual communications and image processing, pp 1333–1340
Zhang E, Zhang Y, Zhang T (2002) Automatic retinal image registration based on blood vessels feature point. In: Proceedings of the international conference on machine learning and cybernetics, pp 2010–2015
Zhou P, Wang M, Cao H (2005) Research on features of retinal images associated with hypertension and diabetes. In: Proceedings of IEEE’s international conference on engineering in medicine and biology society, pp 6415–6417
Acknowledgments
We would like the thank NSERC and the Ontario Government for funding this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khademi, A., Krishnan, S. Shift-invariant discrete wavelet transform analysis for retinal image classification. Med Bio Eng Comput 45, 1211–1222 (2007). https://doi.org/10.1007/s11517-007-0273-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-007-0273-z