Abstract
The relevance of the complexity of fetal heart rate fluctuations with regard to the classification of fetal behavioural states has not been satisfyingly clarified so far. Because of the short behavioural states, the permutation entropy provides an advantageous complexity estimation leading to the Kullback–Leibler entropy (KLE). We test the hypothesis that parameters derived from KLE can improve the classification of fetal behaviour states based on classical heart rate fluctuation parameters (SDNN, RMSSD, ln(LF), ln(HF)). From measured heartbeat sequences (35 healthy fetuses at a gestational age between 35 and 40 completed weeks) representative intervals of 256 heartbeats were visually preclassified into fetal behavioural states. Employing discriminant analysis to separate the states 1F, 2F and 4F, the best classification result by classical parameters was 80.0% (SDNN). After additionally considering KLE parameters it was improved significantly (p<0.0005) to 94.3% (ln(LF), KLE_Mean). It could be confirmed that KLE can improve the state classification. This might reflect the consideration of different physiological aspects by classical and complexity measures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
With increasing gestational age, the human fetus develops progressively synchronised patterns of neurovegetative ‘behaviour’ [21, 23]. The patterns have been categorised into four determined fetal behavioural states from around 34 weeks of gestation, namely, by means of heart rate patterns, the occurrence of eye movements and of gross body movements in accordance with observations in the newborn [9, 21, 28]. In general, a shifting window of 3 min length is used for polygraphic analysis [12, 21, 28]. The determination of the fetal states is based on the evaluation of cardiotocography (CTG) recording accompanied by continuous B-mode ultrasound observation as the gold standard procedure. The determination of the actual state of activity is essential in almost any case when developmental or behavioural aspects of the fetus are subjects of research. Table 1 summarises the features of the distinct states.
The fetal magnetocardiographic signal contains spaciotemporal properties and a general signal quality that is unrivalled by any other modality throughout the third trimester of gestation [11, 22, 32, 40]. Magnetocardiography (MCG) provides the necessary resolution of 1 ms and a reasonable signal-to-noise ratio in pregnant women beyond 24 weeks of gestation to assess fetal heart rate fluctuations [11, 13, 22, 32, 35]. Under this circumstance, the temporal resolution of ultrasound based CTG is insufficient.
As can be seen from Table 1, heart rate patterns account for a reasonable state determination particularly towards term when developmental synchronisation is high [ 9, 21, 23].
The respective impact of heart rate fluctuation analyses in children and adults was shown and standardised for time and frequency domain measures [34]. In the last decade, several complexity measures were introduced which have improved the heart rate analysis. Their impact on the assessment of fetal behavioural states is an outstanding question. One aspect, namely the complexity estimation of short states, is the subject of the present work.
The power spectrum depends on maturation of the autonomic nervous system [10, 19, 36] and fetal behavioural states [8]. Furthermore, the respiratory sinus arrhythmia [39] and the sympatho-vagal balance [41] were assessed by fetal MCG based heart rate power spectra. The established complexity measure of approximate entropy (ApEn) [25, 26] shows an overall increase and divergence with increasing gestational age and, hence, fetal maturation [35]. In this sample the apparent fetal behavioural state was not accounted for. The observed ApEn increase during maturation was lost in ApEn of binary heart rate pattern [7].
The resulting task is the search for a complexity measure and corresponding estimator appropriate for fetal behavioural state assessment. This includes the search for appropriate estimation parameters such as data length, embedding dimension, time scale, and coding. The data length is limited by the short duration of the behavioural states. An optimal embedding dimension can be found by a minimisation procedure [27]. Arbitrary time scales can lead to misleading or wrong complexity measures [16]. Complexity measures as function of the time scale of the different autonomic nervous system controllers were introduced in 1998 [14]. Here, the mutual information function was applied to a piglet’s heart rate during active and quiet sleep as well as during anesthesia, hypoxia, and vagal blockade. In the meantime, this approach representing the autonomic information flow (AIF) was confirmed both by experimental [16] and clinical [15] studies. The relevance of time scales was also confirmed by the concept of multiscale entropy [6]. The coding of heart rate by a few symbols, which improves the statistics for a given data length, may inflict an oversimplification leading to a loss of the physiologically relevant pattern [7] or accentuate relevant dynamics [37].
In order to overcome some of the shortcomings mentioned above, in the present work, the permutation entropy is investigated in the context of limited data length and in dependency on the time scale. The resulting complexity measures are evaluated with regard to the fetal behavioural state classification in connection with the conventional time and frequency domain measures of heart rate fluctuations. We test the hypothesis that parameters derived from permutation entropy can improve the classification of fetal behaviour states based on classical heart rate fluctuation parameters. Representative approximate entropy results are shown as reference.
2 Methods
2.1 Data acquisition and processing
From the database of fetal MCGs that was built up between 1999 and 2003 containing 129 recordings of apparently healthy fetuses, a sample was drawn of those subjects with fetuses between 35 and 40 completed weeks of gestation. Neither of the subjects received medication with known cardiac side effects. The studies on fetal MCG were approved by the local ethics committee of the Medical Faculty of the Friedrich Schiller University of Jena, Germany, and written informed consent was obtained from each subject prior to investigation. A total number of 39 recordings were selected containing at least one 5 min sequence (needed for visual classification) with less than 3% artifacts.
All measurements were taken in the magnetically shielded room (AK 3b, Vakuumschmelze Hanau, Germany) of the Biomagnetic Centre, Friedrich Schiller University Jena. The facilities provide a 31 channel SQUID Biomagnetometer (Philips), consisting of first order gradiometers (coil diameter 20 mm, baseline 70 mm, diameter of the array 145 mm). The pregnant women were positioned supine or with a slight twist to either side to prevent compression of the inferior vena cava by the pregnant uterus. The dewar was positioned with its curvature above the fetal heart after sonographic localisation as close as possible to the maternal abdominal wall without contact.
The SQUID signal was recorded over a period of 5 min with a sampling rate of 1,000 Hz using a filtering bandpass between 0.3 and 500 Hz (CURRY, Neuroscan, Neurosoft, Sterling, VG, USA). Simultaneously, a single lead ECG of the mother was recorded to distinguish between maternal and fetal cardiac activity.
The initial raw data underwent smoothing by a polynomial filter (Savitzky–Goolay, see [29]) to increase the performance of an algorithm described in detail by Schneider et al. [30]. Detection of the heartbeats was based on an algorithm of maximum coherence matching (MCM) on the basis of a representative QRS complex in the smoothed data. MCM was then performed in the maternal ECG first to determine the time instants of all maternal heartbeats, to average the maternal excitation cycles in each of the magnetic channels of the raw data set and, eventually, to subtract the maternal MCG from the raw data.
MCM of the fetal signals was, consecutively, performed in the magnetic channel with the highest signal-to-noise ratio following the same basic principles. Matching criteria were in some cases varied in order to obtain a close-to-complete list of beat-to-beat intervals. The beat-to-beat variability plot was used for plausibility control indicating false inter-beat intervals in the case of positive artefact matching [11, 30, 33].
2.2 Visual determination of the fetal state
Sonographic observation of the fetus is impossible during fetal MCG [38]. Therefore, under the assumption of high temporal coincidence of the state variables between 35 and 40 completed weeks of gestation, the actual fetal state was visually determined by classifying the heart rate pattern. The heart rate pattern was visualised from the train of beat-to-beat intervals and presented in a CTG like fashion (Fig. 1). State classification was based on visual integration of the pattern information over 5 min continuous CTG applying the criteria stated in Table 1 (‘the clinician’s eye’) by an obstetrician with daily routine experience of
non-stress test assessment. From each train, a representative sequence of 256 heartbeats was chosen. The necessity to proceed in such fashion is illustrated in the examples in Fig. 1. If the pattern changed during the 5 min of recording, the 256 beats sequence was taken from only one of the patterns. The number of fetal heartbeat sequences in each state is shown in Fig. 1.
2.3 Permutation entropy
Permutation entropy is a complexity measure for time series operating on an ordinal level, i.e., only the ranks of the data in the time series are regarded, not the distances (metric) of the data. Its main features are (1) robustness with respect to some noise possibly corrupting the data, and (2) its easy computation (estimation). Permutation entropy measures the entropy of sequences of ordinal patterns derived from m-dimensional delay embedding vectors. In the following, we briefly summarize the definition of the permutation entropy. A more detailed introduction can be found in [3, 5].
The scalar time series {x(t)} T t=1 is embedded into an m-dimensional space X t = [x(t), x(t + L),..., x(t + (m − 1)L)], where m is called the embedding dimension and L the embedding delay time. For m=2, there are two possible ordinal patterns of X t , namely π 1 = x(t) < x(t + L) and π 2 = x(t + L) < x(t). (For this moment, we suppose that there are no equal values in X t , i.e., no tied ranks.) For m=3, X t can attain one of six different order patterns,
In general, there are just m! possible order patterns, which is the number of permutations of the m coordinates in X t . Now, let p(π) denote the relative frequency of order pattern π,
Then, for fixed embedding dimensions m≥2, and fixed delay L, permutation entropy is defined as
where the sum runs over all m! patterns π.
Equal values in the time series, which can occur because of the limited accuracy of measurement, will be treated as follows. In case X t contains two equal values x a (t + aL) = x b (t + bL), a, b = 0,1,...,(m − 1), the relative frequency of the permutations which correspond to the cases x a < x b and x a > x b is increased by 1/2. For n equal values the respective n! permutations are increased by 1/n!. Practically, this can be done by adding a random number to the data, which is smaller than the accuracy of measurements.
For convenience we normalize H (m, L) by its maximum value log2 m!
Now we introduce the (normalized) Kullback–Leibler entropy (KLE, [20])
which is an information measure for the distance between the probability distribution of the ordinal patterns (permutations) and the uniform distribution. With increasing complexity of the time series, KLE decreases until it reaches zero for noise [independent and identically distributed (iid) process] that corresponds to a uniform distribution of all patterns. Note that due to our handling of tied ranks, a constant series would also provide KLE=0.
We have to choose appropriate values for m and L. The value of m should be at least three, the maximum is limited by the length of time series. For an accurate estimation of KLE, the length of the time series must be considerably larger than the factorial of the embedding dimension. This allows for short series of 256 heartbeats only embedding dimensions m=3 and m=4. We tried both values and could not find significant differences in the discriminatory impact of the respective entropy measures. On account of computation rate and memory requirements we chose m=3. To get an equidistant time scale, all data were resampled with a sampling frequency of 10 Hz; in this way, the time base of the autonomic modulations is suitably considered [16].
Figure 2 shows the KLE of order 3 for varying delay time L, grouped by the visual classified behavioural states 1F, 2F and 4F.
2.4 Discrimination parameters
For the classification of the behavioural states, the following complexity parameters were calculated for each 256-heartbeat sequence: The term KLE_1 indicates the KLE for a delay time of 0.4 s. That corresponds to about one heartbeat period in the NN-interval time series reflecting beat-to-beat variability. The term KLE_Mean indicates mean KLE, averaged over embedding delays in the short-term range up to 2 s reflecting the structure of the KLE(L) curves around the first maximum (Fig. 2).
As reference we include two parameters from the approximate entropy, which has been employed in the past to infant heart rate data [26].
The ApEn was calculated from heartbeat interval series by means of the software of Kaplan [18] setting the embedding lag to 1 and the embedding dimension to 2. Analogous to Pincus [26], we determined the parameters ApEn_sub and ApEn_pop. For the calculation of ApEn_sub, the filter factor r, which sets the length scale over which to compute the approximate entropy, is adjusted to 0.2 of the standard deviation of each individual data set. ApEn_pop is determined with the same r for all data taken from 0.2 of the population average standard deviation.
In addition to these complexity parameters, the classical parameters for the measurement of heart rate fluctuation [34] have been determined. We consider the following four parameters in the time and frequency domain corresponding to different time scales: In the time domain SDNN, the standard deviation of the NN-intervals, reflects the overall variability in the time series, RMSSD is the square root of mean squared differences of successive NN intervals (both values in ms). In the frequency domain, LF is the power in the range between 0.04 and 0.15 Hz and HF describes the power in the range between 0.15 and 0.4 Hz. We consider the natural logarithm of these values (in ms2).
2.5 Statistical analysis
The discrimination parameters have been evaluated with different statistical methods contained in the statistical software package SPSS [31].
Firstly, we visualised the discriminating power of each parameter by means of boxplots (Fig. 3).
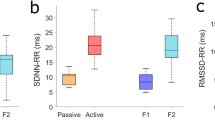
Boxplots for the parameters SDNN, RMSSD, ln(LF), ln(HF), KLE_Mean, KLE_1, ApEn_sub, ApEn_pop of the data of 35 healthy fetuses, grouped by behavioural states (1F, 2F, 4F). Each grey box represent the interquartil range which contains 50% of values with a line at the median. The whiskers show overall data range, excluding outliers and extreme values. Outliers (circle) are cases with values between 1.5 and 3 box lengths below 25th or above 75th percentile. Extreme values (asterisk) are cases with values more than 3 box lengths below 25th or above 75th percentile
Secondly, statistical tests have been applied to each pair of parameters (Table 2). Because the t-test can only be applied in case of homogeneous variances we used Levene’s test to check the homogeneity of variances (p>0.1) for each pair of states and all parameters. In case, the homogeneity assumption has been met, the t-test, otherwise Welch’s test, which can deal heterogeneous variances also, was used to decide if the respective parameter value allows a significant discrimination (p<0.05) of the states.
The crucial evaluation of the discriminating power was done by means of linear discriminant analysis [1, 4, 31]. All results have been cross-validated. That means, each case is classified by functions derived from all cases other than that case (leave-one-out method). We accomplished for each parameter and for each possible bivariate parameter set a statistical discriminant analysis. Furthermore, we have performed a stepwise analysis. In the latter case, variables are added to the discriminant functions one by one until it is found that adding extra variables does not give significantly better discrimination. The selection of the variables is based on Wilks’ lambda (entry criteria p<0.05). Using a Bayesian approach the software package SPSS permits prior probabilities of group memberships to be taken into account. Prior probabilities for the states have been taken from Table 1.
Finally, the parameter space of the best separating parameters is graphically presented.
3 Results
Because of the small sample size of 3F data (n=4), the 3F data sets have been excluded from statistical state classification. Concerning states 1F (n=11), 2F (n=16), and 4F (n=8), Fig. 2 shows that KLE allows assessment of the complexity of heart rate fluctuations on different time scales. KLE shows significant changes with L, especially in the short-term range (up to about 5 heartbeat periods or 2 s). Starting from a delay time that suits the mean heartbeat interval (0.42 s), all curves raise to a local maximum at about 1.2 s. (For smaller values of the delay time L→0 the curves approach KLE(L=0)=1. This is a consequence of the definition of KLE and has no physiological meaning.) For large delay times all curves approach zero, that means, all ordinal patterns comply with a uniform distribution. The curves corresponding to active states 2F and 4F are clearly above the curves corresponding to quiet state 1F.
Boxplots in Fig. 3 give an overview on how the selected parameters might discriminate between the fetal states. All parameters appear to be able to separate at least some of the states. SDNN and ln(LF) seem to separate between the states 1 and 2 whereas the complexity measures between states 2 and 4.
The results of t-test (Table 2) indicate that none of the linear parameters alone is able to separate all states. Only the complexity parameters (KLE_Mean, KLE_1) discriminate all states considered.
The crucial method used for state discrimination was discriminant analysis. Results are shown in Table 3. Employing univariate analysis SDNN (80.0%) and KLE_Mean (77.1%) show the best prediction rates. Employing bivariate analysis the best prediction rate has been achieved using the pair KLE_Mean and ln(LF) (94.3%), followed by the pairs KLE_Mean and RMSSD (88.6%), KLE_Mean and ln(HF) (88.6%). A multivariate analysis with more than two parameters could not improve the best result.
The significance of these differences between prediction rates has been evaluated by the entry criteria of the stepwise discrimination procedure. Starting from linear variables (SDNN or pairs of linear parameters) and adding the parameter KLE_Mean, this parameter gives a significantly better discrimination for all cases (p<0.0005). The other way around, starting from the variables KLE_Mean and ln(LF), none of the linear parameters SDNN (p=0.067), RMSSD (p=0.875), ln(HF) (p=0.906) give a significant improvement.
For the best parameter pair in the discriminant analysis KLE_Mean and ln(LF) the classification results are shown in more detail in Table 4, a graphical representation is given in Fig. 4. Classification errors occurred for one of the 2F data and one of the 4F data.
Graphical representation of the states classification in the parameter space of ln(LF) and KLE_Mean. The visual estimated state classification is indicated by symbols. Boundaries between the classification regions are shown by solid lines. Two classification errors occurred (2F state in 1F region, 4F state in 2F region)
4 Discussion
The focus of the present work was the improvement of fetal state classification by complexity measures in comparison to classical HRV measures under particular consideration of appropriate time scales. For this purpose, permutation entropy has been used because of its methodological advantages. A main challenge of heart rate complexity estimation of sleep states is their short duration. The permutation entropy and, hence, the derived KLE is able to handle short data sequences and different time scales. In Fig. 2 it is shown that the prediction over special time horizons plays a crucial role for the classification and offers possibilities of physiological interpretation. Higher values of KLE for the active states 2F and 4F than for the quiet state 1F reflect the better predictability of heart rate fluctuations in 2F and 4F. This is pronounced at the local maxima of the curves between 0.5 and 2 s. It points to sympathetic modulations of slow rhythms, which are less pronounced for shorter prediction horizons (beat-to-beat). The KLE of the probability distribution of ordinal patterns (order m=3) with respect to the uniform distribution, averaged over embedding delays between 0.1 and 2 s (KLE_Mean), is found to be a suitable complexity measure with high discriminating value on the behavioural states.
The active states 2F and 4F are characterised by accelerations and decelerations, which lead to higher bandwidths and decreased complexity. State 4F is distinguishable from State 2F only by the length and frequency of accelerations. The relative number of monotone ordinal patterns increases with increasing length and frequency of accelerations and decelerations. Therefore, all complexity measures (KLE_Mean, KLE_1, ApEn_sub, ApEn_pop) discriminate clearly between states 2F and 4F whereas classical measures cannot discriminate between these both states.
Best classification result for the states could be reached by means of a bivariate discriminant analysis combining the complexity parameter KLE_Mean with the classical parameter ln(LF) (Table 3). The results of univariate analysis show that parameters describing slow heart rate variations (SDNN, ln(LF), KLE_Mean) contribute better to the discrimination than parameters describing fast variations (RMSSD, ln(HF), KLE_1). This shows the importance of selection of appropriate time scales that could be the reason of the observed advantages of KLE in comparison with ApEn. However, a systematical comparison of both methods was not the subject of this work.
The presented technique of fetal behavioural state classification by including a short window assessment of heart rate complexity provides a remarkable development of the established methods. Previous attempts of sleep state classification from HRV patterns were based on CTG recordings and were done using linear HRV measures [17]. As far as we know, complexity of fetal heart rate fluctuations was assessed for longer data sets only and by disregarding the inevitably shorter sleep states [24, 26, 35]. A corresponding first approach of complexity estimation of sleep state dynamics in a newborn piglet is described by Hoyer et al. [14]. The different discriminatory value of different time scales of KLE, found in the study presented here, confirm the physiological and discriminatory relevance of time scale dependent complexity measures such as found in the prognosis of multiple organ dysfunction syndrome patients [15].
In the present study, the subjective visual state classification was done based on objective criteria and it was reproducible within the investigator groups of the Friedrich Schiller University. Furthermore, it was done prior the statistical analysis and, therefore, did not influence it.
Because of the small sample size, the 3F state was excluded from the statistical discrimination. Reasons are the small overall amount of data in connection with the distribution of the states 1F–4F, which correspond to findings of other authors. Also Arabin and Riedewald have neglected the 3F state because of its low appearance [2]. The analysis of data sets representative for the 3F state require the acquisition of much more data which might require a multicenter study. Otherwise, the low appearance of 3F state makes its importance in clinical routine diagnostic questionable.
We conclude that the KLE of heart rate fluctuations contributes to improved automatic behavioural state classification and the analysis of fetal behavioural dynamics.
References
Afifi AA, Clark V (1996) Computer-aided multivariate analysis. Chapman and Hall, London
Arabin B, Riedewald S (1992) An attempt to quantify characteristics of behavioral states. Am J Perinatol 9:115–119
Bandt C, Pompe B (2002) Permutation entropy: a natural complexity measure for time series. Phys Rev Lett 88:174102
Brosius F (2002) SPSS 11. mitp, Bonn
Cao Y, Tung WW, Gao JB, Protopopescu VA, Hively LM (2004) Detecting dynamical changes in time series using the permutation entropy. Phys Rev E Stat Nonlin Soft Matter Phys 70:046217
Costa M, Goldberger AL, Peng CK (2002) Multiscale entropy analysis of complex physiologic time series. Phys Rev Lett 89:068102
Cysarz D, van Leeuwen P, Bettermann H (2000) Irregulaties and nonlinearities in fetal heart period time series in the course of pregnancy. Herzschr Elektrophys 11:179–183
Davidson SR, Rankin JH, Martin CB Jr, Reid DL (1992) Fetal heart rate variability and behavioral state: analysis by power spectrum. Am J Obstet Gynecol 167:717–722
DiPietro JA, Hodgson DM, Costigan KA, Hilton SC, Johnson TR (1996) Fetal neurobehavioral development. Child Dev 67:2553–2567
Ferrazzi E, Pardi G, Setti PL, Rodolfi M, Civardi S, Cerutti S (1989) Power spectral analysis of the heart rate of the human fetus at 26 and 36 weeks of gestation. Clin Phys Physiol Meas 10(Suppl B):57–60
Grimm B, Haueisen J, Huotilainen M, Lange S, Van Leeuwen P, Menendez T, Peters MJ, Schleussner E, Schneider U (2003) Recommended standards for fetal magnetocardiography. Pacing Clin Electrophysiol 26:2121–2126
Groome LJ, Bentz LS, Singh KP (1995) Behavioral state organization in normal human term fetuses: the relationship between periods of undefined state and other characteristics of state control. Sleep 18:77–81
Horigome H, Takahashi MI, Asaka M, Shigemitsu S, Kandori A, Tsukada K (2000) Magnetocardiographic determination of the developmental changes in PQ, QRS and QT intervals in the foetus. Acta Paediatr 89:64–67
Hoyer D, Bauer R, Pompe B, Palus M, Zebrovski J, Rosenblum M, Zwiener U (1998) Nonlinear Analysis of the Cardiorespiratory Coordination of a Newborn Piglet. In: Kantz K, Kurths J, Mayer-Kress G (eds) Nonlinear analysis of physiological data, springer series of synergetics. Springer, Berlin Heidelberg New York, pp 167–190
Hoyer D, Friedrich H, Zwiener U, Pompe B, Baranowski R, Werdan K, Müller-Werdan U, and Schmidt H (2005a) Prognostic Impact of Autonomic Information Flow in Multiple Organ Dysfunction Syndrome Patients. Int J Cardiology (in press). online: doi: 10.1016/j.ijcard.2005.1005.1031
Hoyer D, Pompe B, Chon KH, Hardraht H, Wicher C, Zwiener U (2005b) Mutual information function assesses autonomic information flow of heart rate dynamics at different time scales. IEEE Trans Biomed Eng 52:584–592
Jongsma HW, Nijhuis JG (1986) Classification of fetal and neonatal heart rate patterns in relation to behavioural states. Eur J Obstet Gynecol Reprod Biol 21:293–299
Kaplan D (1998) HRV software. http://www.macalester.edu/~kaplan/hrv/doc/Feb3snap.tar
Karin J, Hirsch M, Akselrod S (1993) An estimate of fetal autonomic state by spectral analysis of fetal heart rate fluctuations. Pediatr Res 34:134–138
Kullback S (1968) Information theory and statistics. Dover Publications, Inc, Mineola
Nijhuis JG, Prechtl HF, Martin CB Jr, Bots RS (1982) Are there behavioural states in the human fetus? Early Hum Dev 6:177–195
Peters M, Crowe J, Pieri JF, Quartero H, Hayes-Gill B, James D, Stinstra J, Shakespeare S (2001) Monitoring the fetal heart non-invasively: a review of methods. J Perinat Med 29:408–416
Pillai M, James D (1990) The development of fetal heart rate patterns during normal pregnancy. Obstet Gynecol 76:812–816
Pincus SM, Viscarello RR (1992) Approximate entropy: a regularity measure for fetal heart rate analysis. Obstet Gynecol 79:249–255
Pincus SM, Gladstone IM, Ehrenkranz RA (1991) A regularity statistic for medical data analysis. J Clin Monit 7:335–345
Pincus SM, Cummins TR, Haddad GG (1993) Heart rate control in normal and aborted-SIDS infants. Am J Physiol 264:R638–646
Porta A, Baselli G, Liberati D, Montano N, Cogliati C, Gnecchi-Ruscone T, Malliani A, Cerutti S (1998) Measuring regularity by means of a corrected conditional entropy in sympathetic outflow. Biol Cybern 78:71–78
Prechtl HF (1974) The behavioural states of the newborn infant (a review). Brain Res 76:185–212
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1992) Numerical recipes in C. Cambridge University Press, Cambridge
Schneider U, Schleussner E, Haueisen J, Nowak H, Seewald HJ (2001) Signal analysis of auditory evoked cortical fields in fetal magnetoencephalography. Brain Topogr 14:69–80
SPSS for Windows R (2002) SPSS Inc., Chicago
Stinstra J, Golbach E, van Leeuwen P, Lange S, Menendez T, Moshage W, Schleussner E, Kaehler C, Horigome H, Shigemitsu S, Peters MJ (2002) Multicentre study of fetal cardiac time intervals using magnetocardiography. BJOG 109:1235–1243
Sturm R, Muller HP, Pasquarelli A, Demelis M, Erne SN, Terinde R, Lang D (2004) Multi-channel magnetocardiography for detecting beat morphology variations in fetal arrhythmias. Prenat Diagn 24:1–9
Taskforce (1996) Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Task force of the European society of cardiology and the North American Society of pacing and electrophysiology. Eur Heart J 17:354–381
Van Leeuwen P, Lange S, Bettermann H, Gronemeyer D, Hatzmann W (1999) Fetal heart rate variability and complexity in the course of pregnancy. Early Hum Dev 54:259–269
Van Leeuwen P, Geue D, Lange S, Cysarz D, Bettermann H, Gronemeyer DH (2003) Is there evidence of fetal-maternal heart rate synchronization? BMC Physiol 3:2
Voss A, Kurths J, Kleiner HJ, Witt A, Wessel N, Saparin P, Osterziel KJ, Schurath R, Dietz R (1996) The application of methods of non-linear dynamics for the improved and predictive recognition of patients threatened by sudden cardiac death. Cardiovasc Res 31:419–433
Wakai RT (2004) Assessment of fetal neurodevelopment via fetal magnetocardiography. Exp Neurol 190(Suppl 1):S65–S71
Wakai RT, Wang M, Pedron SL, Reid DL, Martin CB Jr (1993) Spectral analysis of antepartum fetal heart rate variability from fetal magnetocardiogram recordings. Early Hum Dev 35:15–24
Wakai RT, Wang M, Martin CB (1994) Spatiotemporal properties of the fetal magnetocardiogram. Am J Obstet Gynecol 170:770–776
Zhuravlev YE, Rassi D, Mishin AA, Emery SJ (2002) Dynamic analysis of beat-to-beat fetal heart rate variability recorded by SQUID magnetometer: quantification of sympatho-vagal balance. Early Hum Dev 66:1–10
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (HO 1634/9-1, KA 1726/2-1). The authors like to thank Barbara Grimm and Alina Schneider for their assistance in obtain the raw data and the team of the Biomagnetic Center, Department of Neurology, Friedrich Schiller University Jena for their technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Frank, B., Pompe, B., Schneider, U. et al. Permutation entropy improves fetal behavioural state classification based on heart rate analysis from biomagnetic recordings in near term fetuses. Med Bio Eng Comput 44, 179–187 (2006). https://doi.org/10.1007/s11517-005-0015-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-005-0015-z