Abstract
Purpose
Changes in microbial communities during natural succession in semiarid areas have been widely studied but their association with plant and soil properties remains elusive. In the present study, we investigated plant characteristics, rhizosphere soil variables, and microbial communities along a chronosequence of grasslands forming on abandoned farmland on the Chinese Loess Plateau.
Materials and methods
Rhizosphere samples were collected from the early-stage dominant plant Artemisia capillaris from farmland abandoned for 5, 10, and 15 years and from the late-stage dominant plant Artemisia sacrorum from farmland abandoned for 10, 15, 20, and 30 years. Microbial community composition, including bacteria and fungi, was determined by high-throughput sequencing. Microbial succession rates represented by temporary turnover were assessed using the slope (w value) of linear regressions, based on log-transformed microbial community similarity over time.
Results and discussion
Cover and aboveground biomass of A. capillaris tended to decrease, whereas those of A. sacrorum increased during the succession. Although the rhizosphere bacteria of A. capillaris transitioned from Proteobacteria-dominant to Actinobacteria-dominant, the bacteria of A. sacrorum exhibited the opposite trend. Bacterial and fungal community diversity tended to increase logarithmically with increasing plant aboveground biomass, indicating that an increase in plant biomass could lead to enhanced rhizosphere microbial diversity, but the rate of enhancement decreased gradually. A lower temporary turnover rate of bacterial and fungal communities in the rhizosphere than that in the bulk soil indicated a higher successional rate of the rhizosphere microbial community. Levels of soil nutrients, such as organic carbon, nitrate nitrogen, and ammonium nitrogen, were closely associated with the abundance and diversity of bacterial and fungal communities, indicating their critical role in shaping the rhizosphere microbial community.
Conclusions
Our results indicate a close association between plant succession and rhizosphere microbial succession in a semiarid area. Plants affect the microbial communities possibly by changing the nutrient input into the rhizosphere.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Secondary succession is an efficient approach used to restore the ecological functions of degraded grasslands, including aboveground diversity and biomass, soil structure, and nutrient level Lankau and Lankau 2014; Lozano et al. 2014; Zhang et al. 2015). Plant succession is an interactive process between plants and soil microorganisms (Walker et al. 2007). In this process, plants affect microorganisms and determine soil status, including physical, chemical, and biological attributes, by the decomposition of litter, turnover of roots, and release of exudates (Jones et al. 2009). In turn, the soil microbiota plays critical roles in the degradation of organic matter, fixation of nitrogen, and transformation of elemental forms and hence greatly affects plant growth and establishment (Klironomos 2003; Kulmatiski et al. 2008). Interactions between plants and microorganisms play essential roles in maintaining the stability of ecosystem structure and function (Kardol et al. 2013). Some studies have reported spatial and temporal variation in microbial communities and associations with plant communities during the succession of different ecosystems, such as secondary forests (Zhang et al. 2018), abandoned mining land (Harantová et al. 2017), and fire-managed grassland (Munoz-Rojas et al. 2016). However, the association between plants and microorganisms is not fully understood, especially in the semiarid and arid ecosystems where plant diversity and composition change rapidly despite the increasing interest in studying this type of ecosystem (Foster and Tilman 2000; Lozano et al. 2014).
Although the aboveground plant community during succession is readily distinguishable and can be predictable, the dynamics of belowground microbial communities are more difficult to determine (e.g., require modern analytical techniques) and therefore remain poorly understood. There is little information regarding microbial communities (Knelman et al. 2015; Zhang et al. 2016); therefore, some important questions related to the succession process remain poorly understood, such as the successional trajectory of microbial communities over time and the factors driving microbial succession. The dynamics of soil microbial communities have recently been reported along successional gradients, including in grasslands and forests, with shifts from early-stage dominant taxa to late-stage dominant taxa (Kuramae et al. 2010; Cline and Zak 2015). Microbial turnover has been reported in some studies as succession proceeds and some species that survive well in nutrient-poor environments were found to be replaced by those assumed to be good competitors in soils rich in nutrients (Blaalid et al. 2012; Davey et al. 2015). Succession without substitution has also been reported in other studies, in which early-stage microbial populations were not necessarily replaced by late-stage populations, thereby increasing the overall diversity as the succession developed (Peay et al. 2011). Although successional soil microbial communities have been widely investigated, the association between plants and microbial communities remains elusive, especially in the rhizosphere zone of plants where microorganisms are diverse and specific due to the influence of root exudates. Rhizosphere microorganisms perform essential roles in the soil ecosystem that contribute to soil nutrient cycling, healthy root growth, and the establishment of plant communities (Marschner et al. 2001). These important functions have led to considerable interest in understanding the interactions between rhizosphere microbes, soil properties, and plant characteristics in various ecosystems.
Plant succession is characterized by the substitution of different species through time, and the changes in plant species and plant growth alter their rhizosphere conditions, e.g., by decreasing soil pH (Tscherko et al. 2004), increasing nutrient levels (Siciliano et al. 2014), and reducing reduction potential (Spohn et al. 2015), which consequently influences the survival, growth, and activity of microorganisms. For instance, Tscherko et al. (2004) found that a decrease in soil pH and soil C/N ratio mediated the shifts in the rhizosphere microbial community composition of Poa alpina across an alpine chronosequence and that the soil environment determined microbial community composition in the initial stage, while the host plant selected for specific microbes in the later mature stage. Our previous studies have also observed a distinct successional trend in the rhizosphere microbial diversity of different successional species communities (Zhang et al. 2015) and soil C supply plays an important role in shaping microbial communities. To better understand how shifts in plant communities affect rhizosphere microbial communities, study of the relationships between microbial and plant communities is required.
The Chinese Loess Plateau, a typical semiarid area, has undergone land use change in the past 30 years (Deng et al. 2014). Most sloped farmlands in this region have been converted to grassland or forest to restore the degraded soil environment. Among them, the abandonment of sloped farmlands above 15° for natural restoration has been considered to be an efficient measure for improving soil nutrient levels and aggregate status (Wang et al. 2009). The natural grasslands of different ages that were formed in the 30 years of secondary succession offered an opportunity to evaluate the succession of plants and microorganisms, as well as their relationship during the natural restoration process. In this study, we investigated the rhizosphere microbial communities (bacterial and fungal) of two species that are dominant during succession along a 30-year succession of abandoned farmland on the Loess Plateau of China. Bacterial and fungal communities were detected using 16S rRNA and internal transcribed spacer (ITS) high-throughput sequencing based on Illumina MiSeq PE300 platform. The objectives of the present study were to (i) evaluate the succession trend of the two dominant plants and their rhizosphere microbial communities during the succession, (ii) determine the association between plant succession and microbial community diversity, and (iii) verify the edaphic factors driving the succession of bacterial and fungal communities.
2 Materials and methods
2.1 Description of experimental area
Our study was carried out at the Zhifanggou catchment (109° 16′ E, 36° 46′ N) located on the Loess Plateau, northern Shaanxi Province, China. The mean annual precipitation is 509 mm and the average temperature is 10.0 °C. A minimum of − 6.0 °C occurred in February and a maximum of 37.5 °C in August. The climate in this region is a mixture of semiarid and arid. The parental soil is a gypsum siltstone derived from wind-blown deposits and is classified as a loessial soil (a Calcaric Cambisol in the FAO classification). The vegetation coverage in the catchment is about 30% and the predominant vegetation is Stipa bungeana Trin, Artemisia sacrorum Ledeb, Lespedeza davurica (Laxm.) Schindl, Caragana korshinskii Kom, and Robinia pseudoacacia L.
2.2 Chronosequence selection
The Chinese government launched the “Green for Grain” project in the 1980s to restore the degraded environments of the Loess Plateau. Most of the sloped cropland in this area was abandoned for secondary succession without human disturbance. After 30 years of natural restoration, a chronosequence of grasslands developed from the abandoned croplands. In our study, five grasslands with different durations of time since abandonment (5, 10, 15, 20, and 30 years) were selected in September 2014. The ages of the grasslands, i.e., the number of years since abandonment, were determined by consulting local residents and governmental land records. These sites were similar in terms of gradient and elevation and had been subjected to similar farming practices prior to abandonment. Rhizosphere soil samples were collected from the roots of the early-successional grass species Artemisia capillaris from farmland abandoned for 5, 10, and 15 years and from the roots of the late-successional grass species A. sacrorum from farmland abandoned for 10, 15, 20, and 30 years.
2.3 Plant and soil sampling
In September 2014, three 400-m2 (20 m × 20 m) plots were established in each grassland. Six 1 m × 1 m subplots were randomly set up in each plot to survey the plant communities, and the percentage coverage of each species was calculated. Twelve plants of each species (A. capillaris and A. sacrorum) were randomly selected from each plot for biomass measurement and soil collection. After digging up the plant, the aboveground parts were clipped and dried at 60 °C for 36 h to obtain the aboveground biomass, rhizosphere soil was gathered by separating the soil adhered to the roots using forceps, and the roots were washed with tap water and subsequently dried at 60 °C for 48 h to measure root biomass. Rhizosphere soil samples were collected from each plot, homogeneously mixed, and sieved through a 2-mm mesh to form one composite sample. After excluding the stones and plant material, the soil samples were sieved through a 2-mm mesh and divided into two pats. One part (~ 100 g) was stored at − 80 °C for microbial DNA analysis, and another (~ 400 g) was air-dried for the measurement of physicochemical properties. All analyses were completed within 1 week of collection.
2.4 Soil chemical analyses
Organic carbon (OC) content was determined using dichromate oxidation (Nelson and Sommers 1982); total nitrogen (TN) content was measured using the Kjeldahl method (Bremner and Mulvaney 1982); total phosphorus (TP) content was determined by melt-molybdenum, antimony, and scandium colorimetry; and available P content was determined by the Olsen method (Olsen and Sommers 1982). NH4+-N and NO3−-N contents were determined using a continuous-flow autoanalyzer (Alpkem, OI Analytical, USA) after the extraction of fresh soil with 2 M KCl for 18 h (Zhang et al. 2016). Soil pH was determined using an automatic titrator (Metrohm 702, Herisau, Switzerland) in suspensions of 1:2.5 soil/water.
2.5 Soil microbial composition: high-throughput sequencing
2.5.1 DNA extraction, purification, and quantification
DNA was extracted from 0.5 g of homogenized soil samples using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Solon, USA) following the manufacturer’s instructions and was purified by extraction from a 2% agarose gel. The quality of the DNA extracts was determined using a NanoDrop® ND-1000 spectrophotometer (Thermo Scientific, Wilmington, USA) after being diluted tenfold. The final soil DNA concentrations were quantified with PicoGreen using a FLUOstar Optima fluorometer (BMG Labtech, Jena, Germany).
2.5.2 Amplification, sequencing, and data processing
The V4 hypervariable region of the bacterial 16S rRNA gene and the fungal internal transcribed spacer (ITS) gene were amplified by PCR. The primers, PCR procedures, and data processing, including high-quality sequencing, calculation of a-diversity, and taxonomic identification based on operational taxonomic units (OTUs), were carried out as per Zhang et al. (2016, 2017). Purified amplicons were sequenced using an Illumina MiSeq PE300 platform (Illumina Corporation, San Diego, USA). The raw sequences were deposited in the NCBI Sequence Read Archive (accession numbers SRP093941 and SRP093942). To compare the soil samples at the same sequencing depth, we selected 37,003 16S rRNA sequences and 28,192 ITS sequences by random sampling.
2.6 Data analysis
One-way analysis of variance was performed to evaluate the differences in soil properties, microbial diversity, and relative abundance of the bacterial and fungal taxa between the successional stages; the significance level was set at 0.05. Post hoc comparisons were conducted using Fisher’s least significant difference (LSD) test. Results are expressed as means ± standard error. Similarity among successional stages in microbial community composition was evaluated by an analysis of similarity (ANOSIM) with Bray-Curtis dissimilarity. The successional rate of the microbial community represented by the temporal turnover of species in the rhizosphere and bulk soils (data pertaining to the bacteria and fungi in bulk soils is from Zhang et al. 2016, 2017) was investigated. The time-decay relationship model for assessing the succession rate of microbes in the form: ln(Ss) = constant − w ln (T), where Ss is the pairwise similarity in community composition, T is the time interval, and w is the turnover rate of the microbial species over time (Horner-Devine et al. 2004; Liang et al. 2015). Pearson correlation coefficient was used to determine the correlations between the relative abundance of microbial communities, plants, and soil variables. We also performed a redundancy analysis (RDA) to identify the effects of plants and soils on microbial community composition, and the significance was testified using Monte Carlo with 999 permutations. The ANOSIM, Pearson correlation, and RDA were conducted using the vegan package of R-3.0.1 (R Development Core Team 2013).
3 Results
3.1 Vegetation characteristics
During the 30 years of succession, the pioneer species A. capillaris dominated in terms of cover and aboveground biomass in the first 15 years after abandonment, whereas the middle-late-successional species A. sacrorum dominated after 15 years (Table 1). Coverage and above and root biomass decreased for A. capillaris over time. Coverage was 45.2% and aboveground biomass was 181.4 g m−2 at the 5-year site, which decreased to 2.4% and 19.5 g m−2, respectively, at the 15-year site, after which A. capillaris disappeared. Some perennial species, such as Heteropappus altaicus, Salsola collina, and Phragmites australis, appeared and coexisted with A. capillaris (data not shown). A. sacrorum appeared in the ecosystem after 10 years as a companion species increasing in abundance and replacing A. capillaris to become the dominant species in the 15-year site and thereafter continuing to increase.
3.2 Rhizosphere soil properties
The dynamics of rhizosphere soil nutrient content differed between the two dominant species along the successional gradient (Table 1). The contents of organic C, NO3−-N, and NH4+-N in the rhizosphere of A. capillaris significantly decreased with increasing time of succession (P < 0.05), whereas the organic C, total N, and NO3−-N contents in the rhizosphere of A. sacrorum increased (P < 0.05). The rhizosphere pH for both species did not differ significantly among the stages (P > 0.05). The available P content of both plants decreased from the 5-year site to the 15-year site.
3.3 Rhizosphere microbial community
3.3.1 Microbial community diversity
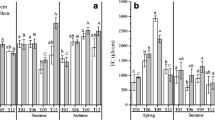
An average of 37,003 bacterial sequences and 28,192 fungal sequences per sample were obtained from the complete data set, of which 2161 and 485 OTUs were identified, respectively. All rarefaction curves for the bacterial and fungal OTUs tended to approach saturation at a similarity level of 97% (Fig. S1, Electronic Supplementary Material—ESM), suggesting that sufficient sequences were collected, and the bacterial and fungal communities were sampled completely. Two indices of rhizosphere bacterial diversity, Chao1 and Shannon, behaved similarly for A. capillaris (Table 2) decreasing with increasing time of succession. In contrast, the bacterial diversity of A. sacrorum increased along the successional gradient. The indices of fungal community diversity decreased with time of succession for A. capillaris. The highest Chao1, species number, and Shannon index of A. sacrorum occurred at the 30-year site.
3.3.2 Microbial community composition
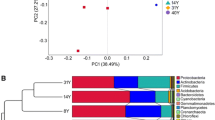
The dominant bacterial phyla in the rhizospheres of A. capillaris and A. sacrorum were Proteobacteria (24.4 and 36.5%, respectively), Actinobacteria (46.0 and 27.0%), Acidobacteria (11.8 and 15.9%), Chloroflexi (8.5 and 8.2%), Gemmatimonadetes (5.1 and 4.9%), and Bacteroidetes (2.4 and 2.7%) (Figs. 1 and 2). The abundance of the dominant group in the rhizosphere of A. capillaris, Proteobacteria, decreased with time of succession (Fig. 1a), whereas the abundance of the other dominant group, Actinobacteria, increased (P < 0.05). This was evident in Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria (Fig. 1b). The relative abundance of Chloroflexi, Acidobacteria, and Gemmatimonadetes did not differ significantly (P > 0.05). In contrast to A. capillaris, the relative abundance of Proteobacteria in the rhizosphere of A. sacrorum increased with time of succession, whereas the abundance of Actinobacteria decreased (P < 0.05) (Fig. 2a). This was significant in Alphaproteobacteria, Gammaproteobacteria, and Actinobacteria (Fig. 2b). The fungal communities of the A. capillaris and A. sacrorum rhizospheres consisted mainly of Ascomycota (78.4 and 72.1%, respectively), Zygomycota (6.8 and 7.4%), Basidiomycota (5.8 and 6.9%), and Glomeromycota (1.1 and 1.0%) (Figs. 3 and 4). No obvious trend was found in the rhizosphere fungal communities at the phylum level for the two plants over time. However, at the class level, the abundance of Dothideomycetes significantly decreased with successional time in the rhizosphere of A. capillaris (Fig. 3b), while it increased with time in the rhizosphere of A. sacrorum (Fig. 4b). Results from the ANOSIM (Table 3) clearly identified variation in the microbial communities among the stages. For both A. capillaris and A. sacrorum, the bacterial and fungal community composition was significantly separated by successional stage. Between A. capillaris and A. sacrorum, bacterial and fungal communities in the rhizospheres of A. capillaris and A. sacrorum did not differ at the 10-year site but differed significantly at the 15-year site (P < 0.05); this was confirmed in a Venn plot showing that the rhizospheres of the two plants shared the most OTUs at the 10-year site but only a few OTUs at the 15-year site (Fig. S2. ESM).
Figure 5 shows the successional rate of the soil microbial community during the succession. Compared to the successional rates of bacterial (w = 0.040, P < 0.001) and fungal (w = 0.041, P < 0.001) communities in the bulk soil, significantly steeper slopes of bacterial (w = 0.054, P < 0.001) and fungal (w = 0.067, P < 0.001) communities were observed in the rhizosphere of A. capillaris (Fig. 5a, b). Similarly, the w values of the bacterial (w = 0.070, P < 0.001) and fungal (w = 0.071, P < 0.001) communities were higher in the rhizosphere of A. sacrorum than those of the bulk soil (Fig. 5c, d). These results suggested a higher microbial turnover in the rhizosphere compared with the bulk soil.
3.4 Rhizosphere microbial succession associated with plants and soils
Figure 6 shows the relationship between microbial diversity and plant succession. Evidently, there was a close association between rhizosphere microbial diversity and aboveground biomass. The diversity of the bacterial and fungal communities increased logarithmically with increasing biomass. We determined the relationships between the plant characteristics, soil variables, and rhizosphere microbial communities to further explore the effect of plants and soils on microbial communities (Tables S1 and S2—ESM). Soil pH was excluded from the analysis of correlation and RDA because of its narrow range (8.39–8.52). For both A. capillaris and A. sacrorum, the coverage, aboveground biomass, and root biomass and the organic C, total N, NO3−-N, and NH4+-N contents were correlated positively with bacterial and fungal diversity (r = 0.62–0.94, P < 0.05). The abundance of Proteobacteria, Dothideomycetes, and Sordariomycetes was negatively correlated with Actinobacteria abundance (P < 0.05). Available P content was correlated positively with Proteobacteria abundance (r = 0.69, P < 0.05) for A. capillaris; however, rhizosphere available P content for A. sacrorum was correlated negatively with Proteobacteria abundance (r = − 0.79, P < 0.05) and positively with Actinobacteria abundance (r = − 0.81, P < 0.05). The RDA supported the results of the correlation analysis and further supported the driving force of plants and soil on bacterial and fungal communities at the class level (Fig. 7). The first two axes explained 76.1 and 77.3% of the total variance for A. capillaris and A. sacrorum, respectively, indicating that organic C, total N, NO3−-N, NH4+-N, and available P contents were the influential factors (P < 0.05) driving the changes in the composition and diversity of the bacterial communities.
Redundancy analysis (RDA) illustrating the relationships among the microbial populations (indicted by blue arrows), plant characteristics, and rhizosphere properties (indicated by red arrows). aA. capillaries. bA. sacrorum. AB, aboveground biomass; RB, root biomass; OC, organic C; TN, total N; AP, available P; Alphapro, Alphaproteobacteria; Gammapro, Gammaproteobacteria; Betapro, Betaproteobacteria; Deltapro, Deltaproteobacteria; Actinoba, Actinobacteria; Thermole, Thermoleophilia, Acidimic, Acidimicrobiia; Rubroba, Rubrobacteria; Acidobac, Acidobacteria-6; Chloracid, Chloracidobacteria; Chlorofl, Chloroflexi; Gemmatim, Gemmatimonadetes; Saprosp, Saprospirae; Dothideo, Dothideomycetes; Sordario, Sordariomycetes; Pezizomy, Pezizomycetes; Leotiomyc, Leotiomycetes; Eurotiom, Eurotiomycetes; Agaricom, Agaricomycetes; Glomerom, Glomeromycetes; SB, Shannon diversity of bacteria; SF, Shannon diversity of fungi
4 Discussion
4.1 Changes in soil properties during the succession
Changes in aboveground characteristics of dominant plants led to significant variation in the rhizosphere soil nutrient properties. Evidently, rhizosphere organic C, total N, NO3-N, and NH4+-N content of A. capillaris decreased with increasing time of succession, whereas those of A. sacrorum greatly increased. This is because soil C and N fractions are derived from the decomposition of litter (Zhou et al. 2015), the release of root exudates, and rhizodeposition (Jones et al. 2009). Reduction of aboveground cover and biomass of A. capillaris could decrease the quantity and quality of litter and roots, thus lowering the soil nutrient content and vice versa. The observed positive relationship between soil nutrients and aboveground characteristics confirmed this argument (Tables S1 and S2—ESM). The rhizosphere pH of the two plants did not vary significantly along the succession; this is possibly due to the organic acids from the root exudates and needs further investigation.
4.2 Changes of rhizosphere microbial communities during the succession
4.2.1 Bacterial community
In our study, changes in plants resulted in significant changes in rhizosphere microbial communities. The successional patterns of microbial communities are still debated. A significant transition in the composition of the bacterial communities for the two plants along the chronosequence indicated a clear pattern of succession in the bacterial community. This is in agreement with the results of Poosakkannu et al. (2017), who reported succession in the rhizosphere bacterial communities of Deschampsia flexuosa across two sand dune successional stages. Interestingly, the bacterial communities belonging to A. capillaris and A. sacrorum did not differ significantly at the 10-year site, suggesting that the two plants at this stage harbored similar bacterial assemblages. This was confirmed by the 85% of microbial OTUs shared by the two plants at this stage (Fig. S2—ESM). A possible reason could be similarities in soil nutrient levels (mainly C and N) (Table 1) in the rhizospheres of these two plants at the 10-year site, which provided similar capacities to offer nutrients to the microorganisms. This phenomenon, however, disappeared as succession proceeded, and C and N contents were much higher for A. sacrorum than A. capillaris at the 15-year site, which would support more microbial growth and lead to higher diversity.
The bacterial communities of the two species consisted mostly of Proteobacteria, Acidobacteria, Actinobacteria, Chloroflexi, Gemmatimonadetes, and Bacteroidetes. Actinobacteria and Proteobacteria were the two most abundant phyla during the 30 years of succession corresponding with previous findings that soils are usually dominated by these two ubiquitous bacterial taxa (Mukherjee et al. 2013; Lagos et al. 2014; Bell et al. 2015). The bacterial community of A. capillaris shifted from Proteobacteria-dominant (including Alpha- and Gemma-) to Actinobacteria-dominant, and the A. sacrorum community presented the opposite trend. This result suggested a directional replacement successional pattern for the bacterial communities in the specific rhizospheres. Recent sequence analyses have characterized Proteobacteria as a fast-growing copiotrophic group that becomes abundant when available substrates increase (Zeng et al. 2017; Goldfarb et al. 2011); however, Actinobacteria are oligotrophic and are abundant in nutrient-poor environments. Thus, it is likely that the lower rhizosphere C and N supplies for A. capillaris were responsible for the decrease in Proteobacteria and increase in Actinobacteria and also for the opposite trend observed in A. sacrorum. This finding was further confirmed by the results of the correlation and RDA analyses. The contents of organic C, total N, and NO3−-N for both A. capillaris and A. sacrorum were positively associated with the abundance of Alpha- and Gammaproteobacteria and negatively with associated with the abundance of Actinobacteria. Additionally, the coverage and biomass of the two plants were positively correlated with the abundance of Proteobacteria and bacterial diversity and negatively with the abundance of Actinobacteria. This indicates that the plants may have had a large influence on the soil microbial communities, which is consistent with the results of Lima et al. (2015) who reported that the quantity and type of microbes in the rhizosphere were largely dependent upon the species of plant. The dynamics of soil microbial communities are likely determined by plants and the mediation of soil nutrient availability. The ways in which plants establish their rhizosphere microbiota can be considered to be a selective process linked with the intrinsic nutrient requirements of the plant.
4.2.2 Fungal community
Successional age greatly influenced the fungal community structure. Variation in fungal richness and diversity between the plants supported previous observations of differences in fungal colonization and responsiveness (Siciliano et al. 2014). The fungal communities associated with the two dominant species of plant consist mostly of Ascomycota, Zygomycota, Basidiomycota, or Glomeromycota, which is consistent with previous observations from different soil systems (He et al. 2016; Franke-Whittle et al. 2015). Most of the fungal taxa detected in our investigation were saprophytic and less than 1% were mycorrhizal such as Glomeraceae (Welc et al. 2014). Mycorrhizal fungi usually exist in forest ecosystems, such as pine forests, and less exist in grasslands especially in arid areas. The fungal communities presented a distinctive succession despite no obvious succession occurring at the phylum level. This was confirmed by the significant difference in fungal communities between successional stages as suggested by the results of the ANOSIM. Dothideomycetes and Sordariomycetes, the two largest classes of Ascomycota, exhibited similar succession patterns in terms of coverage and biomass of plants, and their abundance was associated with soil C and N contents. This is because these two fungal groups are typically saprotrophic in soils (Xiong et al. 2014) and their growth rate is particularly vulnerable to nutrient levels. Zhou et al. (2016), however, found no significant effect of nitrogen fertilization on relative abundance. This inconsistency might be due to the different soil conditions causing differences in microbial response. For example, the soil in our study was alkaline (pH 8.4), but the soil investigated by Zhou et al. (2016) was acidic (pH 4.7–6.3). Different soils can produce different responses in microbial communities. Secondly, the particular soil environment investigated will affect observations of microbial response. We focused mainly on the rhizosphere environment, which was highly influenced by plant roots, whereas Zhou et al. (2016) investigated the bulk soil, which has different characteristics.
Our results suggest that Ascomycota species are capable of early-stage colonization as pioneer species, but they can also survive well under increasing competitive pressure unlike other pioneer species, such as Glomeromycota and Basidiomycota, in the rhizosphere of A. sacrorum that undergo replacement in late-successional stages. Fierer et al. (2010) postulated that successional models of plant communities could apply to microbial communities. Two contrasting hypotheses have been proposed for the successional trajectory of fungal communities in natural ecosystems, namely, succession with or without directional replacement. Our results support the first hypothesis that no obvious replacement occurred in the rhizosphere fungal community, which is in contrast with that of the bacterial community that shifted from Proteobacteria-dominant to Actinobacteria-dominant for A. capillaris and vice versa for A. sacrorum. Models of plant succession suggest that increased environmental resistance does not lead to the replacement of species along successional gradients due to weaker competition but rather that low resistance tends to cause succession with directional replacement (Davey et al. 2015). Our results support this model for the belowground fungal community, suggesting a varied successional trajectory for rhizosphere fungal communities.
4.2.3 Comparison of microbial succession between rhizosphere and bulk soil
High microbial turnover and activity are found in the rhizosphere because it is adjacent to plant roots, usually making its chemical and microbial properties distinct from those of the bulk soil (Paredes and Lebeis 2016). In our study, the bacteria and fungi, whether from the A. capillaris or A. sacrorum rhizosphere, exhibited higher successional rates than those from the bulk soil. This result suggests higher microbial turnover in the rhizosphere compared with the bulk soil. A possible reason for this could be the abundant C substrates in this zone. It has been documented that most C in soils is derived from plants with individual plants releasing 5–15% of their photosynthetically fixed C via their roots (Marschner et al. 2001). The C released is a combination of active secretions of specific root exudates and the passive release of plant debris from both shoots and roots. This process creates a C-rich environment in the rhizosphere, while the surrounding bulk soils are assumed to be C limited (Lambers et al. 2009). As a result, there are higher levels of microbial activity and turnover in this region as compared to the bulk soil and a distinct bacterial taxonomic profile. Furthermore, C fixed by the plant via photosynthesis is directly incorporated by specific bacterial taxa in the rhizosphere and that this assimilation is dependent upon proximity to the root.
4.3 Rhizosphere microbial communities associated with plant and soil properties
Previous studies on secondary succession have reported a remarkable difference in the composition of plant and microbial communities between successional stages (Zeng et al. 2017). Lozano et al. (2014) reported that microbial composition was significantly different between the successional stages in a semiarid abandoned cropland in Spain and that these variations were correlated with changes in aboveground productivity, diversity, and nutrient accumulation as the succession progressed. These findings indicated that variation in plant community composition could be a good predictor of changes in microbial succession. Our results confirmed the hypothesis that changes in aboveground biomass were significantly correlated with the diversity of bacterial and fungal communities. This is consistent with the observations of Millard and Singh (2010), who stated that microbial diversity was largely dependent upon the quality and type of soil organic matter derived from the decomposition of plant biomass. Interestingly, we also found a logarithmical rise in bacterial and fungal community diversity with increasing biomass, indicating that elevated aboveground biomass could lead to enhanced rhizosphere microbial diversity, but the rate of enhancement decreased gradually. This phenomenon was probably due to the growth of fast-growing copiotrophic taxa as a result of increasing organic substances during the succession (Freedman et al. 2016). Most soil organic matter (mainly C and N) in natural ecosystems is derived from the decomposition of aboveground biomass (Cline and Zak 2015), and the quantity and quality of these soil components are directly determined by plant biomass. As the succession progressed, an increase in aboveground biomass led to an increase in the accretion of organic materials such as litter and roots and consequently promoted the accumulation of soil OC. These organic components provided abundant substrate for the copiotrophic microbes (r-living strategies), such as Alphaproteobacteria (Zhang et al. 2016), and promoted their growth, thereby causing a quick rise in microbial diversity. As more organic matter entered the soil, however, excessive copiotrophic microbes had a negative effect on microbial community diversity due to their high resource-use ability, which decreases the availability of resources for other microbial groups, especially oligotrophic microbes (k-living strategies) (Fierer et al. 2007). Accumulation of OC stabilizes soil temperatures and increases water-holding capacity (Filep et al. 2015). Plant biomass, by its effects on OC content and microclimatic conditions, affected the microbial growth and community diversity. Therefore, we assumed that plants probably induce a succession of rhizosphere microbes by the mediation of nutrient status mainly caused by changes in aboveground biomass. Furthermore, microorganisms that interact positively with plants include rhizobia, mycorrhiza, endophytes, and epiphytes. These interactions afford plants some benefits, such as protection against biotic and abiotic stress, growth promotion, and increased nutrient availability (Paredes and Lebeis 2016). For example, soil microbes are often considered to be pioneering during the process of succession. Their establishment affects soil physicochemical properties, indirectly facilitates pedogenesis, and paves the way for later colonization by plants (Chagas et al. 2018).
Our results are in agreement with previous studies that have suggested a significant correlation between N components and microbial community diversity (McHugh et al. 2017; Jach-Smith and Jackson 2018). N components, including TN, NO3-N, and NH4+-N, were strongly correlated with aboveground biomass, diversity, and the abundance of dominant taxa in the bacterial communities. TN and NO3−-N content affected fungal diversity and NH4+-N content had little effect. These results suggested that only some forms of N contributed to the succession of the fungal communities. Spohn et al. (2015) reported that bacterial communities in the rhizospheres of Hordeum vulgare depended on the availability of P and that P-fertilization could decrease the abundance of Firmicutes and increase the abundance of Beta- and Gammaproteobacteria. We also identified a close association between available P content and specific bacterial taxa, suggesting the importance of P in structuring bacterial communities. However, the correlation between rhizosphere available P content and the abundance of most bacterial groups differed between the two dominant species being positive for A. capillaris and negative for A. sacrorum, indicating a varied influence of soil P on bacterial composition along the succession. Loessial soil has very low P content; therefore, P fertilizer is required to meet the demands of crop growth. Consequently, when the cropland was abandoned, the P content decreased dramatically. The decrease in abundance of the pioneer species A. capillaris during succession could lead to decreases in the absorption of P and the decrease in rhizosphere P content. The increase in abundance of the replacement species A. sacrorum could increase the P content to support the higher demands of the plants. More P would be absorbed from the soil by the roots and transported to the tissues, thereby decreasing the rhizosphere P content (Becquer et al. 2014). However, further study is needed to verify this hypothesis.
5 Conclusions
Along the grassland succession, the early-successional plant species A. capillaris exhibited decreasing ground cover and biomass, while those of the late-successional plant species A. sacrorum increased. These changes resulted in differences in the rhizosphere microbial communities. Bacterial and fungal community diversity exhibited a logarithmical relationship with aboveground biomass, indicating that increasing plant biomass could lead to enhanced rhizosphere microbial diversity. However, the enhancement rate was found to decrease gradually. Rhizosphere microbes showed a higher succession rate than those from the bulk soil during the succession. Rhizosphere organic C, total N, NO3−-N, and NH4+-N played significant roles in shaping the bacterial communities, suggesting that plants affect rhizosphere bacterial communities by mediating soil nutrients. Our results contribute to our understanding of plant-microbe interactions during secondary succession.
References
Becquer A, Trap J, Irshad U, Ali MA, Claude P (2014) From soil to plant, the journey of P through trophic relationships and ectomycorrhizal association. Front Plant Sci 5:548
Bell CW, Asao S, Calderon F, Wolk B, Wallenstein MD (2015) Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol Biochem 85:170–182
Blaalid R, Carlsen T, Kumar S, Halvorsen R, Ugland KI, Fontana G, Kauserud H (2012) Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol Ecol 21:1897–1908
Bremner JM, Mulvaney CS (1982) Nitrogen-total. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis. Part 2. Chemical and microbial properties. Agronomy Society of America Agronomy Monograph 9, Madison, pp 595–624
Chagas FO, Pessotti RC, Caraballo-Rodriguez AM, Pupo MT (2018) Chemical signaling involved in plant-microbe interactions. Chem Soc Rev 47:1652–1704
Cline LC, Zak DR (2015) Soil microbial communities are shaped by plant-driven changes in resource availability during secondary succession. Ecolgy 96(12):3374–3385
Davey M, Blaalid R, Vik U, Carlsen T, Kauserud H, Eidesen PB (2015) Primary succession of Bistorta vivipara (L.) Delabre (Polygonaceae) root-associated fungi mirrors plant succession in two glacial chronosequences. Environ Microbiol 17(8):2777–2790
Deng L, Shangguan ZP, Sweeney S (2014) “Grain for Green” driven land use change and carbon sequestration on the Loess Plateau, China. Sci Rep 4:7039
Development Core Team R (2013) R version 3.0.1: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Fierer N, Nemergut DR, Knight R, Craine JM (2010) Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161:635–642
Filep T, Draskovits E, Szabo J, Koos S, Laszlo P, Szalai Z (2015) The dissolved organic matter as a potential soil quality indicator in arable soils of Hungary. Environ Assess 187(7):479
Foster BL, Tilman D (2000) Dynamic and static views of succession: testing the descriptive power of the chronosequence approach. Plant Ecol 146:1–10
Franke-Whittle IH, Manici LM, Insam H, Stres B (2015) Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 395:317–333
Freedman ZB, Upchurch RA, Zak DR, Cline LC (2016) Anthropogenic N deposition slows decay by favoring bacterial metabolism: insights from metagenomic analyses. Frontier Microbiol 7:259
Goldfarb KC, Karaoz U, Hanson CA, Santee CA, Bradford MA, Treseder KK, Wallenstein MD, Brodie EL (2011) Differential growth responses of soil bacterial taxa to carbon substrates of varying chemical recalcitrance. Frontier Microbiol 2:94
Harantová L, Mudrák O, Kohout P, Elhottová D, Frouz J, Baldrian P (2017) Development of microbial community during primary succession in areas degraded by mining activities. Land Degrad Dev 28:2574–2584
He F, Yang BS, Wang H, Yan QL, Cao YN, He XH (2016) Changes in composition and diversity of fungal communities along Quercus mongolica forests developments in Northeast China. Appl Soil Ecol 100:162–171
Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM (2004) A taxa-area relationship for bacteria. Nature 432:750–753
Jach-Smith LC, Jackson RD (2018) N addition undermines N supplied by arbuscular mycorrhizal fungi to native perennial grasses. Soil Biol Biochem 116:148–157
Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil 321:5–33
Kardol P, De Deyn GB, Laliberte E, Mariotte P, Hawkes CV (2013) Biotic plant-soil feedbacks across temporal scales. J Ecol 101(2):309–315
Klironomos JN (2003) Variation in plant response to native and exotic arbuscular mycorrhizal fungi. Ecology 84:2292–2301
Knelman JE, Grahama EB, Trahand NA, Schmidt SK, Nemerguta DR (2015) Fire severity shapes plant colonization effects on bacterial community structure, microbial biomass, and soil enzyme activity in secondary succession of a burned forest. Soil Biol Biochem 90:161–168
Kulmatiski A, Beard KH, Stevens JR, Cobbold SM (2008) Plant-soil feedbacks: a meta-analytical review. Ecol Lett 11:980–992
Kuramae EE, Gamper HA, Yergeau E, Piceno YM, Brodie EL, DeSantis TZ, Andersen GL, Van veen JA, Kowalchuk GA (2010) Microbial secondary succession in a chronosequence of chalk grasslands. ISME J 4:711–715
Lagos LM, Navarrete OU, Maruyama F, Crowley DE, Cid FP, Mora ML, Jorquera MA (2014) Bacterial community structures in rhizosphere microsites of ryegrass (Lolium perenne var. Nui) as revealed by pyrosequencing. Biol Fertil Soils 50:1253–1266
Lambers H, Mougel C, Jaillard B, Hinsinger P (2009) Plant-microbe-soil interactions in the rhizosphere: an evolutionary perspective. Plant Soil 321:83–115
Lankau EW, Lankau RA (2014) Plant species capacity to drive soil fungal communities contributes to differential impacts of plant–soil legacies. Ecology 95(11):3221–3228
Liang Y, Jiang Y, Wang F, Wen C, Deng Y, Xue K, Qin Y, Yang Y, Wu L, Zhou J, Sun B (2015) Long-term soil transplant simulating climate change with latitude significantly alters microbial temporal turnover. ISME J 9(12):2561–2572
Lima AB, Cannavan FS, Navarrete AA, Teixeira WG, Kuramae EE, Tsai SM (2015) Amazonian dark earth and plant species from the Amazon region contribute to shape rhizosphere bacterial communities. Microbial Ecol 69:855–866
Lozano YM, Hortal S, Armas C, Pugnaire FI (2014) Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol Biochem 78:298–306
Marschner P, Yang CH, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
McHugh TA, Morrissey EM, Mueller RC, Gallegos-Graves LV, Kuske CR, Reed SC (2017) Bacterial, fungal, and plant communities exhibit no biomass or compositional response to two years of simulated nitrogen deposition in a semiarid grassland. Environ Microbiol 19(4):1600–1611
Millard P, Singh BK (2010) Does grassland vegetation drive soil microbial diversity? Nutr Cycl Agroecosyst 88(2):147–158
Mukherjee S, Heinonen M, Dequvire M, Sipilä T, Pulkkinen P, Yrjälä K (2013) Secondary succession of bacterial communities and co-occurrence of phylotypes in oil-polluted Populus rhizosphere. Soil Biol Biochem 58:188–197
Munoz-Rojas M, Erickson T, Martini D, Dixon KW, Merritt DJ (2016) Soil physicochemical and microbiological indicators of short, medium nd long term post-fire recovery in semi-arid ecosystems. Ecol Indic 63:14–22
Nelson DW, Sommers LE (1982) Total carbon, organic carbon, and organic matter. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis. Part 2. Chemical and microbial properties. Agronomy Society of America, Agronomy Monograph 9, Madison, pp 539–552
Olsen SR, Sommers LE (1982) Phosphorous. In: Page AL, Miller RH, Keeney DR (eds) Methods of soil analysis. Part 2. Chemical and microbial properties. Agronomy Society of America, Agronomy Monograph 9, Madison, Wisconsin, pp 403–430
Paredes HS, Lebeis SL (2016) Giving back to the community: microbial mechanisms of plant–soil interactions. Funct Ecol 30:1043–1052
Peay KG, Kennedy PG, Bruns TD (2011) Rethinking ectomycorrhizal succession: are root density and hyphal exploration types drivers of spatial. Fungal Ecol 4:233–240
Poosakkannu A, Nissinen R, Mannisto M, Kytoviita MM (2017) Microbial community composition but not diversity changes along succession in arctic sand dunes. Environ Microbiol 19:698–709
Siciliano SD, Palmer AS, Winsley T, Lamb E, Bissett A, Brown MV, van Dorst J, Ji M, Ferrari BC, Grogan P, Chu HY, Snape I (2014) Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol Biochem 78:10–20
Spohn M, Treichel NS, Cormann M, Schloter M, Fischer D (2015) Distribution of phosphatase activity and various bacterial phyla in the rhizosphere of Hordeum vulgare L. depending on P availability. Soil Biol Biochem 89:44–51
Tscherko D, Ute H, Marie-Claude M, Ellen K (2004) Shifts in rhizosphere microbial communities and enzyme activity of Poa alpina across an alpine chronosequence. Soil Biol Biochem 36:1685–1698
Walker LR, Walker J, Hobbs RJ (2007) Linking restoration and ecological succession. Springer, Amsterdam, pp 5–21
Wang GL, Liu GB, Xu MX (2009) Above and belowground dynamics of plant community succession following abandonment of farmland on the Loess Plateau, China. Plant Soil 316:227–239
Welc ME, Frossard E, Egli S, Else K, Bünema JJ (2014) Rhizosphere fungal assemblages and soil enzymatic activities in a 110-years alpine chronosequence. Soil Biol Biochem 74:21–30
Xiong J, Peng F, Sun H, Xue X, Chu H (2014) Divergent responses of soil fungi functional groups to short-term warming. Microb Ecol 68:708–715
Zeng QC, An SS, Liu Y (2017) Soil bacterial community response to vegetation succession after fencing in the grassland of China. Sci Total Environ 609:2–10
Zhang C, Liu GB, Xue S, Wang GL (2015) Changes in rhizospheric microbial community structure and function during the natural recovery of abandoned cropland on the Loess Plateau China. Ecol Eng 75:161–171
Zhang C, Liu GB, Xue S, Wang GL (2016) Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Boil Biochem 97:40–49
Zhang C, Liu GB, Song ZL, Qu D, Fang LC, Deng L (2017) Natural succession on abandoned cropland effectively decreases the soil erodibility and improves the fungal diversity. Ecol Appl 27(7):2142–2154
Zhang KR, Cheng XL, Shu X, Liu Y, Zhang QF (2018) Linking soil bacterial and fungal communities to vegetation succession following agricultural abandonment. Plant Soil 431:19–36
Zhou WJ, Sha LQ, Schaefer DA, Zhang YP, Song QH, Tan ZH, Deng Y, Deng XB, Guan HL (2015) Direct effects of litter decomposition on soil dissolved organic carbon and nitrogen in a tropical rainforest. Soil Biol Biochem 81:255–258
Zhou J, Jiang X, Zhou BK, Zhao BS, Ma MC, Guan DW, Li J, Chen SF, Cao FM, Shen DL, Qin J (2016) Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol Biochem 95:135–143
Funding
National Natural Sciences Foundation of China (41701556, 41771554), National Key Research and Development Program of China (2016YFC0501707), and Chinese University Scientific Fund (2452017111).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Jizheng He
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 1768 kb)
Rights and permissions
About this article
Cite this article
Song, Z., Liu, G. & Zhang, C. Response of rhizosphere microbial communities to plant succession along a grassland chronosequence in a semiarid area. J Soils Sediments 19, 2496–2508 (2019). https://doi.org/10.1007/s11368-019-02241-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-019-02241-6