Abstract
Purpose
Previous investigations—field samplings and laboratory experiments—support the hypothesis that the degradation of s-triazines is enhanced in previously exposed as compared to pristine soils in terrestrial environments. Despite this, bottlenecks of soil sampling and various soil modification practices in microcosm studies have made it difficult to guarantee that previous contamination history enhances contaminant degradation regardless of soil origin in terrestrial ecosystems. We test the hypothesis that the degradation of simazine (2-chloro-4,6-bis(ethylamino)-s-triazine) is enhanced in previously exposed soils as compared to pristine soils in 10 l buckets at the mesocosm scale.
Materials and methods
We collected soil at three separate sites consisting of a previously exposed and a pristine field. At every field, soil was collected at three separate plots and simazine degradation (days 0 and 65) and the response to atzB degrader gene primers (days 0 and 110) were followed. We analyzed the results using analysis of covariance (ANCOVA). Previous exposure and field site were assessed as fixed factors and initial simazine concentration and abiotic soil conditions as covariates.
Results and discussion
After the 65-day exposure, remaining simazine concentrations depended on previous exposure but not on collection site. The response to atzB gene primers was positive in all mesocosms where simazine degradation had been rapid. Soil moisture, pH, and organic matter content were insignificant. If soil moisture was not included in the ANCOVA model, previous exposure did not appear as a significant factor.
Conclusions
The results support the hypothesis that simazine is degraded more rapidly in previously exposed soils as compared to pristine environments, provided that degradation genes are available. Previously exposed soil might be used to enhance the degradation of simazine in recently contaminated terrestrial soils, supposing that the central requirements for microbial growth are adequate.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
In the soil environment, microbial degradation is a key process that determines the ecological consequences of any anthropogenic pollutant (Sinkkonen et al. 2010). Several field samplings and laboratory studies suggest that if previously exposed soils are exposed later again, the degradation of organic contaminants will be faster than in pristine soils (Gentry et al. 2004; Krutz et al. 2009; Penet et al. 2006; Rhine et al. 2003; Kauppi et al. 2012). The likely explanation is that at least some soil bacteria retain the ability to degrade the organic contaminant. However, in field sampling, it is not easy to exclude the effects of agricultural practices and different soil characteristics, such as fertility and moisture, on contaminant degradation (McGee et al. 1995; Webster 2007). Similarly, soil sieving, manipulations of soil moisture, and pooling of samples collected at different field plots are likely to cause bias in studies made using microcosms, i.e., pots that contain a few hundred grams or less soil. As soil pooling, soil sieving, and moisture adjustments typically change soil texture and reduce soil heterogeneity in microcosms, these practices are likely to change the structure of soil microbial community. They may even aid specific degradation genes to spread in spiked soil. For these reasons, it has been hard to guarantee that previous contamination history enhances contaminant degradation regardless of soil origin in terrestrial ecosystems.
In a number of studies, soil remediation has been advanced when a single previously exposed soil has been mixed with a newly contaminated soil (e.g., Gentry et al. 2004; Laine and Jørgensen 1996). This has been associated with the invasion of microorganisms containing degradation genes and the spread of these genes in the previously unexposed soil (Avery 2006; McGowan et al. 1998; Sarand et al. 2000). Although some of the mechanisms behind the phenomenon are well known (Joner et al. 2004; Sarand et al. 2001), precise tests of the importance of contamination history on the degradation of anthropogenic environmental contaminants in terrestrial soils are rare and typically restricted to microcosm studies where soil sieving, pooling, and moisture manipulations are a common practice (Morán et al. 2006). Although soil texture and properties always change when soil is sampled, it is valuable to study the importance of contamination history at larger-scale laboratory studies—mesocosm studies—where each replicate contains at least 5 kg soil and soil sieving, pooling, and moisture manipulations are avoided, as in our experiment.
Herbicides containing a triazine ring are among the most frequently used and studied organic xenobiotics in the world. As most of the mesocosm experiments investigating the degradation of s-triazines have been done in aqueous environments (e.g., sediment and slurry studies: Bacci et al. 1989; Huggins 1990; Runes et al. 2001; Fairchild and Sappington 2002; Lytle and Lytle 2002; Sapozhnikova et al. 2009), the effects of previous exposure should still be evaluated in terrestrial mesocosms (Lima et al. 2009; Chelinho et al. 2010). S-triazines are often recalcitrant in terrestrial soils, and they have been observed to cause changes in the soil microbial community at concentrations as low as 5 μg g−1 dry weight soil (Mahía et al. 2008). In surface soils, simazine and other triazine herbicides are often mineralized slowly as a result of chemical reactions. As triazine herbicides are poor carbon but moderate nitrogen sources for soil microbes, high levels of inorganic nitrogen are known to slow down microbial triazine degradation in surface soils (Krutz et al. 2010). Today, many triazine-degrading bacteria are known (e.g., in De Souza et al. 1998; Radosevich and Tuovinen 2004; Saijaphan et al. 2010). Despite this, only a couple of microbial degradation pathways have been identified (Behki et al. 1993; Shapir et al. 2007; Vibber et al. 2007). The most efficient and widespread of those pathways is the plasmid-borne gene family atzABCDEF that encodes amidohydrolase-related enzymes (Devers et al. 2004, 2005). The presence of the atzB gene has been associated with faster than average degradation of simazine in two geographically separate agricultural soils that had been repeatedly treated with simazine (Martín et al. 2008).
In this study, we tested the degradation of spiked simazine in 10-l buckets containing soil collected from three different terrestrial sites; every site consisted of a field with a long contamination history and a comparable field without any previous contamination history. We also determined the response of soil bacterial DNA to atzB primers in polymerase chain reaction (PCR) in relation to simazine degradation. We hypothesize that simazine degradation depends on earlier contamination history and the presence of atzB genes but not on soil origin.
2 Materials and methods
2.1 Soil collection sites
Soil collection was performed at three separate sites in Finland during the summer 2007. There was a field with a long s-triazine history at each site. At each site, a pristine control field that had similar soil physicochemical characteristics was selected for the study as well. The minimum distance between an exposed and a control field was at least 200 m. Although field selection was not random, the probability of systematic bias was minimized as local gardeners were asked to show us the exposed and similar but pristine fields. Simazine was used at exposed fields when needed, i.e., once in 1–3 years 1970–1998, and probably also in 1960s. Application rates were between 7 and 20 kg/ha before 1985 and 3–10 kg/ha after 1985.
The first site (VT) is situated in front of Villa Tapanila in Jalkaranta in the northwestern part of the city of Lahti, Finland. The site consisted of two garden fields. The first field was a flower bed that had been planted with rose (Rosa rugosa) ca. 3 years earlier. The flower bed had been treated with simazine and possibly other s-triazine herbicides repeatedly (see above) until 1998. The second garden field (at a distance of ca. 200 m from the first one) was an open field without previous exposure to triazine herbicides. Vegetation on the pristine field consisted of annual and perennial grasses and small herbs, like Elymus repens, Festuca sp., Poa sp., Agrostis sp., Trifolium repens, Trifolium pratense, and Taraxacum sp.
The second site (CG) was a lawn within Lahti City Gardens in the southern part of Lahti, ca. 6 km from the first site. The first field was located within three meters of a hawthorn fence that had been treated regularly with simazine (see above) until 1998. The second field was 200 m apart from the first one. It has never been exposed to triazine herbicides. Vegetation on both fields consisted of annual and perennial grasses and small herbs, like Festuca sp., Poa sp., Agrostis sp., Phleum pratense, T. repens, Taraxacum sp., and Tripleurospermum inodorum.
The third site (Piikkiö) was at the MTT (Agrifood Finland) agricultural research station in Piikkiö, Finland, ca. 200 km southwest from Lahti. Two fields with ca. 1 000 years of agricultural history and with similar soil properties were selected for the study based on previous exposure to triazine herbicides. The first field was a former apple plantation that had been treated at least 15 times with simazine (see above) and casually with atrazine (years 1973, 1977), 2,6-dichlorobenzenecarbothioamide (1978, 1981, 1988), terbuthylazine (1989), 2,6-dichlorobenzonitrile (1990), and glufosinate-ammonium (1997, 1998). In 2007, vegetation consisted of rye (Secale cereale L.) as a cover crop and scattered tiny annual weeds. The second field was ca. 400 m apart from the first field. It had never been exposed to s-triazine herbicides. Soil was sampled at the end of strawberry rows where vegetation consisted mostly of annual and perennial weeds, such as Taraxacum sp., Fumaria officinalis, Cerastium vulgare, and Plantago major.
2.2 Soil collection
In all six fields, soil was collected from three separate (distance >5 m) ca. 1 m2 plots as described in Kauppi et al. (2012). In short, at each plot, living vegetation and plant debris were removed and 15–20 l of surface soil (depth 2–15 cm) was collected and mixed thoroughly. The soil was then randomly divided into two 10-l polyethylene buckets (called hereafter as mesocosm pairs), and the procedure was repeated at each 1-m2 plot. Hence, soil originating from different sampling plots was never pooled. Thereafter, all 36 mesocosms were sealed and stored at 16 ± 1 °C. To avoid the transfer of adapted microorganisms between plots and mesocosms, previously exposed soil was always collected after the collection was performed on a pristine field, and the equipments were carefully cleaned between plots. Sealed mesocosms were exposed to passive air flow through two 10-mm holes (filled with cotton wool) in the lid.
2.3 The degradation experiment
One of each pair of mesocosms was artificially contaminated with simazine (100 μg g−1 dw). As we did not dry experimental soils before the experiment, we had to use a dry weight estimate for each soil for calculating the total amount of simazine per a mesocosm. As soil dry weight in mesocosms varied between 5–8 kg, this resulted in 0.5–0.8 g simazine per mesocosm. Simazine (purity 98.1%, Dr. Ehrenstorfer GmbH) was first dissolved into 10 ml of ethanol that was mixed carefully with 100 ml of sand (blasting sand by Loimaan Hiekka, Loimaa, Finland; grain size 0.1-1.0 mm) and the sand was thoroughly mixed in the mesocosm soil. Control mesocosms received pure ethanol mixed in the sand. Mesocosms were stored at 16 ± 1 °C for 110 days. The experiment thus consisted of three consecutive samplings and two factors (previous exposure, soil origin) with three replicates (n = 3).
Samples for simazine analyses were taken on days 0 and 65 and samples for microbiological analyses on days 0 and 110. Each mesocosm was sampled at three random spots (distance to mesocosms walls was always more than 20 mm): three ca. 10-mm-thick and 15-cm-deep soil cores were taken using 20-cm-long tweezers, and these three cores were combined to a ca. 20–40 g soil sample. The tweezers were sterilized between mesocosms. After sampling, no holes were visible in the mesocosms. Samples were frozen (−18 °C) within 2 h and stored at −18 °C until analyzed.
2.4 Soil physical and chemical characteristics
All sites had been in agricultural use and consisted of fine-textured (silt and clay) soil where separate organic and mineral soil layers did not exist. There were no initial differences in physicochemical characteristics of a pristine and a previously exposed field soils within sites, except in case of pH and organic matter (Table 1). Any possible effects of these soil physicochemical characteristics on simazine degradation were taken into account in statistical analysis. Soil moisture was determined as mass loss of the samples (24 h at 90 °C) and organic matter content as loss on ignition (4 h at 550 °C). Soil pH was determined in 0.01 M CaCl2 (50 g fresh soil in 10 ml solution). For measuring soil NH4 + –N and NO3 − –N concentrations, 10-g subsamples of fresh soil were extracted in 2 M KCl by shaking for 2 h, after which the extract was filtered (S&S 5892, White ribbon) and analyzed colorimetrically using an autoanalyzer (QuikChem 8000, Lachat Instruments, Zellweger Analytics, Inc.). QuikChem® Method 12-107-04-1-E was used for nitrate and QuikChem® Method 10-107-06-1-F for ammonium.
2.5 Pesticide analysis
Frozen soil (2 g) was melted, mixed with 15 ml of methanol:water (3:1 v/v) solution and internal standard, 100 μl of propazine:methanol:water solution added. Propazine concentration in the methanol:water solution was 500 mg l−1 in contaminated and 50 mg l−1 in control samples. The samples were mixed thoroughly with vortex for 15 s, sonicated for 15 min at 20 °C and extracted by shaking (200 rpm) overnight at room temperature. The samples were allowed to settle for 1 to 5 min, and the supernatant was transferred to a centrifuge tube for a 15-min centrifugation at 2000 rpm. Thereafter, the solvent was collected to a 50-ml tube and the soil extraction protocol was repeated twice. The solvent was evaporated in a vacuum centrifuge to dryness, and the samples were stored frozen at −18 °C.
The simazine concentrations were analyzed with HPLC. Standards and samples were filtered through a 0.45-μm GHP membrane (Acrodisc®, Gelman, Pall Corporation Ltd., NY, USA), and 20 μl was analyzed using an HPLC equipped with Shimadzu Prominence (Shimadzu, Kyoto, Japan) SIL-20A auto sampler, LC-20AT solvent delivery module, DGU-20A5 online degasser, and SPD-20A UV/VIS detector using a wavelength of 225 nm for simazine and propazine. Waters SunFire column (C18, 3.5 μm, 3.0 × 150 mm, Waters, MA, USA) was used with a flow rate of 0.6 ml min−1, and the system was controlled with Shimadzu LC Solution software. The mobile phase was acetonitrile and filtered water, and the chromatographic separation was carried out using the following gradient profile: first acetonitrile concentration at 30% for 2.5 min, then to 65% for 5 min, and finally back to 30% for 3.5 min. Standards in methanol: water (3:1 v/v) contained simazine at six concentrations ranging from 1.0 to 40.0 ng μl−1 with 14.2 ng μl−1 of propazine as an internal standard. In the analysis, the linear standard responses and detection limit were comparable to those presented by Talja et al. (2008).
2.6 DNA extraction and PCR protocols
DNA from the soil was extracted using PowerSoil™ DNA Isolation Kit (MO BIO Laboratories, Inc., USA). To approach the maximal DNA yield, the manufacturer’s protocol was modified as follows: 150 μl of C2 Solution was utilized instead of 250 μl, 100 μl of C3 Solution was utilized instead of 200 μl, and finally, 2 min centrifuge followed by 2 min air drying.
PCR was performed as in Martín et al. (2008) using atzB primers. PCR for denaturing gel electrophoresis (DGGE) was done with 16S-rRNA universal primers MF341GC and MR907 (Muyzer and Smalla 1998), using the protocol by Kauppi et al. (2011).
DGGE was used for generating a fingerprint image of the soil bacterial community. The DGGE (6% acrylamide-N,N′-methylenebisacrylamide) concentration (denaturing gradients 35–65%) was performed as in Kauppi et al. (2011) with exception of electrophoresis, which was performed at 150 V for 5 h with DCode™ System (Bio-Rad Inc., Hercules, USA). Gels contained 1 × TAE buffer, N,N,N′,N′-tetramethylenediamine, and ammonium persulfate as in manufacturer’s instructions (Bio-Rad Inc., Hercules, USA). Ethidium bromide (two drops of 10 mg/ml) solution was used to dye DNA in the DGGE gel.
2.7 Statistical analyses
Effects of previous exposure (pristine or previously exposed) and soil origin (three separate sites) on simazine concentrations were analyzed using analysis of covariance (ANCOVA) and Pearson correlation analysis in PASW Statistics 18.0 (SPSS Inc. Chicago, Illinois). The measured initial simazine concentrations (day 0) were used as a covariate. In addition, we had to control random noise caused by arbitrarily varying soil characteristics. Therefore, moisture at day 65, organic matter concentration, and soil pH were used as covariates. The other reason was that there was a difference in each of these factors within at least one site, i.e., between a previously exposed and a pristine soil (see Table 1). We also tested other ANCOVA models: we added or dropped out covariates, but as this did not significantly change the outcome as far as soil moisture was included, we show only the original ANCOVA model (Table 2). In particular, the inclusion of a positive response of soil bacterial DNA to atzB gene primers as a dummy variable did not change the significance of previous exposure or other factors. The assumptions of the analyses were met in Box’s test of equality of covariance matrices, Levene’s test of equality of error variances, and Shapiro–Wilk’s Test of Normality.
To fully understand the importance of factors other than soil origin and previous exposure, we dismissed these factors from statistical analysis, and made a t test for the effect of a positive response to atzB gene primers on simazine concentrations. We also performed Pearson correlation analyses between simazine concentrations (Day 65) and physicochemical soil characteristics (moisture day 0, moisture day 65, pH, organic matter, nitrate, ammonium). We did the same correlation analyses between the relative change in simazine concentrations (simazine concentration Day 0 / simazine concentration Day 65) and the soil characteristics, but as significant differences were similar, the data are not shown.
3 Results
3.1 Chemical results
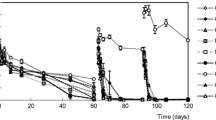
Compared to pristine soils, simazine degradation was enhanced in soils that had been earlier exposed to triazine herbicides (Fig. 1, Table 2). At day 65, simazine concentrations were lower in previously exposed as compared to pristine mesocosms. Soil origin (i.e., field site) had no effect on simazine concentration, and the interaction term previous exposure × soil origin was insignificant (see Fig. 1, Table 2). Tiny concentrations (0.1–0.2 μg g−1 dw) of simazine were found in two soil samples that had not received simazine in our experiment. These samples represented previously exposed soils from the Lahti City Gardens.
Simazine concentrations (in micrograms per gram dw, mean ± SD) in spiked 10-l buckets at days 0 and 65. Abbreviations are in Table 1
When soil origin and previous exposure were not considered in the statistical analysis, soil moisture at day 0 correlated with simazine concentration at day 65 (r = −0.85, p < 0.0005), and soil moisture at day 65 correlated with simazine concentration at day 65 (r = −0.65, p = 0.018). Soil nitrate concentration correlated negatively with simazine concentration at day 65 (r = −0.68, p = 0.002). However, there was also a correlation between soil nitrate concentration and soil moisture (Day 0: r = 0.74, p < 0.005; Day 65: r = 0.56, p = 0.015). No other correlations were found between the measured physicochemical soil characteristics (p > 0.10).
3.2 Microbiological results
PCR with atzB primers ended up in blank gels at day 0. At day 110, bands became visible in four soil samples (Fig. 2); all three preexposed and experimentally contaminated mesocosms and one pristine and experimentally contaminated mesocosm from Villa Tapanila indicated the presence of bacteria harboring an atzB degrader gene. No bands became visible in pristine soil samples that we had not spiked with simazine. If we included the response of soil bacterial DNA to atzB gene primers into our ANCOVA model, the effect of the response on simazine degradation was almost significant (p = 0.07). When we excluded soil origin and previous exposure from the analysis, a positive response of soil bacterial DNA to atzB gene primers was associated with the remaining simazine concentrations at day 65 (t = 3.9, df = 16, p = 0.001).
Response of different soil samples to AtzB gene primers at days 0 and 110 in PCR. Left: All previously exposed and spiked replicates of Villa Tapanila (VT) had a positive response. Right: A single pristine and spiked replicate had a positive response. The other soil origins are not shown as the response to atzB gene primers was always negative
In the DGGE analysis, the use of universal primers resulted in an equally strong smear in all samples, i.e., a high, undistinguishable number of bands. Individual fragments could not be detected, even though different voltage and longer gels were tested in DGGE, indicating a bacterial community with a high diversity in all samples.
4 Discussion
The results of our study support the hypothesis that previous exposure advances the degradation of an anthropogenic s-triazine regardless of soil origin. This underlines the major role of previous contamination history in the adaptation of soil microbial communities to biodegradable chemical stressors (Morán et al. 2006; Morgante et al. 2010); soil bacteria are serving as adapting organisms and organic contaminants are the novel but potentially utilizable stressors (Sinkkonen et al. 2010). The positive response to atzB gene primers is in accordance with the hypothesis that the bacteria carrying degrader genes are selected under contaminant exposure (van Veen et al. 1997). An aspect that requires further testing is whether multi-trophic consequences of simazine exposure are similar in previously exposed versus pristine soils (Rohr et al. 2007). Nevertheless, the results of this study are valuable when the importance of previous exposure is evaluated in bioremediation of s-triazine contaminated soils (De Souza et al. 1998; Hamer and Marschner 2002; Devers et al. 2005).
As our study consisted of three separate soil collection sites and three independent sampling plots within every field, we were able to separate the consequences of soil origin from the effect of previous exposure (Webster 2007). In previous terrestrial studies, the aim has been different, the number of within-site or between-site replicates has been smaller, or abiotic soil characteristics, at least soil moisture, have been standardized before microcosm experiments (e.g., Rhine et al. 2003; Morán et al. 2006; Chelinho et al. 2010). In field studies, on the other hand, it has not been possible to exclude the effects of major abiotic soil characteristics in statistical testing (McGee et al. 1995; Webster 2007). Even though mesocosm studies have been quite common in aqueous environments (Sapozhnikova et al. 2009), our study aids to fill the gap between field sampling and ecologically less heterogeneous microcosm studies in terrestrial environments (Lima et al. 2009; Chelinho et al. 2010).
If simazine degradation is estimated visually (see Fig. 1), there seems to be a remarkable difference in simazine degradation between Villa Tapanila and the other two soil origins. In Villa Tapanila soils, simazine degradation was more rapid in buckets containing previously exposed soil as compared to buckets containing pristine soil, while there seems to be hardly any (Piikkiö) or even an inverse difference (Lahti City Gardens) in simazine degradation between previously exposed and pristine buckets in case of the other two soils. Often this kind of visual impressions may only refer to differences caused by covariates, like moisture and initial simazine concentration. In our study, initial (day 0) simazine levels look higher in pristine Lahti City Gardens soils as compared to previously exposed Lahti City Gardens soils (see Fig. 1), and abiotic soil characteristics were not the same in pristine and previously exposed Piikkiö soils (see Table 1). Even more, there was variation in these covariates at the within-site level, as can be seen from the standard deviations in Table 1 and Fig. 1. Notably, always when soil moisture (day 0 or day 65) and initial simazine concentration were included in an ANCOVA model, previous exposure was the only significant factor. However, when soil moisture was not included in the analysis as a covariate, the ANCOVA model showed soil origin and the interaction previous exposure × soil origin as significant factors explaining simazine degradation. In other words, when random noise caused by initial simazine concentration and soil moisture was taken into account, previous exposure did enhance simazine degradation in our non-homogenized (not sieved, no moisture standardization) mesocosms, but when the noise caused by abiotic covariates was not taken into account, simazine degradation only depended on soil origin. This supports the view on the importance of soil moisture and other abiotic soil characteristics affecting the degradation of simazine, but it does not defeat the result that previous contamination history affected simazine degradation regardless of soil origin. As our statistical model took into account also initial simazine levels, we were able to distinguish the effect of previous exposure in a way that is not possible in microcosm experiments where abiotic soil properties, such as soil moisture, have been standardized before the experiment.
Our counterintuitive result that previous exposure enhances simazine degradation despite the lack of a universal visual effect (see Fig. 1) has implications on attempts to use bioaugmentation (inoculation with microorganisms) and biostimulation (enhancement of abiotic growth conditions) in the remediation of recently contaminated field soils. If the degradation of simazine depends on abiotic covariates, the decrease of simazine concentrations in previously contaminated field soils may not always be faster than the decrease in newly contaminated field soils. Instead, despite the accelerating effect of previous exposure, abiotic conditions seem to have a crucial role in simazine degradation in heterogeneous environments, such as mesocosms and in situ remediation. Hence, contact with previously exposed soil may not result in distinguishably faster degradation in a pristine soil as compared to intact pristine soils if these have more favorable abiotic conditions for contaminant degradation (Koivula et al. 2004; Kauppi et al. 2012; Sinkkonen et al. 2013).
In our study, the positive response to atzB gene primers was restricted to previously exposed and spiked Villa Tapanila mesocosms (see Fig. 2), indicating a strong role of the atz pathway in simazine degradation. Even though we could not find a statistically significant connection between simazine concentrations at day 65 and the positive response of soil bacterial DNA to atzB gene primers (day 110) in our ANCOVA models (p value was 0.07), the positive response to atzB gene primers explained simazine degradation in a t test that obviously did not include soil origin and previous exposure. It is possible that previous exposure enhances simazine degradation if and only if specific degradation genes are present in soil microbial community. To conclusively prove or disprove this, future studies should contain a high number of replicates at the site level, i.e., several fields should be included, each of which contains at least three sampling points (see Section 2).
The positive response of soil microbial community to atzB gene primers in four simazine-spiked samples at day 110 together with negative responses to atzB gene primers in all non-spiked control samples at day 110 indicate that the atz pathway was present in at least some of the soils. As the response to atzB gene primers was negative in all samples at day 0, it is likely that bacteria carrying these genes became enriched during the experiment in the presence of simazine. Presuming that the selected sites have not been treated with simazine after the ban of the compound in 2002 in Finland, we assume that heterotrophic bacterial communities can retain a genetic capacity to degrade anthropogenic chemical stressors for several years if the stressor can also be utilized as a nutrient or a carbon source. Interestingly, as visible difference between spiked and non-spiked mesocosms could not be distinguished in our DGGE analysis (data not shown), simazine degraders appear not to have been dominant strains in the total microbial community.
We also found that soil nitrate concentration was negatively correlated with the remaining simazine levels which is in contrast with previous knowledge that simazine is a poor carbon but a more competitive nitrogen source for soil microorganisms (García-González et al. 2003; Saijaphan et al. 2010). As we also found a correlation between moisture and nitrate levels in our soils, we assume that the observed negative correlation between soil nitrate level and simazine degradation is related to the connection between moisture and nitrate levels in our experiment. Therefore, we do not see a reason to doubt earlier findings that nitrogen scarcity enhances the degradation of s-triazines (Abdelhafid et al. 2000).
5 Conclusions
Our findings have implications on practical soil remediation attempts. As discussed above, biostimulation may be of utmost importance. Supposing that the central requirements for microbial growth are adequate, the mixing of previously exposed soil into recently polluted soil could serve as a strategy to spread the degradation genes of s-triazines into pristine but recently contaminated soils. This strategy might speed up the degradation and thus minimize the risks for severe environmental hazards. A similar strategy has enhanced the degradation of other organic contaminants, such as diesel oil and polyaromatic hydrocarbons (Joner et al. 2004; Kauppi et al. 2012).
In our terrestrial mesocosm experiment, simazine degradation depended on previous exposure and the response of soil bacterial community to atzB gene primers. Importantly, our statistical analysis took into account the crucial roles of soil moisture and initial simazine concentration, which is not typical for degradation studies as those environmental variables tend to be made uniform. This highlights the difference between our bucket-scale experiment and more often published microcosm scale studies. Our results aid to fill a gap between field observations and microcosm studies in terrestrial environments, and they support the hypothesis that previous exposure enhances the degradation of s-triazines, presuming that abiotic conditions favor microbial activity, and that degradation genes are present.
References
Abdelhafid R, Houot S, Barriuso E (2000) How increasing availabilities of carbon and nitrogen affect atrazine behaviour. Biol Fertil Soils 30:333–340
Avery SV (2006) Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4:577–587
Bacci E, Renzoni A, Gaggi C, Calamari D, Franchi A (1989) Models, field studies, laboratory experiments: an integrated approach to evaluate the environmental fate of atrazine (s-triazine herbicide). Agric Ecosyst Environ 27:513–522
Behki R, Topp E, Dick W, Germon P (1993) Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol 59:1955–1959
Chelinho S, Moreira-Santos M, Lima D, Silva C, Viana P, André S, Lopes I, Ribeiro R, Fialho AM, Viegas CA, Sousa JP (2010) Cleanup of atrazine-contaminated soils: ecotoxicological study on the efficacy of a bioremediation tool with Pseudomonas sp. ADP. J Soils Sediment 10:568–578
De Souza ML, Seffernick J, Martinez B, Sadowsky MJ, Wackett LP (1998) The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol 180:1951–1954
Devers M, Soulas G, Martin-Laurent F (2004) Real-time reverse transcription PCR analysis of expression of atrazine catabolisim genes in two bacterial strains isolated from soil. J Microbiol Methods 56:3–15
Devers M, Henry S, Hartmann A, Martin-Laurent F (2005) Horizontal gene transfer of atrazine-degrading genes (atz) from Agrobacterium tumefaciens St96-4 pADP1:Tn5 to bacteria of maize-cultivated soil. Pest Manag Sci 61:870–880
Fairchild JF, Sappington LC (2002) Fate and effects of the triazinone herbicide metribuzin in experimental pond mesocosms. Arch Environ Contam Toxicol 43:198–202
García-González V, Govantes F, Shaw LJ, Burns RG, Santero E (2003) Nitrogen control of atrazine utilization in Pseudomonas sp. Strain ADP. Appl Environ Microbiol 69:6987–6993
Gentry TJ, Josephson KL, Pepper IL (2004) Functional establishment of introduced chlorobenzoate degraders following bioaugmentation with newly activated soil. Biodegradation 15:67–75
Hamer U, Marschner B (2002) Priming effects of sugars, amino acids, organic acids and catechol on the mineralization of lignin and peat. J Plant Nutr Soil Sci 165:261–268
Huggins DG (1990) Ecotoxic effects of atrazine on aquatic macroinvertebrates and its impact on ecosystem structure. University of Kansas, Ph. D. Dissertation, Lawrence, KS
Joner EJ, Hirmann D, Szolar OH, Todorovic D, Leyval C, Loibner AP (2004) Priming effects on PAH degradation and ecotoxicity during a phytoremediation experiment. Environ Poll 128:429–435
Kauppi S, Sinkkonen A, Romantschuk M (2011) Enhancing bioremediation of diesel-fuel-contaminated soil in a boreal climate: comparison of biostimulation and bioaugmentation. Int Biodeterior Biodegrad 65:359–368
Kauppi S, Romantschuk M, Strömmer R, Sinkkonen A (2012) Natural attenuation is enhanced in previously contaminated and coniferous forest soils. Environ Sci Pollut Res. doi:10.1007/s11356-011-0528-y
Koivula TT, Salkinoja-Salonen M, Peltola R, Romantschuk M (2004) Pyrene degradation in forest humus microcosms with or without pine and its mycorrhizal fungus. J Environ Qual 33:45–53
Krutz LJ, Burke IC, Reddy KN, Zablotowitz RM, Price AJ (2009) Enhanced atrazine degradation: evidence for reduced residual weed control and a method for identifying adapted soils and predicting herbicide persistence. Weed Sci 57:427–434
Krutz LJ, Shaner DL, Weaver MA, Webb RMT, Zablotowicz RM, Reddy KN, Huang Y, Thomson SJ (2010) Agronomic and environmental implications of enhanced s-triazine degradation. Pest Manag Sci 66:461–481
Laine MM, Jørgensen KS (1996) Straw compost and bioremediated soil as inocula for the bioremediation of chlorophenol-contaminated soil. Appl Environ Microbiol 62:1507–1513
Lima D, Viana P, André S, Chelinho S, Costa C, Ribeiro R, Sousa JP, Fialho AM, Viegas CA (2009) Evaluating a bioremediation tool for atrazine contaminated soils in open soil microcosms: the effectiveness of bioaugmentation and biostimulation approaches. Chemosphere 74:187–192
Lytle JS, Lytle TF (2002) Uptake and loss of chlorpyrifos and atrazine by Juncus effuses L. in a mesocosm study with a mixture of pesticides. Environ Toxicol Chem 21:1817–1825
Mahía J, Cabaneiro A, Carballas T, Días-Raviña M (2008) Microbial biomass and C mineralization in agricultural soils as affected by atrazine addition. Biol Fertil Soils 45:99–105
Martín M, Gibello A, Lobo C, Nandi M, Garbi C, Fajardo C, Barra-Caracciolo A, Grenni P, Martínez-Iñigo MJ (2008) Application of fluorescence in situ hybridization technique to detect simazine-degrading bacteria in soil samples. Chemosphere 71:703–710
McGee EJ, Keatinge MJ, Synnott HJ, Colgan PA (1995) The variability in fallout content of soils and plants and the design of optimum field sampling strategies. Heal Phys 68:320–327
McGowan C, Fulthorpe R, Wright A, Tiedje JM (1998) Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl Environ Microbiol 64:4089–4092
Morán AC, Müller A, Manzano M, González B (2006) Simazine treatment history determines a significant herbicide degradation potential in soils that is not improved by bioaugmentation with Pseudomonas sp. ADP. J Appl Microbiol 101:26–35
Morgante V, López-López A, Flores C, González M, González B, Vásquez M, Rosselló-Mora R, Seeger M (2010) Bioaugmentation with Pseudomonas sp. strain MHP41 promotes simazine attenuation and bacterial community changes in agricultural soils. FEMS Microbiol Ecol 71:114–126
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Penet S, Vendeuvre C, Bertoncini F, Marchal R, Monot F (2006) Characterisation of biodegradation capacities of environmental microflorae for diesel oil by comprehensive two-dimensional gas chromatography. Biodegradation 17:577–585
Radosevich M, Tuovinen OH (2004) Microbial degradation of atrazine in soils, sediments, and surface water. Pesticde Decontamination and Detoxification ACS Symposium Series 863:129–139
Rhine ED, Fuhrmann JJ, Radosevich M (2003) Microbial community responses to atrazine exposure and nutrient availability: linking degradation capacity to community structure. Microbial Ecol 46:145–160
Rohr JR, Kerby JL, Sih A (2007) Community ecology as a framework for predicting contaminant effects. Trends Ecol Evol 21:606–613
Runes HB, Jenkins JJ, Bottomley BJ (2001) Atrazine degradation by bioaugmented sediment from constructed wetlands. Appl Microbiol Biotechnol 57:427–432
Saijaphan K, Heepngoen P, Sadowsky MJ, Boonkerd N (2010) Arthrobacter sp. strain KU001 isolated from a Thai soil degrades atrazine in the presence of inorganic nitrogen sources. J Microbiol Biotechnol 20:602–608
Sapozhnikova Y, Pennington P, Wirth E, Fulton M (2009) Fate and transport of Irgarol 1051 in a modular estuarine mesocosm. J Environ Monit 11:808–814
Sarand I, Haario H, Jorgensen KS, Romantschuk M (2000) Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate amended soil. FEMS Microbiol Ecol 31:127–141
Sarand I, Skärfstad E, Forsman M, Romantschuk M, Shingler V (2001) Role of the DmpR-mediated regulatory circuit in bacterial biodegradation properties in methylphenol-amended soils. Appl Environ Microbiol 67:162–171
Shapir N, Mongodin EF, Sadowsky MJ, Daugherty SC, Nelson KE, Wackett LP (2007) Evolution of catabolic pathways: genomic insights into microbial s-triazine metabolism. J Bacteriol 189:674–682
Sinkkonen A, Simpanen S, Romantschuk R (2010) An evolutionary perspective to the stimulation of bacterial activity in contaminated soil. In: Płaza G (ed) Trends in bioremediation and phytoremediation. Research Signpost, Kerala, pp 73–81
Sinkkonen A, Kauppi S, Simpanen S, Rantalainen A-L, Strömmer R, Romantschuk M (2013) Layer of organic pine forest soil on top of chlorophenol contaminated mineral soil enhances contaminant degradation. Environ Sci Pollut Res 20:1737–1745
Talja M, Kaukonen S, Kilpi-Koski J, Malin I, Tuominen J, Romantschuk M, Kairesalo T, Kontro M (2008) Atrazine and terbutryn degradation in deposits from groundwater environment within the boreal region in Lahti, Finland. J Agric Food Chem 56:11962–11968
van Veen JA, van Overbeek LS, van Elsas JD (1997) Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev 61:121–135
Vibber LL, Presler MJ, Colores GM (2007) Isolation and characterization of novel atrazine-degrading microorganisms from an agricultural soil. Appl Microbiol Biotechnol 75:921–928
Webster R (2007) Analysis of variance, inference, multiple comparisons and sampling effects in soil research. Eur J Soil Sci 58:74–82
Acknowledgments
Two anonymous reviewers gave constructive comments. Jorma Hellstén, Hannu Neuvonen, and Markku Saari assisted in soil selection and history. Academy of Finland (decision no. 139847, AS), Suomen Luonnonvarain tutkimussäätiö (AS), Marjatta and Eino Kolli Foundation (SK, TP), Elite project funded by ERDF (MR), and the Regional Council of Päijät-Häme (MR) funded the research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Jean-Paul Schwitzguébel
Rights and permissions
About this article
Cite this article
Sinkkonen, A., Kauppi, S., Pukkila, V. et al. Previous exposure advances the degradation of an anthropogenic s-triazine regardless of soil origin. J Soils Sediments 13, 1430–1438 (2013). https://doi.org/10.1007/s11368-013-0742-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-013-0742-y